Small RNAs of Haloferax mediterranei: Identification and Potential Involvement in Nitrogen Metabolism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Bioinformatic Prediction of small RNAs
2.3. RNA Isolation
2.4. Library of Complementary DNA Preparation and Sequencing
2.5. RNA-Sequencing Bioinformatic Analysis
2.6. Validation of sRNAs Using Reverse Transcription Polymerase Chain Reaction
3. Results
3.1. Identification of Putative small RNAs in Haloferax mediterranei
3.2. Verification of Predicted 295 sRNAs Using RNA-Sequencing
3.3. Bioinformatic Analysis of Putative Small RNAs in H. mediterranei
3.3.1. Identification and Classification of small RNAs Verified by RNA-Sequencing
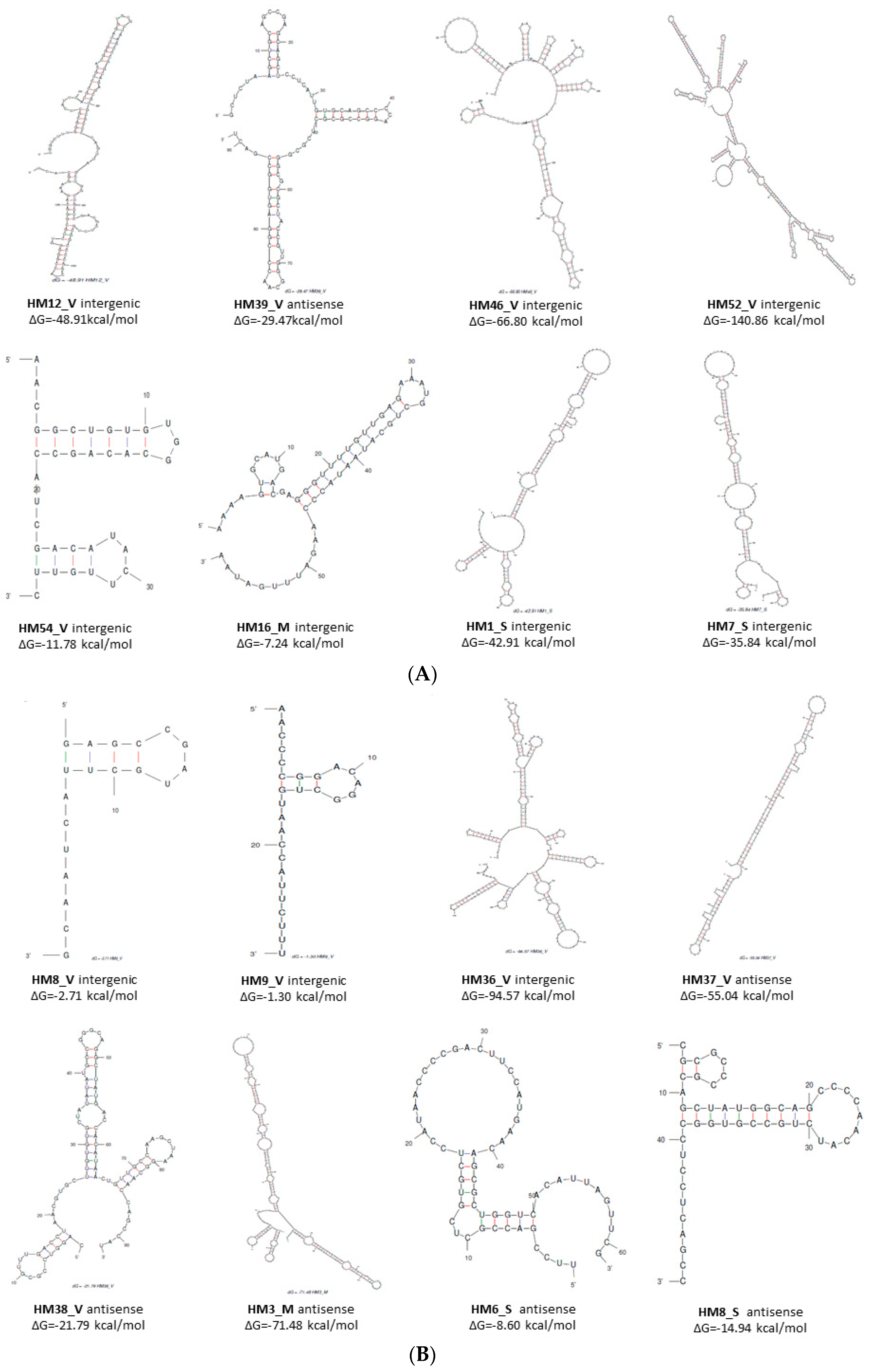
3.3.2. Structure and Targets of 88 small RNAs
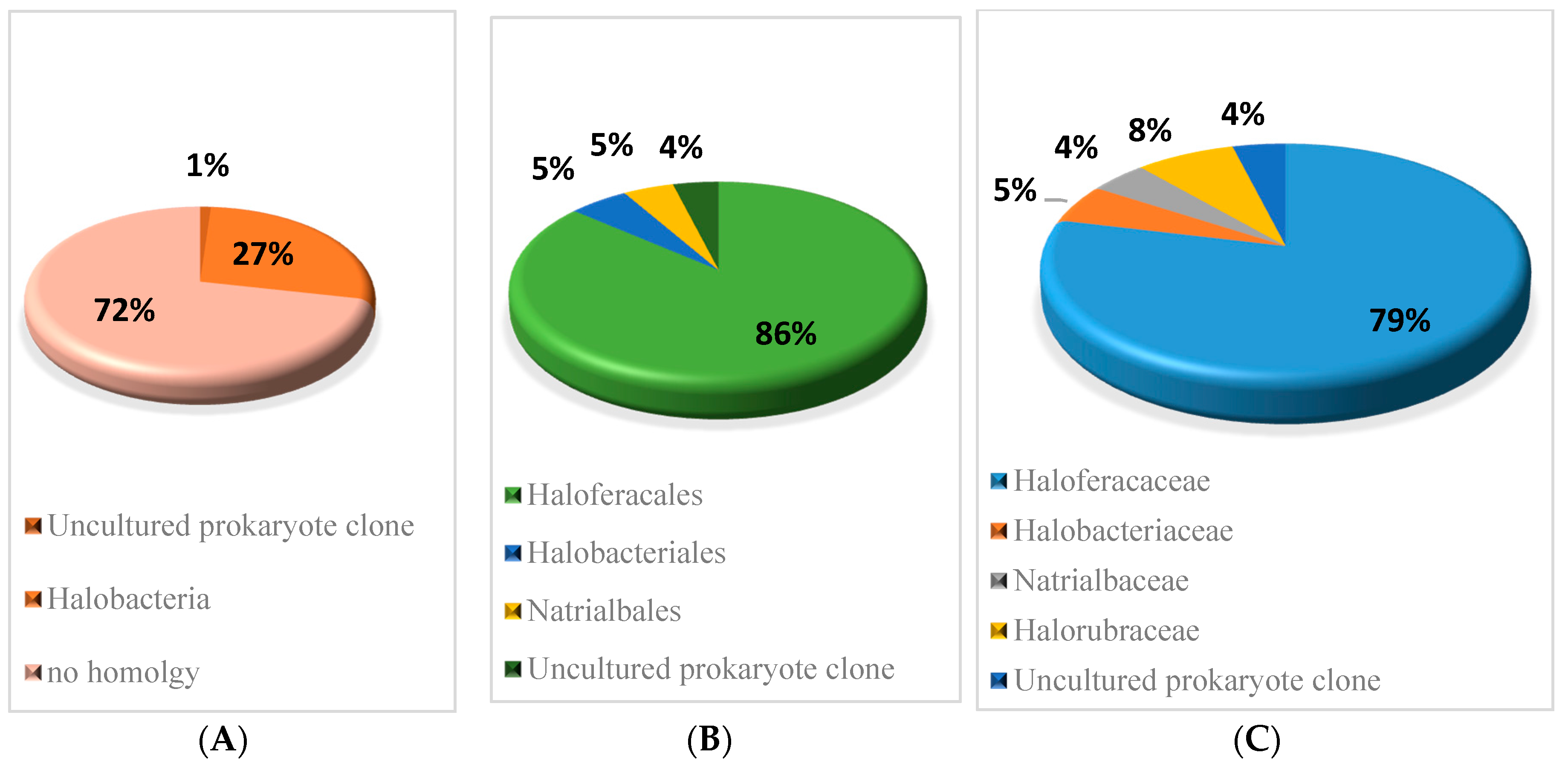
3.3.3. Conservation of small RNAs Verified in H. mediterranei
3.4. Expression Analysis According to the Nitrogen Source
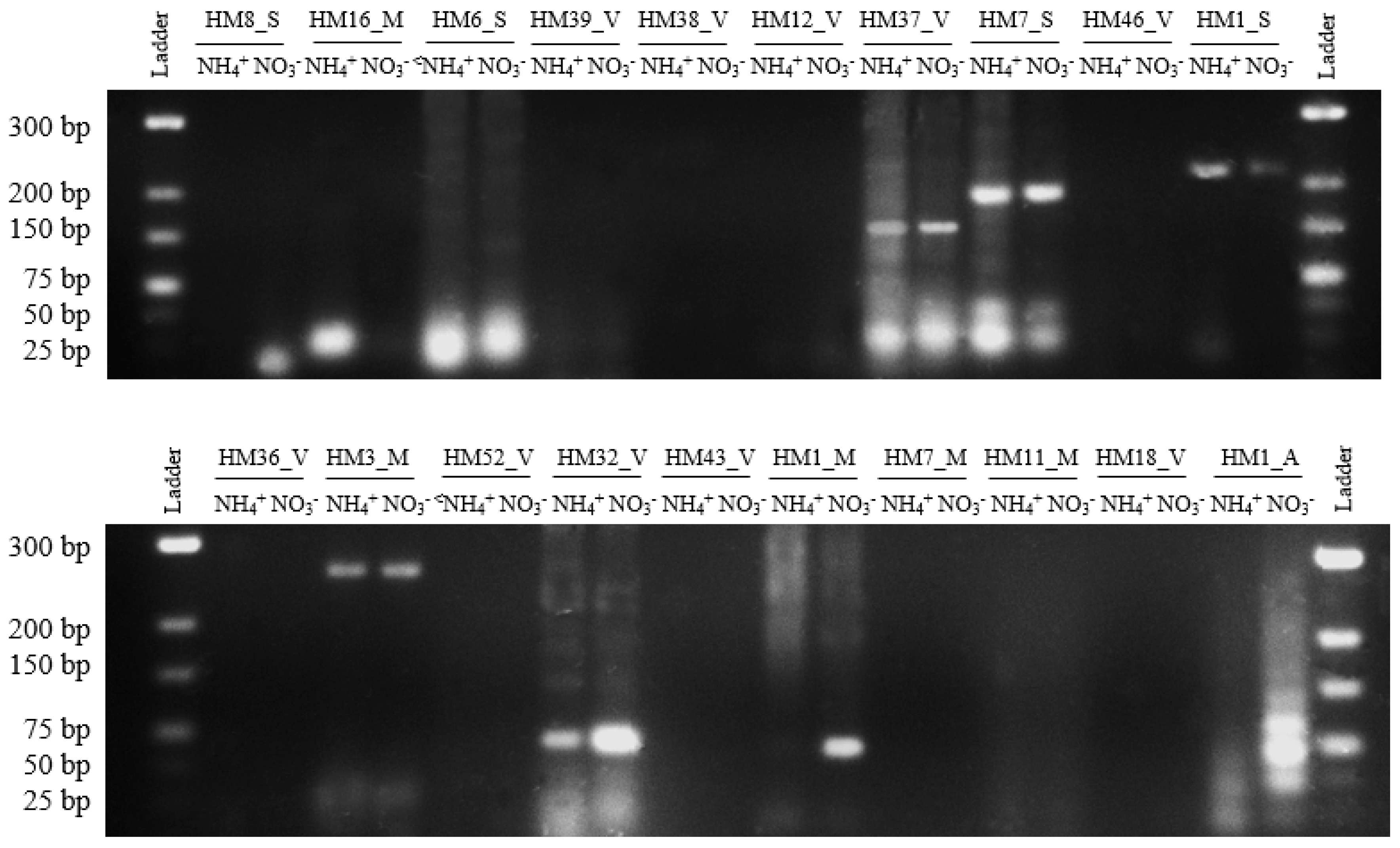
3.5. Validation of Expression of Small RNAs Using Reverse Transcription Polymerase Chain Reaction
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chu, C.Y.; Rana, T.M. Small RNAs: Regulators and guardians of the genome. J. Cell. Physiol. 2007, 213, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Choudhuri, S. Lesser known relatives of miRNA. Biochem. Biophys. Res. Commun. 2009, 388, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Paroo, Z. Biochemical principles of small RNA pathways. Annu. Rev. Biochem. 2010, 79, 295–319. [Google Scholar] [CrossRef] [PubMed]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by small RNAs in bacteria: Expanding frontiers. Mol. Cell. 2011, 43, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Babski, J.; Maier, L.K.; Heyer, R.; Jaschinski, K.; Prasse, D.; Jäger, D.; Randau, L.; Schmitz, R.A.; Marchfelder, A.; Soppa, J. Small regulatory RNAs in Archaea. RNA Biol. 2014, 11, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Michaux, C.; Hartke, A.; Martini, C.; Reiss, S.; Albrecht, D.; Budin-Verneuil, A.; Sanguinetti, M.; Engelmann, S.; Hain, T.; Verneuil, N.; et al. Involvement of Enterococcus faecalis small RNAs in stress response and virulence. Infect. Immun. 2014, 82, 3599–3611. [Google Scholar] [CrossRef] [PubMed]
- Durand, S.; Tomasini, A.; Braun, F.; Condon, C.; Romby, P. sRNA and mRNA turnover in Gram-positive bacteria. FEMS Microbiol. Rev. 2015, 39, 316–330. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, S.; Storz, G. Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- Moll, I.; Leitsch, D.; Steinhauser, T.; Bläsi, U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 2003, 4, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.; Leclerc, F.; Behm-Ansmant, I.; Fourmann, J.B.; Charpentier, B.; Branlant, C. Combined in silico and experimental identification of the Pyrococcus abyssi H/ACA sRNAs and their target sites in ribosomal RNAs. Nucleic Acids Res. 2008, 36, 2459–2475. [Google Scholar] [CrossRef] [PubMed]
