3.1. Translational Symmetry
As we mentioned, some graphs may be embedded in a metric space where vertices are assigned positions. In this case, the symmetries (automorphisms) of a graph are induced by the transformations of coordinates in the embedding space, even if topologically their are simply permutations of vertices. This means that the topological properties of the graph, which are independent of the embedding space, will nonetheless reflect the properties of the latter. For instance, lattices are naturally formed by connecting vertices to their nearest neighbours in some embedding space (see
Figure 1). A simple type of discrete symmetry encountered in (either infinite or periodic) regular lattices is translational symmetry. That is, the fact that the topology of a lattice embedded in some
D-dimensional space does not change after a displacement by an integer multiple of the lattice spacing. Lattices are a particular type of
regular graphs,
i.e. graphs where every vertex has the same number of neighbours. In
Figure 2 we show three examples of regular graphs embedded in different dimensions (
,
and
) and with differently ranged connections (nearest neighbours, nearest and second-nearest neighbours, infinite neighbours).
If the labeling of vertices reflects their position in space, then translational symmetry is reflected in some regularities of the adjacency matrix
A of the network (for undirected graphs, where no orientation is defined on the edges, the adjacency matrix
A is a binary matrix whose entries equal
if a link between vertex
i and vertex
j is present, and
otherwise; here
where
N is the total number of vertices,
i.e. the size of the network). For instance, if the vertices are numbered cyclically along the ring, the adjacency matrices
and
of the graphs shown in
Figure 2a and c read
respectively. Translational symmetry is one of the traditional assumptions used in the theoretical study of discrete (or discretized) dynamical systems, and most of the available analytical results about dynamical processes are only valid under the assumption of the existence of this symmetry.
However, as one moves beyond the simple case of atoms regularly embedded in crystal lattices, virtually all real-world networks strongly violate translational symmetry. An important deviation from lattice-like topology in real networks is signaled by a surprisingly small value of the average
inter-vertex distance,
i.e. the average number of links one needs to traverse along the shortest path connecting two vertices. In most real networks, this quantity increases at most logarithmically with the number
N of vertices, a phenomenon known as the
small-world effect [
2]. This behavior is also encountered in the random graph model mentioned in
Section 1 but not in lattices, where the average distance (if infinite-ranged connections are not allowed, e.g. for the graphs in
Figure 2a and b but not for that in
Figure 2c) grows as
, thus much faster. The breakdown of translational symmetry implies that the wealth of knowledge accumulated in the literature about the outcome of dynamical processes on lattices cannot be applied to the same processes when they take place on real networks [
4]. We already mentioned epidemic spreading processes as an example of the surprising deviation between dynamics on lattices and on more complicated networks. Nonetheless, real networks bear an interesting similarity with regular graphs, namely a large average value of the
clustering coefficient, defined as the number of triangles (loops of length three) starting at a vertex, divided by its maximum possible value.
The simultaneous presence of a small average distance and of a large clustering coefficient (which is sometimes taken as a stronger definition of the
small-world effect) has motivated the introduction of an important and popular network model which is somehow ‘intermediate’ between regular lattices and random graphs. In the model proposed by Watts and Strogatz [
21], one starts with a regular lattice and then, with fixed probability
p, goes through every edge and rewires one of its two end-point connections to a new, randomly chosen vertex. Clearly, when
one has the original lattice (large clustering and large distance), while when
one has a completely random graph (small clustering and small distance). Thus the parameter
p can be viewed as a measure of the deviation from complete translational symmetry in the model. Interestingly, in a broad intermediate range of values one simultaneously obtains a large clustering and a small distance, thus recovering the empirically observed effect. This suggests that real networks may be partially, but surely not completely, affected by translational symmetry (due for instance to the existence of a natural spatial embedding). As we shall discuss in
Section 3.5, translational symmetry, and in general the dependence of structural properties on the vertices’ positions in some embedding space, is an example of a more general situation where vertices are characterised by some non-topological quantity that may determine or condition their connectivity patterns.
