SARS-CoV-2: A Glance at the Innate Immune Response Elicited by Infection and Vaccination
Abstract
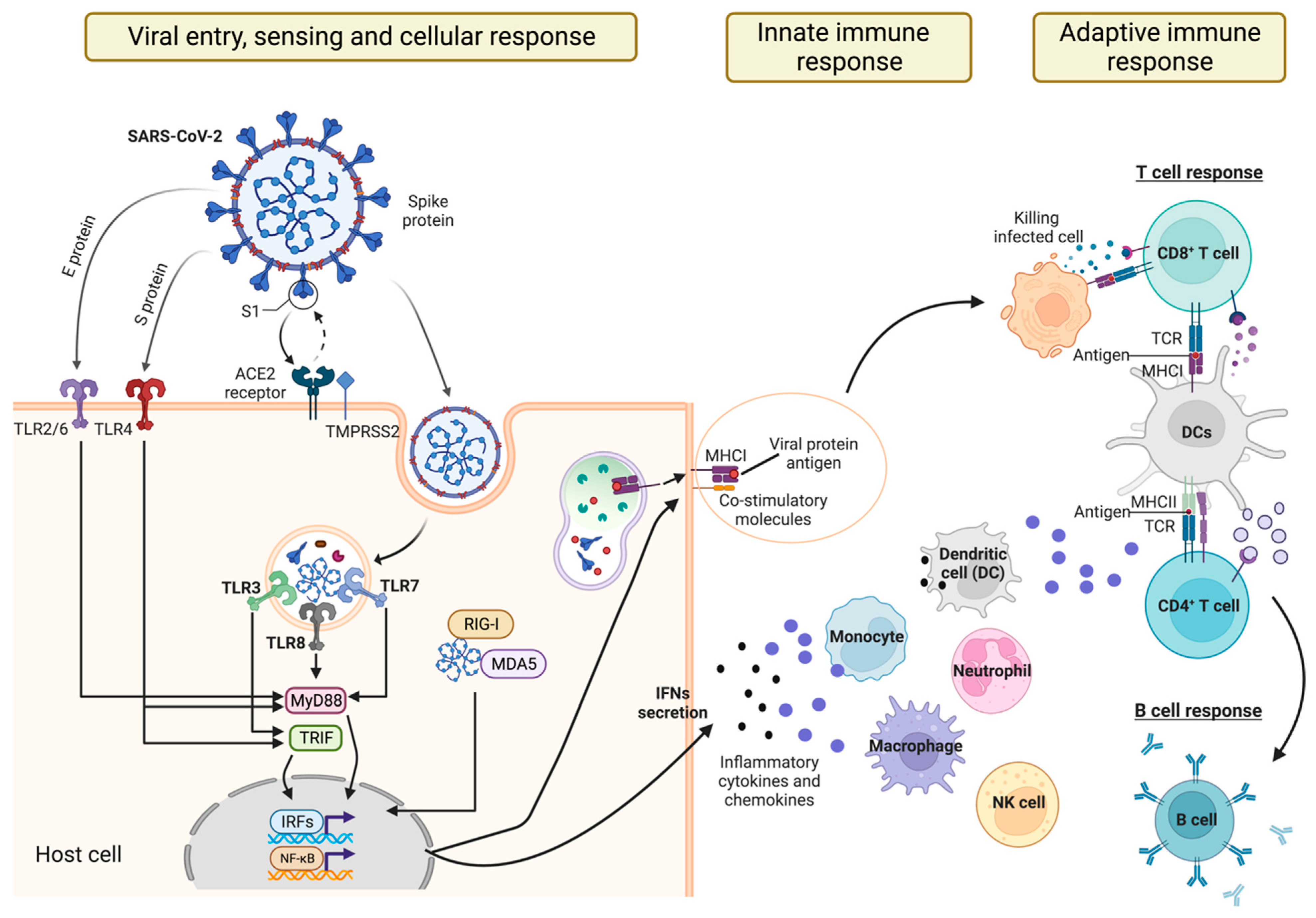
:1. Introduction
2. Sensing SARS-CoV-2 Infection
2.1. Recognition of SARS-CoV-2 by TLRs
2.2. Recognition of SARS-CoV-2 by RLRs
2.3. Recognition of SARS-CoV-2 by NLRs
3. Innate Cell Responses to SARS-CoV-2 Infection
3.1. Macrophages, Monocytes, and SARS-CoV-2
3.2. Dendritic Cells and SARS-CoV-2
3.3. Natural Killer Cells and SARS-CoV-2
4. Innate Immunity and COVID-19 Vaccines
4.1. Differences in Innate Immune Responses Induced by Different Vaccines
4.2. Innate and Adaptive Immune Response Cross-Talk upon COVID-19 Vaccination
5. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Nalla, L.V.; Sharma, M.; Sharma, N.; Singh, A.A.; Malim, F.M.; Ghatage, M.; Mukarram, M.; Pawar, A.; Parihar, N.; et al. Association of COVID-19 with Comorbidities: An Update. ACS Pharmacol. Transl. Sci. 2023, 6, 334–354. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Cai, Z.; Xiao, X.; Rao, J.; Chen, J.; Hu, N.; Yang, M.; Xing, X.; Wang, Y.; Li, M.; et al. The architecture of the SARS-CoV-2 RNA genome inside virion. Nat. Commun. 2021, 12, 3917. [Google Scholar] [CrossRef] [PubMed]
- Mousavizadeh, L.; Ghasemi, S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2021, 54, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Troyano-Hernáez, P.; Reinosa, R.; Holguín, Á. Evolution of SARS-CoV-2 Envelope, Membrane, Nucleocapsid, and Spike Structural Proteins from the Beginning of the Pandemic to September 2020: A Global and Regional Approach by Epidemiological Week. Viruses 2021, 13, 243. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef] [PubMed]
- Nejat, R.; Torshizi, M.F.; Najafi, D.J. S Protein, ACE2 and Host Cell Proteases in SARS-CoV-2 Cell Entry and Infectivity; Is Soluble ACE2 a Two Blade Sword? A Narrative Review. Vaccines 2023, 11, 204. [Google Scholar] [CrossRef]
- Ashraf, U.M.; Abokor, A.A.; Edwards, J.M.; Waigi, E.W.; Royfman, R.S.; Hasan, S.A.-M.; Smedlund, K.B.; Hardy, A.M.G.; Chakravarti, R.; Koch, L.G. SARS-CoV-2, ACE2 expression, and systemic organ invasion. Physiol. Genom. 2021, 53, 51–60. [Google Scholar] [CrossRef]
- Miluzio, A.; Cuomo, A.; Cordiglieri, C.; Donnici, L.; Pesce, E.; Bombaci, M.; Conti, M.; Fasciani, A.; Terracciano, L.; Manganaro, L.; et al. Mapping of functional SARS-CoV-2 receptors in human lungs establishes differences in variant binding and SLC1A5 as a viral entry modulator of hACE2. EBioMedicine 2023, 87, 104390. [Google Scholar] [CrossRef]
- Arrindell, J.; Abou Atmeh, P.; Jayet, L.; Sereme, Y.; Mege, J.-L.; Desnues, B. Vimentin is an important ACE2 co-receptor for SARS-CoV-2 in epithelial cells. iScience 2022, 25, 105463. [Google Scholar] [CrossRef] [PubMed]
- Fraser, B.J.; Beldar, S.; Seitova, A.; Hutchinson, A.; Mannar, D.; Li, Y.; Kwon, D.; Tan, R.; Wilson, R.P.; Leopold, K.; et al. Structure and activity of human TMPRSS2 protease implicated in SARS-CoV-2 activation. Nat. Chem. Biol. 2022, 18, 963–971. [Google Scholar] [CrossRef] [PubMed]
- Takeda, M. Proteolytic activation of SARS-CoV-2 spike protein. Microbiol. Immunol. 2022, 66, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, T.; Mukai, K. Innate immunity signalling and membrane trafficking. Curr. Opin. Cell Biol. 2019, 59, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Weber, F. Antiviral Innate Immunity: Introduction. In Encyclopedia of Virology; Elsevier: Amsterdam, The Netherlands, 2021; pp. 577–583. [Google Scholar] [CrossRef]