- Phok, K.; Moisan, A.; Rinaldi, D.; Brucato, N.; Carpousis, A.J.; Gaspin, C.; Clouet-d’Orval, B. Identification of CRISPR and riboswitch related RNAs among novel noncoding RNAs of the euryarchaeon Pyrococcus abyssi. BMC Genom. 2011, 12, 312. [Google Scholar] [CrossRef] [PubMed]
- Heyer, R.; Dörr, M.; Jellen-Ritter, A.; Späth, B.; Babski, J.; Jaschinski, K.; Soppa, J.; Marchfelder, A. High throughput sequencing reveals a plethora of small RNAs including tRNA derived fragments in Haloferax volcanii. RNA Biol. 2012, 9, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Soppa, J.; Straub, J.; Brenneis, M.; Jellen-Ritter, A.; Heyer, R.; Fischer, S.; Granzow, M.; Voss, B.; Hess, W.R.; Tjaden, B.; et al. Small RNAs of the halophilic archaeon Haloferax volcanii. Biochem. Soc. Trans. 2009, 37, 133–136. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.G.; Romby, P. Small RNAs in bacteria and archaea: Who they are, what they do, and how they do it. Adv. Genet. 2015, 90, 133–208. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.H.; Bachellerie, J.P.; Rozhdestvensky, T.; Bortolin, M.L.; Huber, H.; Drungowski, M.; Elge, T.; Brosius, J.; Hüttenhofer, A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. USA 2002, 99, 7536–7541. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.-H.; Polacek, N.; Zywicki, M.; Huber, H.; Brugger, K.; Garrett, R.; Bachellerie, J.P.; Hüttenhofer, A. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol. Microbiol. 2005, 55, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Jäger, D.; Sharma, C.M.; Thomsen, J.; Ehlers, C.; Vogel, J.; Schmitz, R.A. Deep sequencing analysis of the Methanosarcina mazei Gö1 transcriptome in response to nitrogen availability. Proc. Natl. Acad. Sci. USA 2009, 106, 21878–21882. [Google Scholar] [CrossRef] [PubMed]
- Bernick, D.L.; Dennis, P.P.; Höchsmann, M.; Lowe, T.M. Discovery of Pyrobaculum small RNA families with atypical pseudouridine guide RNA features. RNA 2012, 18, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Jäger, D.; Förstner, K.U.; Sharma, C.M.; Santangelo, T.J.; John, N.; Reeve, J.N. Primary transcriptome map of the hyperthermophilic archaeon Thermococcus kodakarensis. BMC Genom. 2014, 15, 684. [Google Scholar] [CrossRef] [PubMed]
- Torreblanca, M.; Rodriguez-Valera, F.; Juez, G.; Ventosa, A.; Masahiro, K.; Morris, K. Classification of non-alkaliphilic halobacteria based on numerical taxonomy and polar lipid composition, and description of Haloarcula gen. nov. and Haloferax gen. nov. Syst. Appl. Microbiol. 1986, 8, 89–99. [Google Scholar] [CrossRef]
- Rodríguez-Valera, F.; Juez, G.; Kushner, D.J. Halobacterium mediterranei spec, nov., a new carbohydrate-utilizing extreme halophile. Syst. Appl. Microbiol. 1983, 4, 369–381. [Google Scholar] [CrossRef]