3.2. Scale Invariance
As we mentioned, one of the most striking and ubiquitous features of real networks is the power-law form
of the degree distribution. This property means that vertices are extremely heterogeneous in terms of their number of connections: many vertices have a few links, and a few vertices (the
hubs) have incredibly many links. An example of a small network with highly heterogeneous degree distribution is shown in
Figure 3. Importantly, most of the empirically observed values of the exponent
are found to be in the range
, where the variance of the distribution diverges. This implies that there is no typical scale for the degree
k in the system, and motivates the expression
scale-free network [
2].
The above property is an example of a remarkable type of symmetry, precisely scale invariance. It is found across different domains [
22], and in particular in fractal objects. In fractals, scale invariance is manifest in the fact that iterated magnifications of an object all have the same shape,
i.e. the system ‘looks the same’ at all scales. Similarly, in networks one finds that if the scale of the observation is changed (e.g. one switches from degree
k to degree
, with
a positive), the number of vertices with given degree only changes by a (magnification) factor, from
to
. This is very different from exponential distributions, characterized by a strong variation in the number of counts as the scale is changed. In networks, power laws have also been found to describe the distribution of link weights, of the sum of link weights (the so-called strength) of vertices, and of many more quantities [
2]. They also appear to hold across various coarse-grained levels of description of the same network, if groups of vertices are iteratively merged into ‘supervertices’ and the original connections collapsed into links among these supervertices [
23]. The symmetry group associated to scale invariance,
i.e. the
renormalization group [
24], has therefore been used many times to theoretically understand power-law distributed network properties.
The presence of a scale-free topology across several real-world networks, which is not reproduced by the Erdős-Rényi model and by the Watts-Strogatz one, has led to the introduction of new theoretical mechanisms that could possibly explain the onset of this widespread phenomenon. The earliest (even if analogous mechanisms were already known in different contexts [
22]) and most popular scale-free network model is the one proposed by Barabasi and Albert [
25]. It is based on two key ideas: firstly, networks can grow in time, therefore one can assume that new vertices are continuously added to a preexisting network; secondly, already popular (highly connected) vertices are likely to become more and more popular (‘rich get richer’). The latter idea, known as
preferential attachment, is modeled as a multiplicative process in degree space: the probability that newly introduced vertices establish a connection to a preexisting vertex
i is proportional to the degree
of that vertex. The iteration of this elementary process of growth and preferential attachment eventually generates a power-law degree distribution of the form
. In degree space, preferential attachment is a symmetry-breaking mechanism: vertices are not equally likely to receive new connections as the network grows. Even if all vertices are identical
a priori, preferential attachment determines and amplifies heterogeneities in the degree, and eventually vertices with different degrees become subject to different probabilistic rules. Since in the model there is a tight relationship between the degree of a vertex and the time the same vertex entered the network, one could also say that different injection times imply different expected topological properties. On the other hand, with respect to scale invariance, preferential attachment is symmetry-preserving and gives rise to a stationary process. Indeed, as the network grows infinitely in size over time, its scale-free degree distribution remains unchanged. This highlights how the same network properties may bear different meanings in relation to different symmetries. There are now many alternative models that reproduce scale-free networks with any value of the power-law exponent
, not only
[
2,
3,
8]. In all of them, there is some mechanism that eventually sets on and drives the network to converge to an extremely heterogeneous topology. We shall describe one of these models [
26] in
Section 3.5. Before doing that, in the following
Section 3.3 and
Section 3.4 we shall make a more general discussion about symmetry breaking due to differences in topological properties in a model-free and real-world framework.