- Diamond, M.S.; Kanneganti, T.-D. Innate immunity: The first line of defense against SARS-CoV-2. Nat. Immunol. 2022, 23, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Kanneganti, T. Intracellular innate immune receptors: Life inside the cell. Immunol. Rev. 2020, 297, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Sonnenberg, G.F.; Hepworth, M.R. Functional interactions between innate lymphoid cells and adaptive immunity. Nat. Rev. Immunol. 2019, 19, 599–613. [Google Scholar] [CrossRef]
- Yamada, T.; Takaoka, A. Innate immune recognition against SARS-CoV-2. Inflamm. Regen. 2023, 43, 7. [Google Scholar] [CrossRef]
- Lester, S.N.; Li, K. Toll-Like Receptors in Antiviral Innate Immunity. J. Mol. Biol. 2014, 426, 1246–1264. [Google Scholar] [CrossRef]
- Shen, J.; Fan, J.; Zhao, Y.; Jiang, D.; Niu, Z.; Zhang, Z.; Cao, G. Innate and adaptive immunity to SARS-CoV-2 and predisposing factors. Front. Immunol. 2023, 14, 1159326. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Kawai, T. Toll-Like Receptor Signaling Pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, S.; Oliviero, B.; Varchetta, S.; Renieri, A.; Mondelli, M.U. TLRs: Innate Immune Sentries against SARS-CoV-2 Infection. Int. J. Mol. Sci. 2023, 24, 8065. [Google Scholar] [CrossRef]
- Tyrkalska, S.D.; Martínez-López, A.; Pedoto, A.; Candel, S.; Cayuela, M.L.; Mulero, V. The Spike protein of SARS-CoV-2 signals via Tlr2 in zebrafish. Dev. Comp. Immunol. 2023, 140, 104626. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Karki, R.; Williams, E.P.; Yang, D.; Fitzpatrick, E.; Vogel, P.; Jonsson, C.B.; Kanneganti, T.-D. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021, 22, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Shirato, K.; Kizaki, T. SARS-CoV-2 spike protein S1 subunit induces pro-inflammatory responses via toll-like receptor 4 signaling in murine and human macrophages. Heliyon 2021, 7, e06187. [Google Scholar] [CrossRef]
- Liu, G.; Zhao, Y. Toll-like receptors and immune regulation: Their direct and indirect modulation on regulatory CD4+ CD25 + T cells. Immunology 2007, 122, 149–156. [Google Scholar] [CrossRef]
- Jung, S.; Potapov, I.; Chillara, S.; del Sol, A. Leveraging systems biology for predicting modulators of inflammation in patients with COVID-19. Sci. Adv. 2021, 7, eabe5735. [Google Scholar] [CrossRef]
- Khan, S.; Shafiei, M.S.; Longoria, C.; Schoggins, J.W.; Savani, R.C.; Zaki, H. SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-κB pathway. Elife 2021, 10, e68563. [Google Scholar] [CrossRef] [PubMed]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Khanmohammadi, S.; Rezaei, N. Role of Toll-like receptors in the pathogenesis of COVID-19. J. Med. Virol. 2021, 93, 2735–2739. [Google Scholar] [CrossRef]
- Zhao, J.; Wohlford-Lenane, C.; Zhao, J.; Fleming, E.; Lane, T.E.; McCray, P.B.; Perlman, S. Intranasal Treatment with Poly(I·C) Protects Aged Mice from Lethal Respiratory Virus Infections. J. Virol. 2012, 86, 11416–11424. [Google Scholar] [CrossRef]
- Bortolotti, D.; Gentili, V.; Rizzo, S.; Schiuma, G.; Beltrami, S.; Strazzabosco, G.; Fernandez, M.; Caccuri, F.; Caruso, A.; Rizzo, R. TLR3 and TLR7 RNA Sensor Activation during SARS-CoV-2 Infection. Microorganisms 2021, 9, 1820. [Google Scholar] [CrossRef]