- Bonete, M.J.; Martínez-Espinosa, R.M.; Pire, C.; Zafrilla, B.; Richardson, D.J. Nitrogen metabolism in haloarchaea. Saline Syst. 2008, 4, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esclapez, J.; Bravo-Barrales, G.; Bautista, V.; Pire, C.; Camacho, M.; Bonete, M.J. Effects of nitrogen sources on the nitrate assimilation in Haloferax mediterranei: Growth kinetics and transcriptomic analysis. FEMS Microbiol. Lett. 2014, 350, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Esclapez, J.; Pire, C.; Camacho, M.; Bautista, V.; Martínez-Espinosa, R.M.; Zafrilla, B.; Vegara, A.; Alcaraz, L.A.; Bonete, M.J. Transcriptional profiles of Haloferax mediterranei based on nitrogen availability. J. Biotechnol. 2015, 193, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Esclapez, J.; Camacho, M.; Pire, C.; Bautista, V.; Vegara, A.; Pedro-Roig, L.; Pérez-Pomares, F.; Martínez-Espinosa, R.M.; Bonete, M.J. Recent Advances in The Nitrogen Metabolism in Haloarchaea and Its Biotechnological Applications. In Biotechnology of Extremophiles: Advances and Challenges; Rampelotto, P., Ed.; Springer: Cham, Switzerland, 2016; Volume 1, pp. 273–301. ISBN 978-3-319-13520-5. [Google Scholar]
- Pedro-Roig, L.; Lange, C.; Bonete, M.J.; Soppa, J.; Maupin-Furlow, J. Nitrogen regulation of protein-protein interactions and transcript levels of GlnK PII regulator and AmtB ammonium transporter homologs in Archaea. Microbiologyopen 2013, 2, 826–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Valera, F.; Ruiz-Berraquero, F.; Ramos-Cormenzana, A. Behaviour of mixed populations of halophilic bacteria in continuous cultures. Can. J. Microbiol. 1980, 26, 1259–1263. [Google Scholar] [CrossRef] [PubMed]
- Babski, J.; Tjaden, B.; Voss, B.; Jellen-Ritter, A.; Marchfelder, A.; Hess, W.R.; Soppa, J. Bioinformatic prediction and experimental verification of sRNAs in the haloarchaeon Haloferax volcanii. RNA Biol. 2011, 8, 806–816. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative genomics viewer (IGV): High-performance genomics. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002 30, 207–210. [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Kery, M.B.F.M.; Livny, J.; Tjaden, B. TargetRNA2: Identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Res. 2014, 42, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Mann, M.; Wright, P.R.; Backofen, R. IntaRNA 2.0: Enhanced and customizable prediction of RNA–RNA interactions. Nucleic Acids Res. 2017, 45, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Gaimster, H.; Chalklen, L.; Alston, M.; Munnoch, J.T.; Richardson, D.J.; Gates, A.J.; Rowley, G. Genome-wide discovery of putative sRNAs in Paracoccus denitrificans expressed under nitrous oxide emitting conditions. Front. Microbiol. 2016, 7, 1806. [Google Scholar] [CrossRef] [PubMed]
- Prasse, D.; Förstner, K.U.; Jäger, D.; Backofen, R.; Schmitz, R.A. sRNA154 a newly identified regulator of nitrogen fixation in Methanosarcina mazei strain Gö1. RNA Biol. 2017, 14, 1544–1558. [Google Scholar] [CrossRef] [PubMed]
- Pánek, J.; Bobek, J.; Mikulík, K.; Basler, M.; Vohradský, J. Biocomputational prediction of small non-coding RNAs in Streptomyces. BMC Genom. 2008, 9, 217. [Google Scholar] [CrossRef] [PubMed]
