Dynamic Interplay between Copper Toxicity and Mitochondrial Dysfunction in Alzheimer’s Disease
Abstract
:1. Introduction
2. Role of Copper in AD
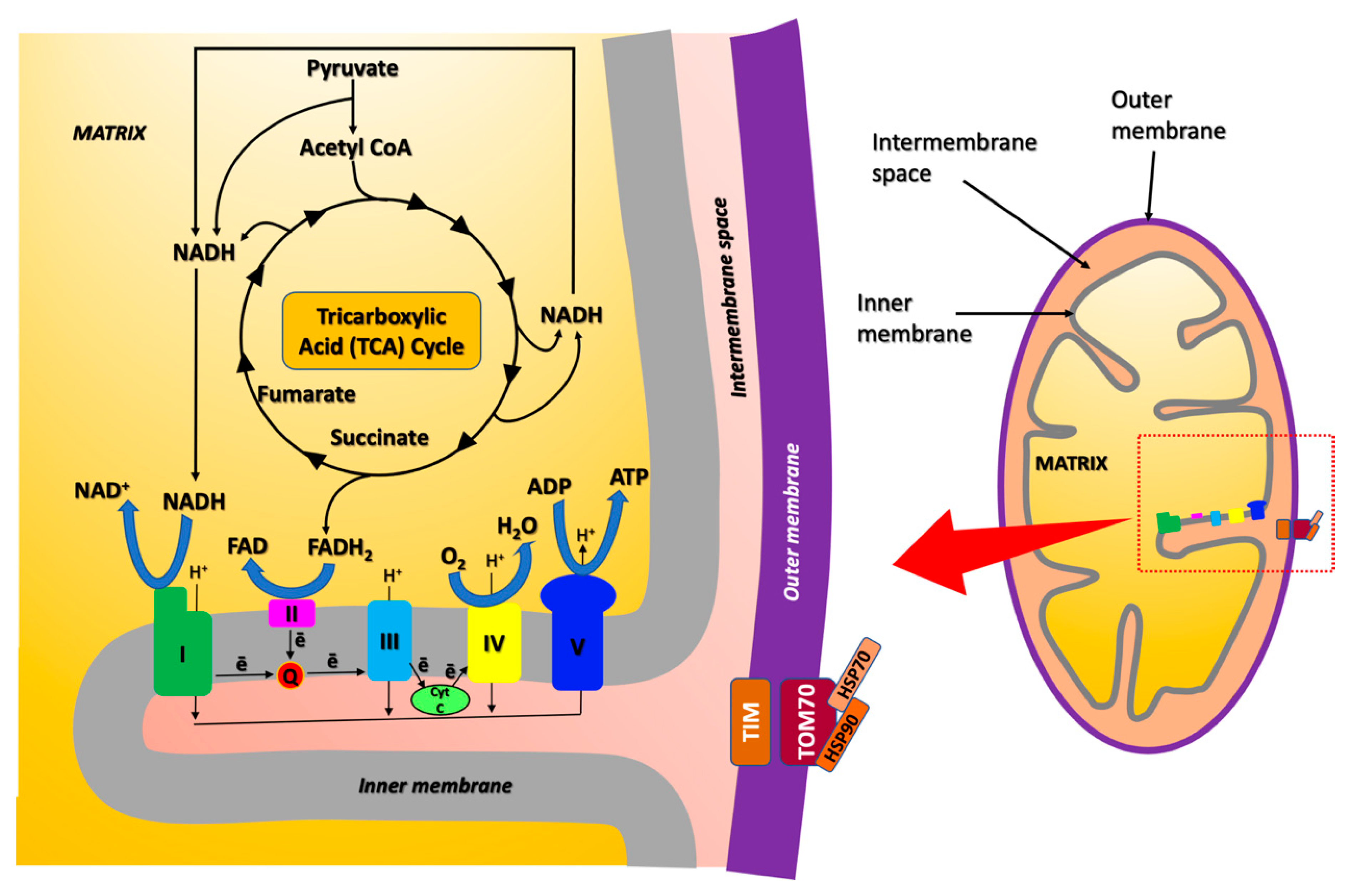
3. Mitochondrial Proteome Altered upon Copper Exposure in AD Identified by Proteomics Approaches
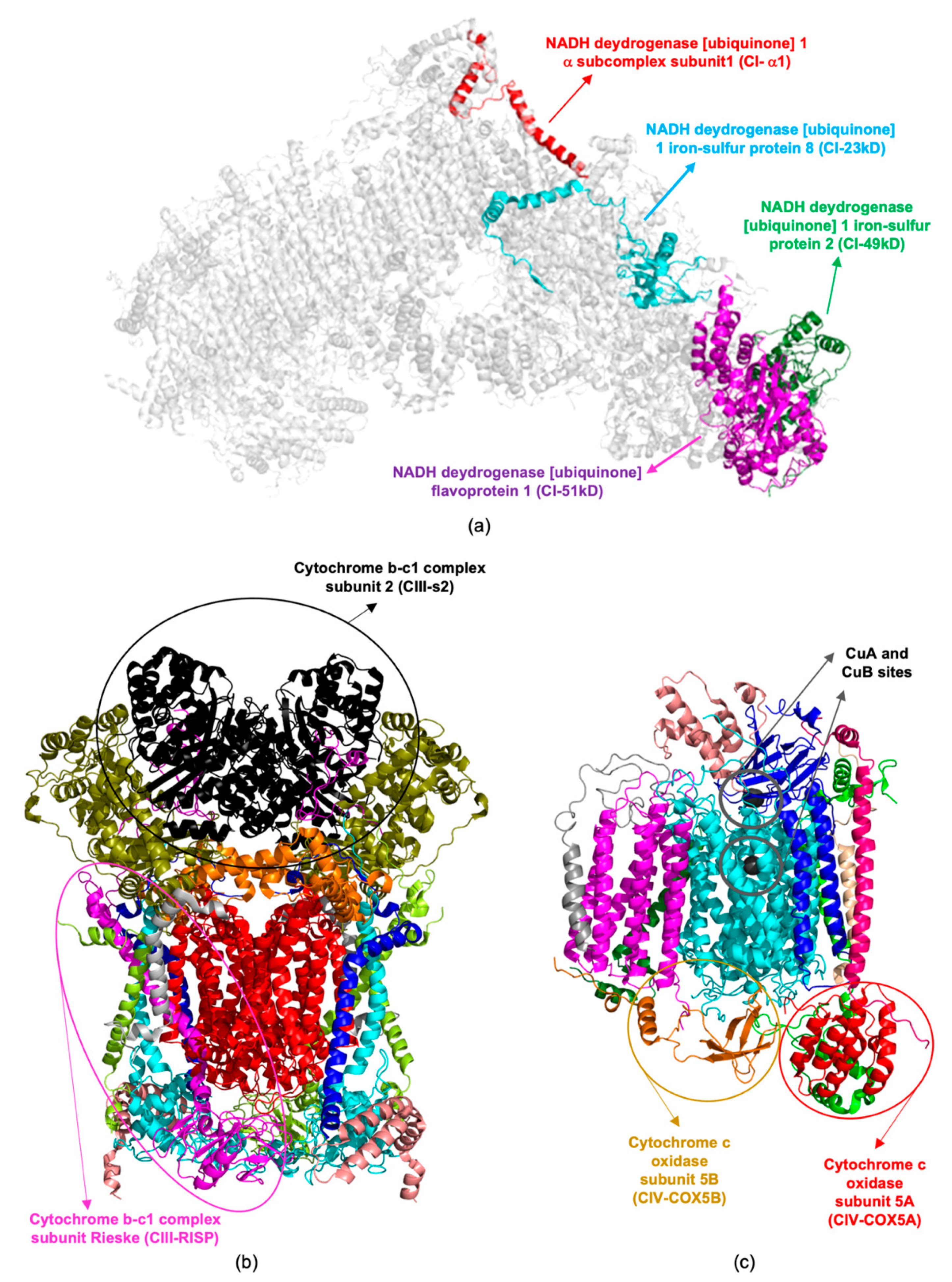
4. Redox Proteomics to Identify Mitochondrial Altered Proteins
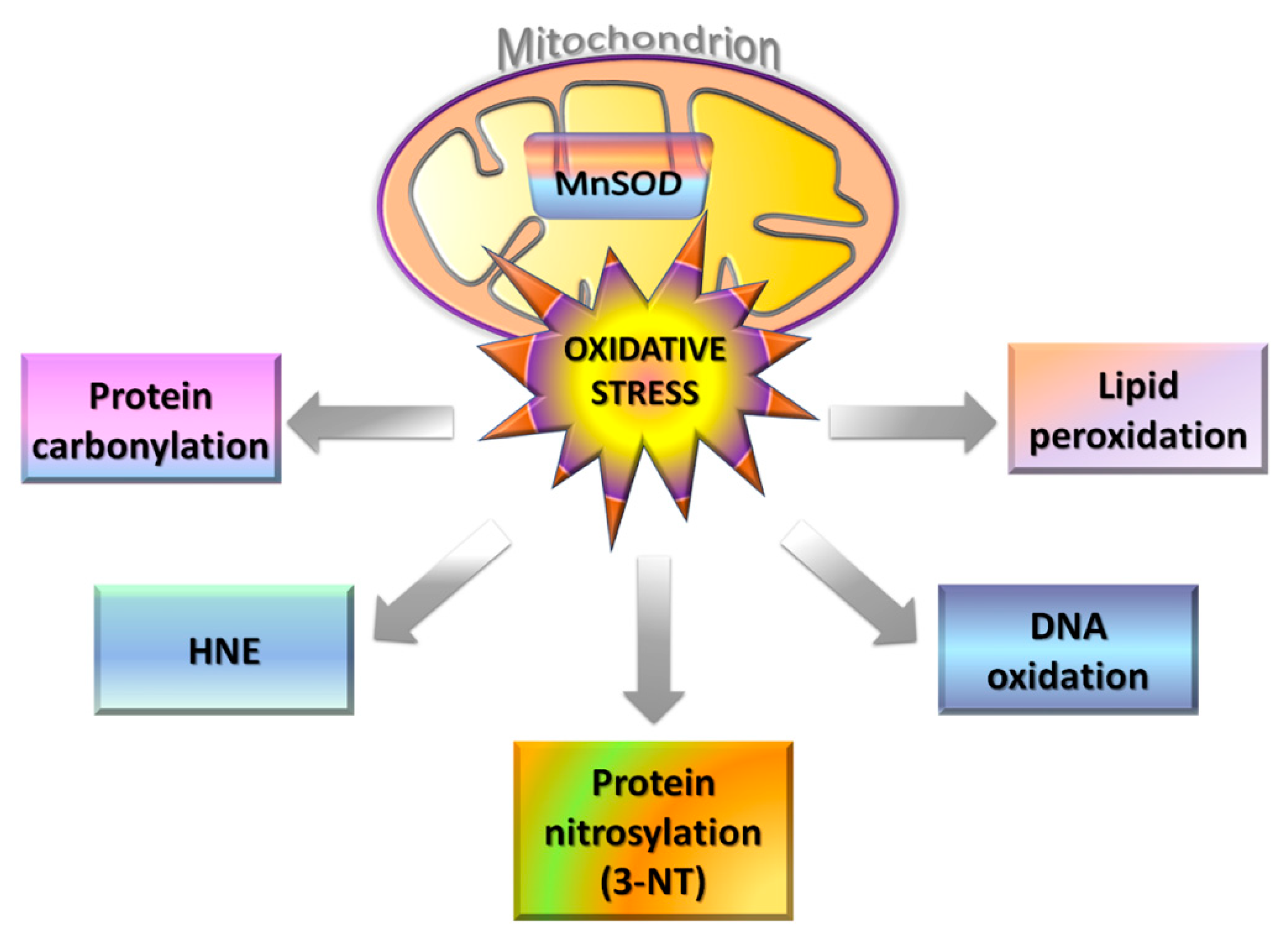
4.1. Oxidative and Nitrosative Stress in Mitochondria
4.2. Redox Proteomics in AD
4.3. Oxidative Modifications of Mitochondrial Proteins in AD
5. Main Protein Targets and Pathways Altered in AD by Copper-Induced and Oxidative Modifications
6. Targeting Mitochondria in AD
6.1. Targeting Mitochondrial ROS Production: Antioxidants
6.2. Targeting of the Mitochondrial ETC Complexes: J147 and Metformin
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Goedert, M.; Spillantini, M.G. A Century of Alzheimer’s Disease. Science 2006, 314, 777–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardy, J.A.; Higgins, G.A. Alzheimer’s disease: The amyloid cascade hypothesis. Science 1992, 256, 184–185. [Google Scholar] [CrossRef]
- Hardy, J.; Selkoe, D.J. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 2002, 297, 353–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchesi, V.T. Alzheimer’s Disease 2012. Am. J. Pathol. 2012, 180, 1762–1767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bezprozvanny, I.; Mattson, M.P. Neuronal calcium mishandling and the pathogenesis of Alzheimer’s disease. Trends Neurosci. 2008, 31, 454–463. [Google Scholar] [CrossRef] [Green Version]
- Stefani, M.; Liguri, G. Cholesterol in Alzheimer’s disease: Unresolved questions. Curr. Alzheimer Res. 2009, 6, 15–29. [Google Scholar] [CrossRef]
- Fraser, T.; Tayler, H.; Love, S. Fatty Acid Composition of Frontal, Temporal and Parietal Neocortex in the Normal Human Brain and in Alzheimer’s Disease. Neurochem. Res. 2010, 35, 503–513. [Google Scholar] [CrossRef]