- Jung, H.E.; Lee, H.K. Current Understanding of the Innate Control of Toll-like Receptors in Response to SARS-CoV-2 Infection. Viruses 2021, 13, 2132. [Google Scholar] [CrossRef]
- Yoneyama, M.; Fujita, T. Function of RIG-I-like Receptors in Antiviral Innate Immunity. J. Biol. Chem. 2007, 282, 15315–15318. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Pattern Recognition Receptors and Inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Thorne, L.G.; Reuschl, A.; Zuliani-Alvarez, L.; Whelan, M.V.X.; Turner, J.; Noursadeghi, M.; Jolly, C.; Towers, G.J. SARS-CoV-2 sensing by RIG-I and MDA5 links epithelial infection to macrophage inflammation. EMBO J. 2021, 40, e107826. [Google Scholar] [CrossRef] [PubMed]
- Rebendenne, A.; Chaves Valadão, A.L.; Tauziet, M.; Maarifi, G.; Bonaventure, B.; McKellar, J.; Planès, R.; Nisole, S.; Arnaud-Arnould, M.; Moncorgé, O.; et al. SARS-CoV-2 Triggers an MDA-5-Dependent Interferon Response Which Is Unable To Control Replication in Lung Epithelial Cells. J. Virol. 2021, 95, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Riva, L.; Pu, Y.; Martin-Sancho, L.; Kanamune, J.; Yamamoto, Y.; Sakai, K.; Gotoh, S.; Miorin, L.; De Jesus, P.D.; et al. MDA5 Governs the Innate Immune Response to SARS-CoV-2 in Lung Epithelial Cells. Cell Rep. 2021, 34, 108628. [Google Scholar] [CrossRef] [PubMed]
- Refolo, G.; Vescovo, T.; Piacentini, M.; Fimia, G.M.; Ciccosanti, F. Mitochondrial Interactome: A Focus on Antiviral Signaling Pathways. Front. Cell Dev. Biol. 2020, 8, 8. [Google Scholar] [CrossRef]
- Wack, A.; Terczyńska-Dyla, E.; Hartmann, R. Guarding the frontiers: The biology of type III interferons. Nat. Immunol. 2015, 16, 802–809. [Google Scholar] [CrossRef]
- Horner, S.M.; Liu, H.M.; Park, H.S.; Briley, J.; Gale, M. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc. Natl. Acad. Sci. USA 2011, 108, 14590–14595. [Google Scholar] [CrossRef] [PubMed]
- Loo, Y.-M.; Gale, M. Immune Signaling by RIG-I-like Receptors. Immunity 2011, 34, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.-M.; Geng, T.-T.; Harrison, A.G.; Wang, P.-H. Differential roles of RIG-I like receptors in SARS-CoV-2 infection. Mil. Med. Res. 2021, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Thiel, V.; Ivanov, K.A.; Putics, Á.; Hertzig, T.; Schelle, B.; Bayer, S.; Weißbrich, B.; Snijder, E.J.; Rabenau, H.; Doerr, H.W.; et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003, 84, 2305–2315. [Google Scholar] [CrossRef] [PubMed]
- Ran, X.-H.; Zhu, J.-W.; Chen, Y.-Y.; Ni, R.-Z.; Mu, D. Papain-like protease of SARS-CoV-2 inhibits RLR signaling in a deubiquitination-dependent and deubiquitination-independent manner. Front. Immunol. 2022, 13, 947272. [Google Scholar] [CrossRef] [PubMed]
- Loske, J.; Röhmel, J.; Lukassen, S.; Stricker, S.; Magalhães, V.G.; Liebig, J.; Chua, R.L.; Thürmann, L.; Messingschlager, M.; Seegebarth, A.; et al. Pre-activated antiviral innate immunity in the upper airways controls early SARS-CoV-2 infection in children. Nat. Biotechnol. 2022, 40, 319–324. [Google Scholar] [CrossRef]
- Bastard, P.; Rosen, L.B.; Zhang, Q.; Michailidis, E.; Hoffmann, H.-H.; Zhang, Y.; Dorgham, K.; Philippot, Q.; Rosain, J.; Béziat, V.; et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 2020, 370, abd4585. [Google Scholar] [CrossRef]
- Yamada, T.; Sato, S.; Sotoyama, Y.; Orba, Y.; Sawa, H.; Yamauchi, H.; Sasaki, M.; Takaoka, A. RIG-I triggers a signaling-abortive anti-SARS-CoV-2 defense in human lung cells. Nat. Immunol. 2021, 22, 820–828. [Google Scholar] [CrossRef]