- Khoo, J.S.; Chai, S.F.; Mohamed, R.; Nathan, S.; Firdaus-Raih, M. Computational discovery and RT-PCR validation of novel Burkholderia conserved and Burkholderia pseudomallei unique sRNAs. BMC Genom. 2012, 13. [Google Scholar] [CrossRef]
- Panda, G.; Tanwer, P.; Ansari, S.; Khare, D.; Bhatnagar, R. Regulation and RNA-binding properties of Hfq-like RNA chaperones in Bacillus anthracis. Biochim. Biophys. Acta 2015, 1850, 1661–1668. [Google Scholar] [CrossRef] [PubMed]
- Kwenda, S.; Gorshkov, V.; Ramesh, A.M.; Naidoo, S.; Rubagotti, E.; Birch, P.R.; Moleleki, L.N. Discovery and profiling of small RNAs responsive to stress conditions in the plant pathogen Pectobacterium atrosepticum. BMC Genom. 2016, 12, 47. [Google Scholar] [CrossRef] [PubMed]
- Babski, J.; Haas, K.A.; Näther-Schindler, D.; Pfeiffer, F.; Förstner, K.U.; Hammelmann, M.; Hilker, R.; Becker, A.; Sharma, C.M.; Marchfelder, A.; Soppa, J. Genome-wide identification of transcriptional start sites in the haloarchaeon Haloferax volcanii based on differential RNA-Seq (dRNA-Seq). BMC Genom. 2016, 17, 629. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Gottesman, S. sRNA roles in regulating transcriptional regulators: Lrp and SoxS regulation by sRNAs. Nucleic Acids Res. 2016, 44, 6907–6923. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Benz, J.; Späth, B.; Maier, L.K.; Straub, J.; Granzow, M.; Raabe, M.; Urlaub, H.; Hoffmann, J.; Brutschy, B.; et al. The archaeal Lsm protein binds to small RNAs. J. Biol. Chem. 2010, 285, 45, 34429–34438. [Google Scholar] [CrossRef] [PubMed]
- Rivers, A.R.; Burns, A.S.; Chan, L.K.; Moran, M.A. Experimental identification of small non-coding RNAs in the model marine bacterium Ruegeria pomeroyi DSS-3. Front. Microbiol. 2016, 29, 380. [Google Scholar] [CrossRef] [PubMed]
- Wurtzel, O.; Sapra, R.; Chen, F.; Zhu, Y.; Simmons, B.A.; Sorek, R. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010, 20, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Straub, J.; Brenneis, M.; Jellen-Ritter, A.; Heyer, R.; Soppa, J.; Marchfelder, A. Small RNAs in haloarchaea: Identification, differential expression and biological function. RNA Biol. 2009, 6, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Zago, M.A.; Dennis, P.P.; Omer, A.D. The expanding world of small RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 2005, 55, 1812–1828. [Google Scholar] [CrossRef] [PubMed]
| Reference Species | Number of Candidate sRNAs | Identity (%) | Number of Mismatches | Number of Gaps | E-Value |
|---|---|---|---|---|---|
| H. volcanii | 116 | 74.19–100 | <30 | <11 | <0.05 |
| M. mazei | 55 | 82.86–100 | <5 | <2 | <1 |
| S. solfataricus | 47 | 95.83–100 | <3 | <3 | <1 |
| A. fulgidus | 74 | 73.28–100 | <30 | <4 | <1 |
| sRNA Name | Size sRNA (pb) | Position (Start-Stop) | Localisation H. Med | Strand | Classification | Gene Environment |
|---|---|---|---|---|---|---|
| HM1_V | 131 | 2947735-2947604 | CHR | - | Intergenic | HFX_3032 (gbp3)/HFX_0001(cdc6A-1) |