- Wang, X.; Su, B.; Zheng, L.; Perry, G.; Smith, M.A.; Zhu, X. The role of abnormal mitochondrial dynamics in the pathogenesis of Alzheimer’s disease. J. Neurochem. 2009, 109, 153–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanzillotta, C.; di Domenico, F.; Perluigi, M.; Butterfield, D.A. Targeting Mitochondria in Alzheimer Disease: Rationale and Perspectives. CNS Drugs 2019, 33, 957–969. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, R.H. Mitochondria and Mitochondrial Cascades in Alzheimer’s Disease. J. Alzheimer’s Dis. 2018, 62, 1403–1416. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Huang, M.; Shang, D.; Yan, X.; Zhao, B.; Zhang, X. Mitochondrial Behavior in Axon Degeneration and Regeneration. Front. Aging Neurosci. 2021, 13, 103. [Google Scholar] [CrossRef]
- Cheignon, C.; Tomas, M.; Bonnefont-Rousselot, D.; Faller, P.; Hureau, C.; Collin, F. Oxidative stress and the amyloid beta peptide in Alzheimer’s disease. Redox Boil. 2018, 14, 450–464. [Google Scholar] [CrossRef]
- Gaggelli, E.; Kozlowski, H.; Valensin, A.D.; Valensin, G. Copper Homeostasis and Neurodegenerative Disorders (Alzheimer’s, Prion, and Parkinson’s Diseases and Amyotrophic Lateral Sclerosis). Chem. Rev. 2006, 106, 1995–2044. [Google Scholar] [CrossRef]
- Kozlowski, H.; Janicka-Klos, A.; Brasun, J.; Gaggelli, E.; Valensin, D.; Valensin, G. Copper, iron, and zinc ions homeostasis and their role in neurodegenerative disorders (metal uptake, transport, distribution and regulation). Coord. Chem. Rev. 2009, 253, 2665–2685. [Google Scholar] [CrossRef]
- Gromadzka, G.; Tarnacka, B.; Flaga, A.; Adamczyk, A. Copper Dyshomeostasis in Neurodegenerative Diseases—Therapeutic Implications. Int. J. Mol. Sci. 2020, 21, 9259. [Google Scholar] [CrossRef] [PubMed]
- Royer, A.; Sharman, T. Copper toxicity. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Patel, R.; Aschner, M. Commonalities between Copper Neurotoxicity and Alzheimer’s Disease. Toxics 2021, 9, 4. [Google Scholar] [CrossRef]