3.4. Statistical Equivalence
Structural equivalence is a very strict definition of similarity between two vertices. A more relaxed condition that is usually of interest in sufficiently large networks is whether two vertices are
statistically equivalent,
i.e. whether their topological properties are the same in an average or weak sense. For instance, one could ask whether two vertices
i and
j have simply the same degree (irrespective of the identity of their neighbors), and/or the same number of second neighbours, or whether they participate in the same number of triangles and/or longer loops. Similarly, one could be interested in finding two vertices whose neighbours have the same average degree, irrespective of the numbers of neighbours of each vertex, and of the individual values of the degrees of these neighbours (this is explained in more detail below). In all these examples, one focuses on a subset (or some average value) of the possible topological properties involving
i and
j, and defines an equivalence with respect to it only. According to this relaxed condition, a number of statistically equivalent vertices are found in real networks. The structure of the resulting equivalence classes determines the symmetry of a particular network. While permutations of structurally equivalent vertices are exact symmetries of the graph (
i.e. automorphisms), permutations of statistically equivalent vertices are stochastic symmetries in the sense introduced in
Section 2. Such transformations do not map a network to itself, but to another member of the family of networks with the same statistical properties. Importantly, while even small errors such as a missing link in the data have a dramatic effect on structural equivalence, statistical equivalence is more robust to fluctuations in network structure. Moreover, introducing this stochastic type of symmetry gives rise to identify more general patterns than those accessible to the analysis of structural equivalence. We discuss this concept by making some examples of the main scientific questions related to statistical equivalence in networks.
Do all vertices in a network have the same degree? As already discussed in
Section 3.2, this type of symmetry is strongly violated in real networks. A weaker question would be: are the degrees of all vertices
nearly the same? In this case, one could speak of a typical degree of vertices, and interpret the deviations from the average value as finite fluctuations due either to external noise or some intrinsic stochasticity. However, as we mentioned, the majority of real networks are scale-free, with degrees being broadly distributed and wildly fluctuating. There are many vertices with small degree, among which one can in principle find vertices with exactly the same number of neighbors, but also a few vertices with extremely large degree, which strongly break the symmetry.
Is the average degree of the neighbors of all vertices (nearly) the same? After recognizing that some vertices attract many more links than others, one can move one step forward and wonder what is the average degree of the neighbors of a given vertex (the so-called
average nearest neighbor degree, or ANND [
2]). This quantity encodes some information about the matching patterns in the network: if the degree plays no role in deciding whether two vertices are connected, then one expects that the ANND is independent of the degree itself (as we discuss below, this is not completely true). By contrast, one finds the presence of strong correlations between the degrees of neighboring vertices. These correlations can be either positive or negative, and have opposite effects on the ANND. In networks where large-degree vertices are more likely to be connected to each other than to low-degree ones, one observes an increasing trend of the ANND as a function of the degree. This property is known as
assortativity [
30]. In networks where the opposite is true, the ANND decreases with the degree, a situation denoted
disassortativity. Importantly, degree-degree correlations have profound effects on the outcomes of dynamical processes taking place on networks [
4].
Do all vertices have (nearly) the same clustering coefficient? Again, this symmetry is generally not observed, as vertices with different degree also have different values of the clustering coefficient. The latter usually displays a decreasing trend with the degree
k. This behavior has been interpreted as the signature of a hierarchically organised topology, where a simple wiring pattern is repeated at different scales in a bottom-up fashion: first creating modules of vertices, then modules of modules,
etc. [
31]. Since both the clustering coefficient and the ANND strongly depend on the degree, and since the latter is broadly distributed, it appears that real networks are characterised by a high level of complexity, with no characteristic scale associated to any of the simplest topological properties one can define.
However, the last observation also leads to a reverse, possibly simplifying, approach to the problem. Interestingly, it has been shown that some of the correlations mentioned above are partly an unavoidable, ‘spurious’ outcome of enforcing some topological constraints in the network [
32,
33]. That is, exactly because many properties ultimately depend on the degree, a number of structural patterns are automatically generated once the degrees of all vertices are fixed to specified values. For instance, in networks with power-law degree distribution the ANND and the clustering coefficient both decrease with the degree. These patterns do not signal ‘true’ higher-order correlations, as they are natural outcomes due to the presence of simpler constraints. If an explanation from the latter exists, it also automatically explains the former. This highlights the importance of separating low-order effects from more fundamental higher-order structural patterns. This problem leads to the definition of suitable
null models of networks, a point that we shall discuss in
Section 3.6.