| HM3_V | 64 | 602004-601940 | CHR | - | Intergenic | HFX_0639/HFX_640 (aspS) |
| HM4_V | 143 | 2374110-2374253 | CHR | - | Intergenic | HFX_2271(htr18-1)/HFX_2270 |
| HM5_V | 46 | 576412-576366 | CHR | - | Intergenic | HFX_0609(cdc6C)/HFX_0610 |
| HM6_V | 117 | 401997-401880 | CHR | - | Intergenic | HFX_0430(ilvA-1)/HFX_0431 |
| HM8_V | 18 | 310608-310590 | CHR | - | Intergenic | HFX_0331(tRNA)/HFX_0332 (rpoH) |
| HM10_V | 42 | 1913258-1913216 | CHR | - | Intergenic | HFX_2748(rpl7AE)/HFX_2747 |
| HM12_V | 130 | 1928092-1927962 | CHR | - | Intergenic | HFX_2734/HFX_2733 (lsm1) |
| HM15_V | 113 | 2841808-2841695 | CHR | - | Intergenic | HFX_1820(rRNA)/HFX_2934 |
| HM17_V | 81 | 136329-136248 | CHR | - | Intergenic | HFX_0140 (tRNA)/HFX_0141 |
| HM19_V | 111 | 2175910-2175799 | CHR | - | Intergenic | HFX_2478/HFX_2477(tRNA) |
| HM20_V | 104 | 1737798-1737694 | CHR | - | Intergenic | HFX_1813/HFX_1814 |
| HM24_V | 96 | 440309-440213 | CHR | - | Intergenic | HFX_0475/HFX_0476 |
| HM25_V | 90 | 2584538-2584448 | CHR | - | Intergenic | HFX_2084(ydjP-1)/HFX_2083 |
| HM26_V | 102 | 1737680-1737578 | CHR | - | Intergenic | HFX_1813/HFX_1814 |
| HM28_V | 77 | 1298728-1298651 | CHR | - | Intergenic | HFX_1375(aroA)/HFX_1377 |
| HM30_V | 160 | 248344-248184 | CHR | - | Intergenic | HFX_0263/HFX_0264 |
| HM35_V | 168 | 2385624-2385456 | CHR | - | Antisense | HFX_2256 |
| HM36_V | 262 | 27244-26982 | CHR | - | Intergenic | HFX_0025/HFX_0026 |
| HM38_V | 91 | 2836827-2836736 | CHR | - | Intergenic | HFX_1823/HFX_1822 |
| HM39_V | 90 | 2685508-2685418 | CHR | - | Antisense | HFX_1980 (abc22A) |
| HM42_V | 58 | 1769424-1769366 | CHR | - | Intergenic | HFX_2905/HFX_2904(folP) |
| HM46_V | 200 | 2756370-2756170 | CHR | - | Intergenic | HFX_1903/HFX_1902 |
| HM47_V | 124 | 1816376-1816252 | CHR | - | Intergenic | HFX_2862/HFX_2861 |
| HM49_V | 38 | 359173-359135 | pHM500 | - | Intergenic | HFX_6336/HFX_6337 |
| HM50_V | 78 | 1116972-1116894 | CHR | - | Intergenic | HFX_1173 (tRNA)/HFX_1174(tatAE) |
| HM52_V | 457 | 2020941-2020484 | CHR | - | Intergenic | HFX_2638(dkgB)/HFX_2637(pepC) |
| HM53_V | 156 | 397174-397018 | CHR | - | Intergenic | HFX_0425/HFX_0426 |
| HM54_V | 72 | 2591733-2591661 | CHR | - | Intergenic | HFX_2076/2075 (tnp4) |
| HM55_V | 33 | 486185-486152 | CHR | - | Intergenic | HFX_0525(mutS)/HFX_0526(livK) |
| HM56_V | 170 | 2852306-2852136 | CHR | - | Intergenic | HFX_2941(pheS)/HFX_2942 |
| HM2_V | 52 | 681041-681093 | CHR | + | Intergenic | HFX_0721/HFX_0722 |
| HM7_V | 24 | 310803-310827 | CHR | + | Intergenic | HFX_0331 (tRNA)/HFX_0332 (rpoH) |
| HM9_V | 27 | 1913243-1913270 | CHR | + | Intergenic | HFX_2748(rpl7AE)/HFX_2747 |
| HM11_V | 29 | 1927979-1928008 | CHR | + | Intergenic | HFX_2734/HFX_2733 (lsm1) |
| HM13_V | 72 | 1930742-1930814 | CHR | + | Intergenic | HFX_2731(purF)/HFX_2730 |
| HM14_V | 49 | 1931964-1932013 | CHR | + | Intergenic | HFX_2729/HFX_2728 |
| HM16_V | 246 | 136148-136394 | CHR | + | Intergenic | HFX_0140 (tRNA)/HFX_0141 |