- Pal, A.; Prasad, R. Regional Distribution of Copper, Zinc and Iron in Brain of Wistar Rat Model for Non-Wilsonian Brain Copper Toxicosis. Indian J. Clin. Biochem. 2015, 31, 93–98. [Google Scholar] [CrossRef] [Green Version]
- Stokum, J.A.; Kurland, D.B.; Gerzanich, V.; Simard, J.M. Mechanisms of Astrocyte-Mediated Cerebral Edema. Neurochem. Res. 2015, 40, 317–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fulop, T.; Witkowski, J.M.; Bourgade, K.; Khalil, A.; Zerif, E.; Larbi, A.; Hirokawa, K.; Pawelec, G.; Bocti, C.; Lacombe, G.; et al. Can an Infection Hypothesis Explain the Beta Amyloid Hypothesis of Alzheimer’s Disease? Front. Aging Neurosci. 2018, 10, 224. [Google Scholar] [CrossRef] [Green Version]
- Kardos, J.; Héja, L.; Simon, Á.; Jablonkai, I.; Kovács, R.; Jemnitz, K. Copper signalling: Causes and consequences. Cell Commun. Signal. 2018, 16, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Brewer, G.J. Copper toxicity in Alzheimer’s disease: Cognitive loss from ingestion of inorganic copper. J. Trace Elem. Med. Biol. 2012, 26, 89–92. [Google Scholar] [CrossRef]
- Kozlowski, H.; Luczkowski, M.; Remelli, M.; Valensin, D. Copper, zinc and iron in neurodegenerative diseases (Alzheimer’s, Parkinson’s and prion diseases). Coord. Chem. Rev. 2012, 256, 2129–2141. [Google Scholar] [CrossRef]
- Bagheri, S.; Squitti, R.; Haertlé, T.; Siotto, M.; Saboury, A.A. Role of Copper in the Onset of Alzheimer’s Disease Compared to Other Metals. Front. Aging Neurosci. 2018, 9, 446. [Google Scholar] [CrossRef] [PubMed]
- Kitazawa, M.; Hsu, H.-W.; Medeiros, R. Copper Exposure Perturbs Brain Inflammatory Responses and Impairs Clearance of Amyloid-Beta. Toxicol. Sci. 2016, 152, 194–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, P.; Ayton, S.; Bush, A.I. The essential elements of Alzheimer’s disease. J. Biol. Chem. 2021, 296, 100105. [Google Scholar] [CrossRef] [PubMed]
- Navarro, A. Brain mitochondrial dysfunction in aging: Conditions that improve survival, neurological performance and mitochondrial function. Front. Biosci. 2007, 12, 1154–1163. [Google Scholar] [CrossRef] [Green Version]