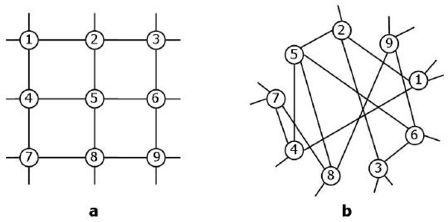
3.5. Invariance under Permutation of External Properties
An important type of permutation symmetry can be defined when some external, non-topological property is attached to vertices (or to edges, or to other subgraphs; but we will consider the case of vertices for simplicity). This situation is particularly relevant when one is interested in studying the relation between the topology and some other property characterising the vertices of a network, and is tightly related (even if in a nontrivial way) to structural and statistical equivalence, as the example in
Figure 5 shows. Note that translational symmetry (described in
Section 3.1) can be viewed as a particular case of this problem, if vertices are assigned positions in some metric space. Translational symmetry is in principle an exact symmetry (the graph is mapped onto itself) since it is the effect of a deterministic graph formation rule. However, symmetries due to external properties are in general stochastic in the sense discussed in
Section 2, since real networks are always best understood as a result of non-deterministic rules. We therefore expect that stochastic symmetry is more powerful in detecting patterns in real networks than exact symmetry, and the following discussion confirms this expectation.
The impact of external factors is an extremely important problem, related to key questions about network formation, for many research areas. Typical examples include: is a social network partly determined by factors such as race, gender, age, etc.? Is wealth or income relevant to the formation of economic networks? In order to answer the above questions, one needs a way to assess the structural impact of properties which are in some sense external to the network.
There have been many attempts in this direction. Social network analysis has a long tradition in dealing with this problem, firmly based on statistical theory. The role of vertex properties is generally inspected through the values of regression parameters used in suitable graph models that are fitted to the real network [
27]. More recently, in the physics community different approaches have been proposed. Techniques have been introduced [
30,
34] in order to capture whether the connections observed in a particular network occur mainly between vertices with similar properties (this is a generalised notion of
assortativity, not necessarily related to vertices’ degrees, also known as
homophily in social science) or between vertices with different properties (
disassortativity). More generally, there have been attempts in understanding whether a specification of vertex properties effectively reduces the available configuration space for a real network [
35] and can thus be interpreted as a structurally important factor. All these different approaches to the same problem could be restated in more general terms as follows:
is the network (stochastically) symmetric under a permutation of the properties attached to vertices? If this is the case, the properties under consideration have no statistically significant impact on network structure. Otherwise, vertex-specific features are symmetry-breaking, as vertices with different properties are no longer equivalent under a somewhat generalised notion of the statistical equivalence described in
Section 3.4. In particular, the overall permutation symmetry of vertex properties is broken and the network is only symmetric under a restricted set of permutations exchanging vertices within the same equivalence classes (sets of vertices with the same external properties). It is therefore clear that the behaviour of a network under the permutations associated to this type of permutation symmetry is determined by, and carries information about, the effects that external quantities have on the topology.