- Kouwaki, T.; Nishimura, T.; Wang, G.; Oshiumi, H. RIG-I-Like Receptor-Mediated Recognition of Viral Genomic RNA of Severe Acute Respiratory Syndrome Coronavirus-2 and Viral Escape From the Host Innate Immune Responses. Front. Immunol. 2021, 12, 700926. [Google Scholar] [CrossRef]
- Harris, J.; Borg, N.A. The multifaceted roles of NLRP3-modulating proteins in virus infection. Front. Immunol. 2022, 13, 987453. [Google Scholar] [CrossRef]
- Zheng, Q.; Hua, C.; Liang, Q.; Cheng, H. The NLRP3 inflammasome in viral infection (Review). Mol. Med. Rep. 2023, 28, 160. [Google Scholar] [CrossRef]
- Swanson, K.V.; Deng, M.; Ting, J.P.-Y. The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019, 19, 477–489. [Google Scholar] [CrossRef]
- Blevins, H.M.; Xu, Y.; Biby, S.; Zhang, S. The NLRP3 Inflammasome Pathway: A Review of Mechanisms and Inhibitors for the Treatment of Inflammatory Diseases. Front. Aging Neurosci. 2022, 14, e879021. [Google Scholar] [CrossRef] [PubMed]
- Pan, P.; Shen, M.; Yu, Z.; Ge, W.; Chen, K.; Tian, M.; Xiao, F.; Wang, Z.; Wang, J.; Jia, Y.; et al. SARS-CoV-2 N protein promotes NLRP3 inflammasome activation to induce hyperinflammation. Nat. Commun. 2021, 12, 4664. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.C.; Soares, V.C.; de Azevedo-Quintanilha, I.G.; da Dias, S.S.G.; Fintelman-Rodrigues, N.; Sacramento, C.Q.; Mattos, M.; de Freitas, C.S.; Temerozo, J.R.; Teixeira, L.; et al. SARS-CoV-2 engages inflammasome and pyroptosis in human primary monocytes. Cell Death Discov. 2021, 7, 43. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.S.; de Sá, K.S.G.; Ishimoto, A.Y.; Becerra, A.; Oliveira, S.; Almeida, L.; Gonçalves, A.V.; Perucello, D.B.; Andrade, W.A.; Castro, R.; et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 2021, 218, e20201707. [Google Scholar] [CrossRef] [PubMed]
- Nieto-Torres, J.L.; Verdiá-Báguena, C.; Jimenez-Guardeño, J.M.; Regla-Nava, J.A.; Castaño-Rodriguez, C.; Fernandez-Delgado, R.; Torres, J.; Aguilella, V.M.; Enjuanes, L. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology 2015, 485, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Aboudounya, M.M.; Heads, R.J. COVID-19 and Toll-Like Receptor 4 (TLR4): SARS-CoV-2 May Bind and Activate TLR4 to Increase ACE2 Expression, Facilitating Entry and Causing Hyperinflammation. Mediat. Inflamm. 2021, 2021, 8874339. [Google Scholar] [CrossRef] [PubMed]
- Rodero, M.P.; Poupel, L.; Loyher, P.-L.; Hamon, P.; Licata, F.; Pessel, C.; Hume, D.A.; Combadière, C.; Boissonnas, A. Immune surveillance of the lung by migrating tissue monocytes. Elife 2015, 4, e07847. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, A.; Das, N.C.; Patra, R.; Mukherjee, S. In silico analyses on the comparative sensing of SARS-CoV-2 mRNA by the intracellular TLRs of humans. J. Med. Virol. 2021, 93, 2476–2486. [Google Scholar] [CrossRef]
- Schiuma, G.; Beltrami, S.; Bortolotti, D.; Rizzo, S.; Rizzo, R. Innate Immune Response in SARS-CoV-2 Infection. Microorganisms 2022, 10, 501. [Google Scholar] [CrossRef]
- Steinmüller, C.; Franke-Ullmann, G.; Lohmann-Matthes, M.-L.; Emmendörffer, A. Local Activation of Nonspecific Defense against a Respiratory Model Infection by Application of Interferon-γ. Am. J. Respir. Cell Mol. Biol. 2000, 22, 481–490. [Google Scholar] [CrossRef]
- Narasimhan, P.B.; Marcovecchio, P.; Hamers, A.A.J.; Hedrick, C.C. Nonclassical Monocytes in Health and Disease. Annu. Rev. Immunol. 2019, 37, 439–456. [Google Scholar] [CrossRef]