- Navarro, A.; del Pino, M.J.S.; Gómez, C.; Peralta, J.L.; Boveris, A. Behavioral dysfunction, brain oxidative stress, and impaired mitochondrial electron transfer in aging mice. Am. J. Physiol. Integr. Comp. Physiol. 2002, 282, R985–R992. [Google Scholar] [CrossRef] [Green Version]
- Saporito-Magriñá, C.; Musacco-Sebio, R.; Acosta, J.M.; Bajicoff, S.; Paredes-Fleitas, P.; Boveris, A.; Repetto, M.G. Rat liver mitochondrial dysfunction by addition of copper(II) or iron(III) ions. J. Inorg. Biochem. 2017, 166, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Behzadfar, L.; Abdollahi, M.; Sabzevari, O.; Hosseini, R.; Salimi, A.; Naserzadeh, P.; Sharifzadeh, M.; Pourahmad, J. Potentiating role of copper on spatial memory deficit induced by beta amyloid and evaluation of mitochondrial function markers in the hippocampus of rats. Metallomics 2017, 9, 969–980. [Google Scholar] [CrossRef]
- Peña-Bautista, C.; Baquero, M.; Vento, M.; Cháfer-Pericás, C. Omics-based Biomarkers for the Early Alzheimer Disease Diagnosis and Reliable Therapeutic Targets Development. Curr. Neuropharmacol. 2019, 17, 630–647. [Google Scholar] [CrossRef]
- Gregersen, N.; Hansen, J.; Palmfeldt, J. Mitochondrial proteomics—A tool for the study of metabolic disorders. J. Inherit. Metab. Dis. 2012, 35, 715–726. [Google Scholar] [CrossRef]
- Lin, X.; Wei, G.; Huang, Z.; Qu, Z.; Huang, X.; Xu, H.; Liu, J.; Yang, X.; Zhuang, Z. Mitochondrial proteomic alterations caused by long-term low-dose copper exposure in mouse cortex. Toxicol. Lett. 2016, 263, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Wang, D.; Zou, L.; Zhang, Z.; Xu, H.; Zhu, F.; Ren, X.; Xu, B.; Yuan, J.; Liu, J.; et al. Proteomic alterations of brain subcellular organelles caused by low-dose copper exposure: Implication for Alzheimer’s disease. Arch. Toxicol. 2018, 92, 1363–1382. [Google Scholar] [CrossRef]
- Szabadkai, G.; Bianchi, K.; Várnai, P.; de Stefani, D.; Wieckowski, M.R.; Cavagna, D.; Nagy, A.I.; Balla, T.; Rizzuto, R. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J. Cell Biol. 2006, 175, 901–911. [Google Scholar] [CrossRef] [Green Version]