In general, the behaviour of a real network under permutation of external properties can be very complicated and lead to a variety of different symmetry properties. However, it is possible to understand the problem clearly in simplified models. Indeed, the idea that vertex properties may be crucial to network formation has led to the definition of an important class of network models known as
fitness or
hidden variable models [
26]. Unlike the Barabasi-Albert model mentioned in
Section 3.2, fitness models are static and do not require the hypothesis of network growth. In these models, one assumes that the probability
that a link is present between vertex
i and vertex
j is a function
of some property
x, or
fitness, attached to these vertices (see
Figure 5). Therefore the model requires the specification of a list of fitness values
, usually assumed to be drawn independently from some probability distribution
, and of the connection function
. All the expected topological properties crucially depend on
. For instance, the expected degree of two vertices
i and
j with different fitness values (
) is in general different. On the other hand, two vertices with
are statistically equivalent. However, due to the probabilistic nature of the model, in a particular realization of the network the statistical equivalence of vertices with equal fitness values does not necessarily reflects in their structural equivalence (see example in
Figure 5). This model specification successfully reproduces the situation mentioned above, as the permutation symmetry of vertex properties is broken down to disjoint equivalence classes represented by sets of vertices with identical hidden values. Moreover, the flexibility in the choice of the fitness values and connection probability allows to reproduce various topological properties of real-world networks. For instance, a power-law distribution of fitness values (mimicking some heterogeneously distributed real-world feature such as individual wealth, country population,
etc.) and a connection probability that linearly depends on the fitness naturally lead to a scale-free network topology [
26]. Besides providing a valid route to network modelling, hidden variable models can also be fitted to real networks and shed light on the presence of external factors case by case [
36,
37]. In particular, inverse methods have been devised in order to extract, only from the topology of a real network, the values of the hidden variables
potentially related to network formation. These values can then be compared with the values of candidate external properties relevant to that particular network, a strategy that has been shown to successfully identify key factors related to structure in real-world cases [
36].
3.6. Ensemble Equiprobability
As we anticipated in
Section 2, there are important symmetries associated not to a single graph, but to a
statistical ensemble of graphs (we will define a graph ensemble rigorously below). If the ensemble is a good model of a real network, these symmetries can then be naturally related to the real network itself. This possibility allows us to illustrate in more detail our idea of stochastically symmetric ensemble, and the definition of stochastically symmetric graph as a network which is well reproduced by a stochastically symmetric ensemble (see
Section 2). Null models automatically come into play when one is interested in understanding whether, in a given network, complicated high-order topological properties can be traced back to simpler low-level constraints. We already mentioned this problem in
Section 3.4. In order to answer this question, it is necessary to consider a null model by generating a collection of graphs having some property in common with the real network (these properties act therefore as constraints), and being completely random otherwise. This amounts to generate an ensemble of graphs that maximizes an
entropy, that we shall define in a moment, under the enforced constraints. Then, one can compare the properties of the real network with the corresponding averages over the randomised ensemble. If there is no statistically significant difference, one can conclude that the constraints considered are indeed enough in order to generate all the other properties of the real network. If differences are significant, then there are other factors shaping the observed topology. We now rephrase this idea more formally, and show how it highlights an intimate and instructive connection between symmetry, entropy and complexity in networks.
A statistical ensemble of graphs [
38] is a collection of
M graphs
, each with an associated occurrence probability
satisfying
We already mentioned examples of graph ensembles, without explicitly noticing it: Erdős-Rényi model (
Section 1 and
Section 3.1), the Watts-Strogatz model (
Section 3.1), the Barabasi-Albert model (
Section 3.2) and the fitness model (
Section 3.5) are all examples of collections of possible graphs generated by probabilistic rules. The Barabasi-Albert model is a non-equilibrium ensemble, as it generates networks growing indefinitely in time; all the other examples mentioned above are instead equilibrium ensembles. In what follows, we restrict ourselves to the equilibrium case. Each graph
G is uniquely specified by its adjacency (or weight) matrix, so we can think of
G as of a matrix. For instance, if one is interested in the ensemble of binary undirected graphs with
N vertices and no self-loops (edges starting and ending at the same vertex), then
G will be a symmetric Boolean matrix with zeroes along the diagonal, and there will be
possible such matrices in the ensemble. In order to generate a maximally random ensemble of graphs with given constraints [
38,
39,
40], one needs to find the form of the probability
that maximises the Shannon-Gibbs entropy
(a standard measure of disorder or uncertainty) under the enforced constraints. The latter are a collection
of
K topological properties, forming a
K-dimensional vector
. Each property
(
) evaluates to
when measured on the particular graph
G. If the ensemble is meant as a null model of an empirical network
, the constraints will be chosen as the properties
evaluated on the particular graph
.