- Rajamanickam, A.; Kumar, N.P.; Pandiarajan, A.N.; Selvaraj, N.; Munisankar, S.; Renji, R.M.; Venkatramani, V.; Murhekar, M.; Thangaraj, J.W.V.; Kumar, M.S.; et al. Dynamic alterations in monocyte numbers, subset frequencies and activation markers in acute and convalescent COVID-19 individuals. Sci. Rep. 2021, 11, 20254. [Google Scholar] [CrossRef]
- Baharom, F.; Rankin, G.; Blomberg, A.; Smed-Sörensen, A. Human Lung Mononuclear Phagocytes in Health and Disease. Front. Immunol. 2017, 8, 499. [Google Scholar] [CrossRef] [PubMed]
- Guilliams, M.; Mildner, A.; Yona, S. Developmental and Functional Heterogeneity of Monocytes. Immunity 2018, 49, 595–613. [Google Scholar] [CrossRef] [PubMed]
- Eskandarian Boroujeni, M.; Sekrecka, A.; Antonczyk, A.; Hassani, S.; Sekrecki, M.; Nowicka, H.; Lopacinska, N.; Olya, A.; Kluzek, K.; Wesoly, J.; et al. Dysregulated Interferon Response and Immune Hyperactivation in Severe COVID-19: Targeting STATs as a Novel Therapeutic Strategy. Front. Immunol. 2022, 13, 888897. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhou, F.; Zhang, L. STING, a critical contributor to SARS-CoV-2 immunopathology. Signal Transduct. Target. Ther. 2022, 7, 106. [Google Scholar] [CrossRef]
- Di Domizio, J.; Gulen, M.F.; Saidoune, F.; Thacker, V.V.; Yatim, A.; Sharma, K.; Nass, T.; Guenova, E.; Schaller, M.; Conrad, C.; et al. The cGAS–STING pathway drives type I IFN immunopathology in COVID-19. Nature 2022, 603, 145–151. [Google Scholar] [CrossRef]
- Benlyamani, I.; Venet, F.; Coudereau, R.; Gossez, M.; Monneret, G. Monocyte HLA-DR Measurement by Flow Cytometry in COVID-19 Patients: An Interim Review. Cytom. Part A 2020, 97, 1217–1221. [Google Scholar] [CrossRef]
- Gatti, A.; Radrizzani, D.; Viganò, P.; Mazzone, A.; Brando, B. Decrease of Non-Classical and Intermediate Monocyte Subsets in Severe Acute SARS-CoV-2 Infection. Cytom. Part A 2020, 97, 887–890. [Google Scholar] [CrossRef]
- Kitamura, H.; Kamon, H.; Sawa, S.; Park, S.-J.; Katunuma, N.; Ishihara, K.; Murakami, M.; Hirano, T. IL-6-STAT3 Controls Intracellular MHC Class II αβ Dimer Level through Cathepsin S Activity in Dendritic Cells. Immunity 2005, 23, 491–502. [Google Scholar] [CrossRef]
- Vremec, D.; O’Keeffe, M.; Hochrein, H.; Fuchsberger, M.; Caminschi, I.; Lahoud, M.; Shortman, K. Production of interferons by dendritic cells, plasmacytoid cells, natural killer cells, and interferon-producing killer dendritic cells. Blood 2007, 109, 1165–1173. [Google Scholar] [CrossRef]
- Sánchez-Cerrillo, I.; Landete, P.; Aldave, B.; Sánchez-Alonso, S.; Sánchez-Azofra, A.; Marcos-Jiménez, A.; Ávalos, E.; Alcaraz-Serna, A.; de los Santos, I.; Mateu-Albero, T.; et al. COVID-19 severity associates with pulmonary redistribution of CD1c+ DCs and inflammatory transitional and nonclassical monocytes. J. Clin. Investig. 2020, 130, 6290–6300. [Google Scholar] [CrossRef] [PubMed]
- Saichi, M.; Ladjemi, M.Z.; Korniotis, S.; Rousseau, C.; Ait Hamou, Z.; Massenet-Regad, L.; Amblard, E.; Noel, F.; Marie, Y.; Bouteiller, D.; et al. Single-cell RNA sequencing of blood antigen-presenting cells in severe COVID-19 reveals multi-process defects in antiviral immunity. Nat. Cell Biol. 2021, 23, 538–551. [Google Scholar] [CrossRef]
- Poli, A.; Michel, T.; Thérésine, M.; Andrès, E.; Hentges, F.; Zimmer, J. CD56bright natural killer (NK) cells: An important NK cell subset. Immunology 2009, 126, 458–465. [Google Scholar] [CrossRef]