- Schon, E.A.; Area-Gomez, E. Mitochondria-associated ER membranes in Alzheimer disease. Mol. Cell. Neurosci. 2013, 55, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Eysert, F.; Kinoshita, P.F.; Mary, A.; Vaillant-Beuchot, L.; Checler, F.; Chami, M. Molecular Dysfunctions of Mitochondria-Associated Membranes (MAMs) in Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 9521. [Google Scholar] [CrossRef] [PubMed]
- Yoo, B.C.; Kim, S.H.; Cairnsb, N.; Fountoulakisc, M.; Lubeca, G. Deranged Expression of Molecular Chaperones in Brains of Patients with Alzheimer’s Disease. Biochem. Biophys. Res. Commun. 2001, 280, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado-Tejedor, M.; Vilariño, M.; Cabodevilla, F.; del Río, J.; Frechilla, D.; Pérez-Mediavilla, A. Enhanced Expression of the Voltage-Dependent Anion Channel 1 (VDAC1) in Alzheimer’s Disease Transgenic Mice: An Insight into the Pathogenic Effects of Amyloid-β. J. Alzheimer’s Dis. 2011, 23, 195–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Turner, R.S.; Gaut, J.R. The Chaperone BiP/GRP78 Binds to Amyloid Precursor Protein and Decreases Aβ40 and Aβ42 Secretion. J. Biol. Chem. 1998, 273, 25552–25555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorbatyuk, M.S.; Gorbatyuk, O.S. The Molecular Chaperone GRP78/BiP as a Therapeutic Target for Neurodegenerative Disorders: A Mini Review. J. Genet. Syndr. Gene Ther. 2013, 4, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Chadha, S.; Behl, T.; Sehgal, A.; Kumar, A.; Bungau, S. Exploring the role of mitochondrial proteins as molecular target in Alzheimer’s disease. Mitochondrion 2021, 56, 62–72. [Google Scholar] [CrossRef]
- Cunnane, S.C.; Trushina, E.; Morland, C.; Prigione, A.; Casadesus, G.; Andrews, Z.B.; Beal, M.F.; Bergersen, L.H.; Brinton, R.D.; de la Monte, S.; et al. Brain energy rescue: An emerging therapeutic concept for neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 2020, 19, 609–633. [Google Scholar] [CrossRef] [PubMed]