There are various possible choices to solve the entropy maximisation problem, and different ensembles that one can define accordingly. If one is interested in matching the constraints
exactly,
i.e. in picking out only the graphs that have exactly the same properties as a given network
, then the solution is given by the probability
where
is the number of graphs matching the constraints
. The above probability is uniform over the set of configurations matching the constraints exactly, and the resulting ensemble is known in statistical physics as the
microcanonical ensemble. With the above choice, the entropy defined in Equation (
3) takes the form
which is known as the
microcanonical entropy and is simply the logarithm of the number of configurations exactly matching the constraints.
A second alternative consists in requiring that the constraints
are matched
on average,
i.e. allowing any graph to occur with non-zero probability, provided that the expected value
of the constraints matches the required value
. This problem can be solved introducing Lagrange multipliers
, each associated to one of the constraints. The solution is the probability distribution
where
(the
graph Hamiltonian) is a linear combination of the constraints
and
Z is the
partition function that properly normalizes the probability:
Thus both
Z and
depend on the
K parameters
. The ensemble generated by the above probability is known in physics as the
canonical ensemble. For a given choice of the parameters
, the expected value of a topological property
X across the ensemble is
(throughout this review, the angular brackets
will denote ensemble averages). In order to match the constraints
on average, the
K parameters
must be set to the particular values
such that
Importantly, the above parameter choice corresponds with what the
maximum likelihood principle would indicate [
36],
i.e. with the values maximising the probability
to obtain the real network
under the model considered. We will indicate the maximum-likelihood parameter choice explicitly in the examples considered later on. It has been shown [
38] that the canonical ensemble of networks coincides with the
exponential random graph models that have been first introduced in social science [
27]. The Hamiltonian
represents the
energy, or
cost, associated with a given configuration, and contains all the information required in order to formally obtain
. This means that any two graphs
and
for which
have the same ensemble probability
. Thus, the symmetries of
are transformations connecting equiprobable graphs in the ensemble. Such transformations map a graph
into a graph
which has a different topology but exactly the same values of the enforced constraints. According to our definition in
Section 2, a canonical graph ensemble is stochastically symmetric under such transformations. If a canonical graph ensemble is a good model of a real network
, the latter is also stochastically symmetric. Maximally random graphs with constraints therefore represent ideal candidates to illustrate the concept of stochastic symmetry. The symmetries of the Hamiltonian, together with the parameter values enforcing the constraints, determine the entropy
S of the ensemble. This entropy is a measure of the residual uncertainty about the detailed topology of a network, once the constraints are fixed.
In statistical physics, there is also a third class of ensembles,
i.e. grandcanonical ensembles. In the latter, the number of particles of the system is also allowed to vary, and it is treated as one of the properties to be matched on average. In the case of networks, the role of particles is played by links [
38], whose number is allowed to vary already in the canonical ensemble, as the examples considered below illustrate. Therefore there is no fundamental difference between the canonical and grandcanonical ensembles of graphs, unless one is interested in networks with different types of links [
14]. For large systems, the microcanonical, canonical and grandcanonical ensembles give very similar results. The canonical and grandcanonical ensembles have the enormous advantage to be analytically treatable, as a consequence of the relaxed requirement on the constraints. For this reason, in what follows we shall consider (grand)canonical ensembles of graphs.