- Xiong, Y.; Liu, Y.; Cao, L.; Wang, D.; Guo, M.; Jiang, A.; Guo, D.; Hu, W.; Yang, J.; Tang, Z.; et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020, 9, 761–770. [Google Scholar] [CrossRef]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef] [PubMed]
- Bortolotti, D.; Gentili, V.; Rizzo, S.; Rotola, A.; Rizzo, R. SARS-CoV-2 Spike 1 Protein Controls Natural Killer Cell Activation via the HLA-E/NKG2A Pathway. Cells 2020, 9, 1975. [Google Scholar] [CrossRef]
- Krämer, B.; Knoll, R.; Bonaguro, L.; ToVinh, M.; Raabe, J.; Astaburuaga-García, R.; Schulte-Schrepping, J.; Kaiser, K.M.; Rieke, G.J.; Bischoff, J.; et al. Early IFN-α signatures and persistent dysfunction are distinguishing features of NK cells in severe COVID-19. Immunity 2021, 54, 2650–2669.e14. [Google Scholar] [CrossRef]
- Kowalczyk, A.; Doener, F.; Zanzinger, K.; Noth, J.; Baumhof, P.; Fotin-Mleczek, M.; Heidenreich, R. Self-adjuvanted mRNA vaccines induce local innate immune responses that lead to a potent and boostable adaptive immunity. Vaccine 2016, 34, 3882–3893. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef]
- Rosa, S.S.; Prazeres, D.M.F.; Azevedo, A.M.; Marques, M.P.C. mRNA vaccines manufacturing: Challenges and bottlenecks. Vaccine 2021, 39, 2190–2200. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Weissman, D. Recent advances in mRNA vaccine technology. Curr. Opin. Immunol. 2020, 65, 14–20. [Google Scholar] [CrossRef]
- Bos, R.; Rutten, L.; van der Lubbe, J.E.M.; Bakkers, M.J.G.; Hardenberg, G.; Wegmann, F.; Zuijdgeest, D.; de Wilde, A.H.; Koornneef, A.; Verwilligen, A.; et al. Ad26 vector-based COVID-19 vaccine encoding a prefusion-stabilized SARS-CoV-2 Spike immunogen induces potent humoral and cellular immune responses. NPJ Vaccines 2020, 5, 91. [Google Scholar] [CrossRef] [PubMed]
- van Doremalen, N.; Lambe, T.; Spencer, A.; Belij-Rammerstorfer, S.; Purushotham, J.N.; Port, J.R.; Avanzato, V.A.; Bushmaker, T.; Flaxman, A.; Ulaszewska, M.; et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 2020, 586, 578–582. [Google Scholar] [CrossRef]
- Kovesdi, I.; Hedley, S.J. Adenoviral Producer Cells. Viruses 2010, 2, 1681–1703. [Google Scholar] [CrossRef]
- Bangaru, S.; Ozorowski, G.; Turner, H.L.; Antanasijevic, A.; Huang, D.; Wang, X.; Torres, J.L.; Diedrich, J.K.; Tian, J.-H.; Portnoff, A.D.; et al. Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate. Science 2020, 370, 1089–1094. [Google Scholar] [CrossRef] [PubMed]
- Stertman, L.; Palm, A.-K.E.; Zarnegar, B.; Carow, B.; Lunderius Andersson, C.; Magnusson, S.E.; Carnrot, C.; Shinde, V.; Smith, G.; Glenn, G.; et al. The Matrix-MTM adjuvant: A critical component of vaccines for the 21st century. Hum. Vaccin. Immunother. 2023, 19, 2189885. [Google Scholar] [CrossRef]
- Dunkle, L.M.; Kotloff, K.L.; Gay, C.L.; Áñez, G.; Adelglass, J.M.; Barrat Hernández, A.Q.; Harper, W.L.; Duncanson, D.M.; McArthur, M.A.; Florescu, D.F.; et al. Efficacy and Safety of NVX-CoV2373 in Adults in the United States and Mexico. N. Engl. J. Med. 2022, 386, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Tirelli, C.; De Amici, M.; Albrici, C.; Mira, S.; Nalesso, G.; Re, B.; Corsico, A.G.; Mondoni, M.; Centanni, S. Exploring the Role of Immune System and Inflammatory Cytokines in SARS-CoV-2 Induced Lung Disease: A Narrative Review. Biology 2023, 12, 177. [Google Scholar] [CrossRef] [PubMed]