| HM18_V | 114 | 2175756-2175870 | CHR | + | Intergenic | HFX_2478/HFX_2477(tRNA) |
| HM21_V | 26 | 27000891-27000917 | CHR | + | Intergenic | HFX_1962/HFX_1961 |
| HM22_V | 21 | 2249860-2249881 | CHR | + | Intergenic | HFX_2397 (tRNA)/HFX_2396 |
| HM23_V | 111 | 257489-257600 | CHR | + | Intergenic | HFX_0274/HFX_0275 (tRNA) |
| HM27_V | 147 | 581307-581454 | CHR | + | Intergenic | HFX_0614(tRNA)/HFX_0615 |
| HM29_V | 258 | 248240-248498 | CHR | + | Intergenic | HFX_0263/HFX_0264 |
| HM31_V | 125 | 1824730-1824855 | CHR | + | Intergenic | HFX_2852/HFX_2851 |
| HM32_V | 102 | 2342182-2342284 | CHR | + | Intergenic | HFX_2304/HFX_2303 |
| HM33_V | 161 | 221020-221181 | CHR | + | Antisense | HFX_0231 (tfb1-1) |
| HM34_V | 95 | 1259873-1259968 | CHR | + | crRNA | HFX_1335/HFX_1336 |
| HM37_V | 149 | 95312-95461 | CHR | + | Antisense | HFX_0090 (HemL) |
| HM40_V | 35 | 1734001-1734036 | CHR | + | Antisense | HFX_1808 (ygcJ) |
| HM41_V | 53 | 2246499-2246552 | CHR | + | Antisense | HFX_2401 (xnuC-1) |
| HM43_V | 100 | 1104687-1104787 | CHR | + | Intergenic | HFX_1163 (tRNA)/HFX_1164 |
| HM44_V | 38 | 979793-979831 | CHR | + | Intergenic | HFX_1024/HFX_1026 |
| HM45_V | 90 | 809546-809636 | CHR | + | Intergenic | HFX_0853/HFX_0854 |
| HM48_V | 195 | 359045-359240 | pHM500 | + | Intergenic | HFX_6336/HFX_6337 |
| HM51_V | 457 | 2020484-2020941 | CHR | + | Intergenic | HFX_2638(dkgB)/HFX_2637(pepC) |
| HM54_V | 35 | 1299991-1300026 | CHR | + | Intergenic | HFX_1375(aroA)/HFX_1377 |
| HM55_V | 44 | 1300050-1300094 | CHR | + | Intergenic | HFX_1375(aroA)/HFX_1377 |
| HM1_M | 92 | 2342183-2342275 | CHR | + | Intergenic | HFX_2304/HFX_2303 (lactoyglutathione lyase) |
| HM2_M | 34 | 56050-56016 | pHM300 | - | Intergenic | HFX_5046 (cyp1_citocromo P450)/ HFX_5047 |
| HM3_M | 292 | 2357250-2357542 | CHR | + | Antisense | HFX_2287 (selR) |
| HM4_M | 35 | 201705-201740 | pHM500 | + | Antisense | HFX_6177 (nahC2_Antiporter Na/H) |
| HM5_M | 18 | 14274-14256 | pHM500 | - | Intergenic | HFX_6013 (nrd-1)/HFX_6014 (sodA) |
| HM6_M | 51 | 338357-338306 | pHM500 | - | Antisense | HFX_6316 (csh2-1_CRISPR-associated csh2 family protein) |
| HM7_M | 83 | 2448865-2448948 | CHR | + | Intergenic | HFX_2200/2199 (imd3) |
| HM8_M | 176 | 804495-804671 | CHR | + | Intergenic | HFX_0847/HFX_0848 (epf2-1) |
| HM9_M | 53 | 1087812-1087865 | CHR | + | Antisense | HFX_1142 |
| HM10_M | 97 | 1087972-1087875 | CHR | - | Intergenic | HFX_1142/1143 |
| HM11_M | 257 | 709963-709706 | CHR | - | Intergenic | HFX_0754 (prot.membr)/HFX_0755 |
| HM12_M | 78 | 862151-862073 | CHR | - | Intergenic | HFX_0905/HFX_0906 |
| HM13_M | 41 | 1287240-1287281 | CHR | + | Intergenic | HFX_1366/HFX_1367 (htlD-1) |
| HM14_M | 41 | 1287281-1287240 | CHR | - | Intergenic | HFX_1366/HFX_1367 (htlD-1) |
| HM15_M | 153 | 2508794-2508641 | CHR | - | Antisense | HFX_2148 (polysaccharide biosynthesis transporter) |
| HM16_M | 57 | 2661554-2661497 | CHR | - | Intergenic | HFX_2006 (paaJ-1)/HFX_2005 (nasD) |