We now discuss some examples. If we consider again the ensemble of all possible undirected graphs with
N vertices, the completely symmetric case is the one where each graph
G has the same energy
where
is a constant. In other words, in this case there are no constraints. Clearly, each of the
M possible graphs has the same probability
and therefore the graph probability is uniformly distributed across the ensemble (in this particular case, the microcanonical and canonical ensembles coincide). Transformations changing a graph
G into any other graph in the ensemble are symmetries of the Hamiltonian, and lead to the same ensemble probability. Thus this ensemble is stochastically symmetric under any transformation. The entropy is the maximum possible, and its value is
A different case is when there is a constraint on the total number of links
. Then
and it can be easily shown that
where
. This shows that, as expected, two graphs
and
with the same number of links
are equiprobable. Graph transformations preserving this number are symmetries of the Hamiltonian, and the ensemble is stochastically symmetric under such transformations. Equation (
16) indicates that, for each of the
pairs of vertices, the probability of an undirected link being there is
p. The probability of exactly
realised edges is
multiplied by the probability
of the complementary number
of missing edges. This case is therefore equivalent the Erdős-Rényi random graph model that we already mentioned in
Section 1, in which each edge is drawn, independently of each other, with probability
p. The entropy of the ensemble now depends on
p, and one can easily see that if
, Equation (
14) is recovered. Indeed, this is the case where each edge is equally likely to be present and absent, which is another way to say that no constraint has been enforced and the entropy is maximum. By contrast, in the two cases
and
the entropy is
as there is no uncertainty about the resulting structure of the network. Indeed, in these cases the ensemble completely shrinks to the only possible network,
i.e., the empty graph and the complete graph respectively. If one wants to use the random graph model as a null model of a real network
, the maximum likelihood principle applied to Equation (
16) indicates [
36] that the parameter
p must be set to the value
which ensures that the expected number of links
, as defined by Equation (
9), reproduces the number of links
of that particular network:
In the random graph model, the expected degree distribution is binomial (in the large network limit with fixed average degree, Poissonian) with mean
. The failure of the random graph model in reproducing the properties of real networks, according to our discussion in
Section 1, can then be restated as the inefficacy of specifying the number of links as the only property of a network. This also means that real networks are generally not stochastically symmetric under transformations preserving the total number of links. A less trivial choice is the so-called
configuration model [
32,
41]. Assuming we are still interested in undirected binary networks, the configuration model is a maximally random graph ensemble where the degrees of all vertices,
i.e. the
degree sequence , are specified. Note that, in terms of the adjacency matrix
A of the graph, the degree of vertex
i is
, and the total number of links is twice the sum of the degrees of all vertices:
. Therefore specifying the degree sequence automatically fixes also the total number of links, which confirms that this model is more constraining than the random graph one. The configuration model naturally comes into play in the problem we described in
Section 3.4, when we stressed the importance of comparing a real network to a null model in order to separate genuine higher-order correlations from mere effects of low-level constraints. The degree sequence is an important constraint to consider, because the widespread occurrence of scale-free architectures implies that major topological differences across real networks must be looked for in other properties beyond the degree distribution. Note that specifying the degree sequence
is different from specifying the degree distribution
. A given degree sequence generates a unique degree distribution, but there are many degree sequences (
permutations) generating the same degree distribution. Therefore fixing the degree distribution is less informative than specifying the entire degree sequence, and we do not consider it here. For directed graphs, the configuration model is naturally extended by simultaneously considering as constraints the number of incoming links (
in-degree) and the number of outgoing links (
out-degree) of all vertices. Similarly, for weighted networks the constraints become the
strength (total edge weight) of all vertices (the
strength sequence), or the corresponding directed quantities when applicable.
In the binary undirected case, the Hamiltonian of the configuration model contains the degrees of all vertices:
and it can be shown [
33] that the form of
determined by the above choice is
where
and
is another way to write the Lagrange multiplier associated to
. In this model, edges are still independent, but have different probabilities.
The probability
of a graph
G only depends on its degree sequence, as evident from Equation (
20). Thus any two graphs
and
with the same degree sequence
are equiprobable in the ensemble specified by Equation (
19). A consequence of this property is illustrated in
Figure 6, where we show two graphs
and
that have exactly the same topology, except for the two edges shown. Graph
can be obtained from
by replacing the two edges
and
with the two edges
and
. Since this transformation preserves the degree sequence, it is a symmetry of the Hamiltonian defined in Equation (
19) and connects equiprobable graphs. According to our definition in
Section 2, the ensemble is stochastically symmetric under such transformation. The equivalence classes of this symmetry are sets of graphs with the same degree sequence.