- Ella, R.; Vadrevu, K.M.; Jogdand, H.; Prasad, S.; Reddy, S.; Sarangi, V.; Ganneru, B.; Sapkal, G.; Yadav, P.; Abraham, P.; et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBV152: A double-blind, randomised, phase 1 trial. Lancet Infect. Dis. 2021, 21, 637–646. [Google Scholar] [CrossRef]
- McMenamin, M.E.; Nealon, J.; Lin, Y.; Wong, J.Y.; Cheung, J.K.; Lau, E.H.Y.; Wu, P.; Leung, G.M.; Cowling, B.J. Vaccine effectiveness of one, two, and three doses of BNT162b2 and CoronaVac against COVID-19 in Hong Kong: A population-based observational study. Lancet Infect. Dis. 2022, 22, 1435–1443. [Google Scholar] [CrossRef] [PubMed]
- Teijaro, J.R.; Farber, D.L. COVID-19 vaccines: Modes of immune activation and future challenges. Nat. Rev. Immunol. 2021, 21, 195–197. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Jeyanathan, M.; Haddadi, S.; Barra, N.G.; Vaseghi-Shanjani, M.; Damjanovic, D.; Lai, R.; Afkhami, S.; Chen, Y.; Dvorkin-Gheva, A.; et al. Induction of Autonomous Memory Alveolar Macrophages Requires T Cell Help and Is Critical to Trained Immunity. Cell 2018, 175, 1634–1650.e17. [Google Scholar] [CrossRef] [PubMed]
- McCoy, K.; Tatsis, N.; Korioth-Schmitz, B.; Lasaro, M.O.; Hensley, S.E.; Lin, S.-W.; Li, Y.; Giles-Davis, W.; Cun, A.; Zhou, D.; et al. Effect of Preexisting Immunity to Adenovirus Human Serotype 5 Antigens on the Immune Responses of Nonhuman Primates to Vaccine Regimens Based on Human- or Chimpanzee-Derived Adenovirus Vectors. J. Virol. 2007, 81, 6594–6604. [Google Scholar] [CrossRef]
- Mendonça, S.A.; Lorincz, R.; Boucher, P.; Curiel, D.T. Adenoviral vector vaccine platforms in the SARS-CoV-2 pandemic. NPJ Vaccines 2021, 6, 97. [Google Scholar] [CrossRef]
- Rydyznski Moderbacher, C.; Kim, C.; Mateus, J.; Plested, J.; Zhu, M.; Cloney-Clark, S.; Weiskopf, D.; Sette, A.; Fries, L.; Glenn, G.; et al. NVX-CoV2373 vaccination induces functional SARS-CoV-2–specific CD4+ and CD8+ T cell responses. J. Clin. Investig. 2022, 132, 19. [Google Scholar] [CrossRef]
- Creech, C.B.; Walker, S.C.; Samuels, R.J. SARS-CoV-2 Vaccines. JAMA 2021, 325, 1318. [Google Scholar] [CrossRef]
- Keech, C.; Albert, G.; Cho, I.; Robertson, A.; Reed, P.; Neal, S.; Plested, J.S.; Zhu, M.; Cloney-Clark, S.; Zhou, H.; et al. Phase 1–2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N. Engl. J. Med. 2020, 383, 2320–2332. [Google Scholar] [CrossRef]
- Diamond, M.S.; Pierson, T.C. The Challenges of Vaccine Development against a New Virus during a Pandemic. Cell Host Microbe 2020, 27, 699–703. [Google Scholar] [CrossRef]
- Del Giudice, G.; Rappuoli, R.; Didierlaurent, A.M. Correlates of adjuvanticity: A review on adjuvants in licensed vaccines. Semin. Immunol. 2018, 39, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Pacheco, M.; Silva, P.L.; Cruz, F.F.; Battaglini, D.; Robba, C.; Pelosi, P.; Morales, M.M.; Caruso Neves, C.; Rocco, P.R.M. Pathogenesis of Multiple Organ Injury in COVID-19 and Potential Therapeutic Strategies. Front. Physiol. 2021, 12, 593223. [Google Scholar] [CrossRef]
- Arish, M.; Qian, W.; Narasimhan, H.; Sun, J. COVID-19 immunopathology: From acute diseases to chronic sequelae. J. Med. Virol. 2023, 95, 28122. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Cao, Z.; Xie, X.; Zhang, X.; Chen, J.Y.-C.; Wang, H.; Menachery, V.D.; Rajsbaum, R.; Shi, P.-Y. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020, 33, 108234. [Google Scholar] [CrossRef]