| HM1_S | 208 | 2948115-2948323 | CHR | + | Intergenic | HFX_3032 (gbp3-1, GTP-binding protein)/HFX_0001 (cell division control protein) |
| HM2_S | 80 | 412151-412231 | CHR | + | Intergenic | HFX_0441 (pyrrolo-quinoline quinone)/HFX_0442 (serine/treonine protein kinase) |
| HM3_S | 98 | 1691336-1691238 | CHR | - | Antisense | HFX_1779 (integrase-like protein) |
| HM4_S | 81 | 310404-310323 | CHR | - | Intergenic | HFX_0329/HFX_0330 (tRNA) |
| HM5_S | 31 | 1104534-1104503 | CHR | - | Antisense | HFX_1161 (tRNA) |
| HM6_S | 60 | 547749-547689 | CHR | - | Antisense | HFX_0587 (ark-6-1, putative signal-transducing histadine kinase) |
| HM7_S | 185 | 2620188-2620373 | CHR | + | Intergenic | HFX_2046/HFX_2045 |
| HM8_S | 48 | 1432982-1432982 | CHR | - | Antisense | HFX_1518 (gdhA1, glutamate dehydrogenase NAD (P)) |
| HM1_A | 141 | 99868-100009 | CHR | + | Antisense | HM_0095 (amt1-1, ammonium transporter) |
| HM2_A | 482 | 792936-793418 | CHR | + | Antisense | HFX_0839 |
| HM3_A | 48 | 1277917-1277965 | CHR | + | Intergenic | HFX_1356/HFX_1357 |
| HM4_A | 19 | 1778748-1778729 | CHR | - | Intergenic | HFX_2897 (fumC-1, fumarate hydratase)/HFX_2896 (carbohydrate ATP-binding cassette (ABC) transporter substrate-binding protein) |
| HM5_A | 22 | 2113156-2113178 | CHR | + | Intergenic | HFX_2528/HFX_2529 (gpm, phosphoglycerate mutase) |
| HM6_A | 127 | 2520322-2520449 | CHR | + | Intergenic | HFX_2140 (cdc6H, cell division control protein cdc 6-like protein)/HFX_2139(galE, nucleoside-diphosphate sugar epimerase) |
| sRNA | sRNA Size (pb) | Gene Target | RNA-Seq Ammonium (+/−) | RNA-Seq Nitrate (+/−) | Target Position | sRNA Position | Energy (Kcal/mol) |
|---|---|---|---|---|---|---|---|
| HM33_V | 161 | tfb1-1 | + | + | 513–662 | 1–150 | −247.734 |
| HM35_V | 168 | HFX_2256 | + | − | 414–468 | 10–88 | −19.201 |
| HM37_V | 149 | HemL | + | + | 971–1120 | 1–149 | −238.470 |
| HM39_V | 90 | abc22A | + | + | 375–412 | 28–62 | −15.561 |
| HM40_V | 35 | ygcJ | + | + | 260–295 | 1–35 | −55.516 |
| HM41_V | 53 | xnuC-1 | + | + | 583–636 | 1–53 | −86.827 |
| HM3_M | 292 | selR | + | + | 316–423 | 186–292 | −162.400 |
| HM4_M | 35 | nahC2, antiporter Na+/H+ | + | + | 1151–1186 | 1–35 | −55.151 |
| HM6_M | 51 | csh2-1_CRISPR-associated csh2 family protein | + | + | – | – | – |
| HM9_M | 53 | HFX_1142 | + | + | 1–24 | 1–24 | −33.421 |
| HM15_M | 153 | Polysaccharide biosynthesis transporter | + | + | 93–110 | 115–129 | −14.191 |
| HM1_A | 141 | amt1-1, ammonium transporter | + | − | 1330–1471 | 1–141 | −229.279 |
| HM2_A | 482 | HFX_0839 | + | + | 513–662 | 301–450 | −245.770 |
| HM3_S | 98 | Integrase-like protein | + | + | 17–29 | 21–33 | −9.201 |
| HM5_S | 31 | tRNA | + | + | 47–54 | 25–32 | −1.222 |
| HM6_S | 60 | ark-6-1, putative signal-transducing histadine kinase | + | + | 1546–1556 | 26–37 | −12.128 |
| HM8_S | 48 | gdhA1, glutamate dehydrogenase NAD (P) | − | + | 230–236 | 2–8 | −6.608 |