This property has been used to constructively define an algorithm that randomises a real network
by iteratively selecting a pair of edges and swapping the end-point vertices exactly as in
Figure 6 [
32]. This procedure, known as the
local rewiring algorithm, ergodically explores the equivalence class where the real network
belongs. Any topological property of interest can be averaged across the set of graphs produced by the algorithm and compared with the value of the same property in the original graph
. This allows to check the effects of the degree sequence alone on the other topological properties. As we mentioned, this null model is restricted to only one equivalence class of the symmetry (it is a
microcanonical ensemble), and requires that averages are numerically performed over the graphs sampled by the local rewiring algorithm. By contrast, the null model defined by Equation (
19) explores the entire set of
undirected graphs (it is a
(grand)canonical ensemble), and allows to obtain the expectation values analytically through Equation (
9). This requires that the parameters
are set to the values
that maximise the likelihood to obtain the real network
[
36,
42]. These values are found by solving the following
N coupled equations
ensuring that the expected degree sequence coincides with the observed one, and thus generalising Equation (
18). As we already anticipated in
Section 3.4, an important conclusion drawn from the analysis of the configuration model is that, if real-world scale-free degree distributions are specified, higher-order patterns are automatically generated. In particular, the average nearest neighbour degree and the clustering coefficient of a vertex with degree
k are both found to decrease with
k [
32,
33,
42]. These patterns should not be interpreted necessarily as the result of additional mechanisms, beyond those required to explain the form of the degree distribution. Note that if a real network is found to be well reproduced by the configuration model, then it is stochastically symmetric under transformations preserving the degree sequence. Also note that any two vertices
i and
j with the same degree
in the real network are statistically equivalent in the sense specified in
Section 3.4. This is because Equation (
22) implies that those vertices would be assigned the same parameter value
, and would therefore have the same expected topological properties as discussed for the fitness model in
Section 3.5. Whereas permutations of structurally equivalent vertices lead to exactly the same topology and are therefore automorphisms (exact symmetries) of the network, permutations of statistically equivalent vertices (here, vertices with the same degree) are stochastic symmetries of the network, if the latter is in accordance with the configuration model. This is an interesting and important relation between ensemble equiprobability, symmetry under permutation of vertex properties, and statistical equivalence. If the ensemble is not a good model of the real network, which signals the presence of mechanisms that break the postulated equiprobability symmetry, then the real network is not stochastically symmetric under transformations preserving the degree sequence, and vertices displaying the same values of the enforced constraints are no longer statistical equivalent.
Note that Equation (
20) generalises Equation (
16), and also that Equation (
21) can be viewed as a particular case of the connection probability
introduced in the fitness model we described in
Section 3.5. Indeed, the configuration model and the fitness model both reduce to the random graph case if
,
i.e. if all vertices have the same properties. In this case, the entropy associated with Equation (
21) coincides with the one associated with Equation (
16). By contrast, if the
’s are heterogeneously distributed, the entropy is significantly decreased. In particular, the values of the
’s required in order to enforce a scale-free degree distribution as observed in real networks are approximately power-law distributed, a result implying a strong reduction of the entropy of the ensemble associated with the degree sequence of real networks. In particular, it was shown that networks with degree distribution
have remarkably small entropy [
39] and can be generated deterministically [
26] like regular graphs. We therefore see that network complexity, as signalled in this example by a scale-free degree distribution, can lead to a decrease in the stochastic symmetry associated with ensemble equiprobability, and to a substantial decrease in the corresponding entropy. From the perspective of the amount of information required in order to reproduce them, real networks (and possibly many real complex systems) turn out to achieve an unsuspected degree of order by following a nontrivial path, which is completely different from that taken by regular structures.