- Tian, Y.; Carpp, L.N.; Miller, H.E.R.; Zager, M.; Newell, E.W.; Gottardo, R. Single-cell immunology of SARS-CoV-2 infection. Nat. Biotechnol. 2022, 40, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, A.; Salvati, L.; Maggi, L.; Annunziato, F.; Cosmi, L. Hallmarks of immune response in COVID-19: Exploring dysregulation and exhaustion. Semin. Immunol. 2021, 55, 101508. [Google Scholar] [CrossRef]
- Darif, D.; Hammi, I.; Kihel, A.; El Idrissi Saik, I.; Guessous, F.; Akarid, K. The pro-inflammatory cytokines in COVID-19 pathogenesis: What goes wrong? Microb. Pathog. 2021, 153, 104799. [Google Scholar] [CrossRef]
- Silva, M.J.A.; Ribeiro, L.R.; Lima, K.V.B.; Lima, L.N.G.C. Adaptive immunity to SARS-CoV-2 infection: A systematic review. Front. Immunol. 2022, 13, 1001198. [Google Scholar] [CrossRef]
- Chen, C.; Wang, X.; Zhang, Z. Humoral and cellular immunity against diverse SARS-CoV-2 variants. J. Genet. Genom. 2023, 50, 934–947. [Google Scholar] [CrossRef]
- Polykretis, P.; Donzelli, A.; Lindsay, J.C.; Wiseman, D.; Kyriakopoulos, A.M.; Mörz, M.; Bellavite, P.; Fukushima, M.; Seneff, S.; McCullough, P.A. Autoimmune inflammatory reactions triggered by the COVID-19 genetic vaccines in terminally differentiated tissues. Autoimmunity 2023, 56, 2259123. [Google Scholar] [CrossRef]
- Fraiman, J.; Erviti, J.; Jones, M.; Greenland, S.; Whelan, P.; Kaplan, R.M.; Doshi, P. Serious adverse events of special interest following mRNA COVID-19 vaccination in randomized trials in adults. Vaccine 2022, 40, 5798–5805. [Google Scholar] [CrossRef] [PubMed]
- Kuhbandner, C.; Reitzner, M. Estimation of Excess Mortality in Germany During 2020–2022. Cureus 2023, 15, e39371. [Google Scholar] [CrossRef] [PubMed]
- Aarstad, J.; Kvitastein, O.A. Is There a Link between the 2021 COVID-19 Vaccination Uptake in Europe and 2022 Excess All-Cause Mortality? Asian Pac. J. Health Sci. 2023, 10, 25–31. [Google Scholar]
- O’Neill, L.A.J.; Netea, M.G. BCG-induced trained immunity: Can it offer protection against COVID-19? Nat. Rev. Immunol. 2020, 20, 335–337. [Google Scholar] [CrossRef] [PubMed]
- Chumakov, K.; Avidan, M.S.; Benn, C.S.; Bertozzi, S.M.; Blatt, L.; Chang, A.Y.; Jamison, D.T.; Khader, S.A.; Kottilil, S.; Netea, M.G.; et al. Old vaccines for new infections: Exploiting innate immunity to control COVID-19 and prevent future pandemics. Proc. Natl. Acad. Sci. USA 2021, 118, e2101718118. [Google Scholar] [CrossRef]
- Netea, M.G.; Ziogas, A.; Benn, C.S.; Giamarellos-Bourboulis, E.J.; Joosten, L.A.B.; Arditi, M.; Chumakov, K.; van Crevel, R.; Gallo, R.; Aaby, P.; et al. The role of trained immunity in COVID-19: Lessons for the next pandemic. Cell Host Microbe 2023, 31, 890–901. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manfrini, N.; Notarbartolo, S.; Grifantini, R.; Pesce, E. SARS-CoV-2: A Glance at the Innate Immune Response Elicited by Infection and Vaccination. Antibodies 2024, 13, 13. https://doi.org/10.3390/antib13010013
Manfrini N, Notarbartolo S, Grifantini R, Pesce E. SARS-CoV-2: A Glance at the Innate Immune Response Elicited by Infection and Vaccination. Antibodies. 2024; 13(1):13. https://doi.org/10.3390/antib13010013
Chicago/Turabian StyleManfrini, Nicola, Samuele Notarbartolo, Renata Grifantini, and Elisa Pesce. 2024. "SARS-CoV-2: A Glance at the Innate Immune Response Elicited by Infection and Vaccination" Antibodies 13, no. 1: 13. https://doi.org/10.3390/antib13010013