| sRNA Name | Localisation | Strand | Position | Size (nt) | Classification | Gene Target 1 | log2 FoldChange | p-Value | p-adj |
|---|---|---|---|---|---|---|---|---|---|
| HM16_M | CHR | - | 2661497-2661554 | 57 | Intergenic | HFX_1537, pgk, HFX_1505, trkA, HFX_1789 | 8.6985 | 5.05 × 10−259 | 8.09 × 10−258 |
| HM7_S | CHR | + | 2620188-2620373 | 185 | Intergenic | HFX_0627, acnA, HFX_1005, arsR, HFX_3006 | 1.7547 | 1.29 × 10−24 | 1.03 × 10−23 |
| HM52_V | CHR | - | 2020941-2020484 | 457 | Intergenic | hflC, HFX_0589, HFX_0355 | 1.3633 | 1.06 × 10−21 | 5.96 × 10−20 |
| HM46_V | CHR | - | 2756370-2756170 | 200 | Intergenic | HFX_0329, psmA, HFX_2148, HFX_1154, graD5 | 1.1966 | 2.59 × 10−6 | 7.25 × 10−5 |
| HM39_V | CHR | - | 2685508-2685418 | 90 | Antisense | ilvB, cbs_4, HFX_0847, gldA, ligA | 0.8498 | 0.006 | 0.0413 |
| HM54_V | CHR | + | 2591733-2591661 | 35 | Intergenic | HFX_0906, HFX_1268, HFX_0562, arsR, nfi | 0.6708 | 0.004 | 0.0338 |
| HM12_V | CHR | - | 1928092-1927962 | 130 | Intergenic | HFX_1575, mutT, HFX_2074, coxB4 | 0.6263 | 0.003 | 0.0333 |
| HM1_S | CHR | + | 2948115-2948323 | 208 | Intergenic | livG, HFX_1497, HFX_2088, HFX_2739, nce1 | 0.5192 | 0.0003 | 0.00086 |
| HM37_V | CHR | + | 95312-95461 | 149 | Antisense | HFX_0565, nifS, HFX_1510, HFX_0591 | −0.4783 | 0.0011 | 0.0158 |
| HM36_V | CHR | - | 27244-26982 | 262 | Intergenic | HFX_2059, rpoF, apa | −0.4845 | 0.0036 | 0.0332 |
| HM38_V | CHR | - | 2836827-2836736 | 91 | Intergenic | menD, hcpE, HFX_1197, HFX_0402 | −0.6120 | 0.008 | 0.0451 |
| HM3_M | CHR | + | 2357250-2357542 | 292 | Antisense | HFX_2088, rnhB, petE, HFX_2672, HFX_2025 | −0.6247 | 0.0112 | 0.0894 |
| HM8_V | CHR | - | 310608-310590 | 18 | Intergenic | no targets | −0.7387 | 0.0079 | 0.0451 |
| HM6_S | CHR | - | 547689-547749 | 60 | Antisense | xthA, dap2, HFX_0771, HFX_0201, yfmJ1 | −0.8373 | 0.0003 | 0.00086 |
| HM8_S | CHR | - | 1432982-1433030 | 48 | Antisense | atpl, HFX_0366, gatD, atpF, gvpJ | −0.8791 | 0.0021 | 0.00413 |
| HM9_V | CHR | + | 1913243-1913270 | 27 | Intergenic | ppiB, HFX_1294, HFX_2540, ispA, HFX_2807 | −1.3253 | 0.0007 | 0.0128 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Payá, G.; Bautista, V.; Camacho, M.; Castejón-Fernández, N.; Alcaraz, L.A.; Bonete, M.-J.; Esclapez, J. Small RNAs of Haloferax mediterranei: Identification and Potential Involvement in Nitrogen Metabolism. Genes 2018, 9, 83. https://doi.org/10.3390/genes9020083
Payá G, Bautista V, Camacho M, Castejón-Fernández N, Alcaraz LA, Bonete M-J, Esclapez J. Small RNAs of Haloferax mediterranei: Identification and Potential Involvement in Nitrogen Metabolism. Genes. 2018; 9(2):83. https://doi.org/10.3390/genes9020083
Chicago/Turabian StylePayá, Gloria, Vanesa Bautista, Mónica Camacho, Natalia Castejón-Fernández, Luís A. Alcaraz, María-José Bonete, and Julia Esclapez. 2018. "Small RNAs of Haloferax mediterranei: Identification and Potential Involvement in Nitrogen Metabolism" Genes 9, no. 2: 83. https://doi.org/10.3390/genes9020083