- Sivanesan, S.; Chang, E.; Howell, M.D.; Rajadas, J. Amyloid protein aggregates: New clients for mitochondrial energy production in the brain? FEBS J. 2020, 287, 3386–3395. [Google Scholar] [CrossRef] [Green Version]
- Adav, S.S.; Park, J.E.; Sze, S.K. Quantitative profiling brain proteomes revealed mitochondrial dysfunction in Alzheimer’s disease. Mol. Brain 2019, 12, 1–12. [Google Scholar] [CrossRef]
- Abate, G.; Vezzoli, M.; Sandri, M.; Rungratanawanich, W.; Memo, M.; Uberti, D. Mitochondria and cellular redox state on the route from ageing to Alzheimer’s disease. Mech. Ageing Dev. 2020, 192, 111385. [Google Scholar] [CrossRef]
- Guo, R.; Zong, S.; Wu, M.; Gu, J.; Yang, M. Architecture of Human Mitochondrial Respiratory Megacomplex I2III2IV2. Cell 2017, 170, 1247–1257.e12. [Google Scholar] [CrossRef] [Green Version]
- Zong, S.; Wu, M.; Gu, J.; Liu, T.; Guo, R.; Yang, M. Structure of the intact 14-subunit human cytochrome c oxidase. Cell Res. 2018, 28, 1026–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Au, H.C.; Seo, B.B.; Matsuno-Yagi, A.; Yagi, T.; Scheffler, I.E. The NDUFA1 gene product (MWFE protein) is essential for activity of complex I in mammalian mitochondria. Proc. Natl. Acad. Sci. USA 1999, 96, 4354–4359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Moreira, D.; Ugalde, C.; Smeets, R.; Rodenburg, R.J.T.; Lopez-Laso, E.; Ruiz-Falco, M.L.; Briones, P.; Martin, M.A.; Smeitink, J.A.M.; Arenas, J. X-linkedNDUFA1gene mutations associated with mitochondrial encephalomyopathy. Ann. Neurol. 2007, 61, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Potluri, P.; Davila, A.; Ruiz-Pesini, E.; Mishmar, D.; O’Hearn, S.; Hancock, S.; Simon, M.; Scheffler, I.E.; Wallace, D.C.; Procaccio, V. A novel NDUFA1 mutation leads to a progressive mitochondrial complex I-specific neurodegenerative disease. Mol. Genet. Metab. 2009, 96, 189–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murai, M.; Inaoka, H.; Masuya, T.; Aburaya, S.; Aoki, W.; Miyoshi, H. Specific Methylation of Asp160 (49 kDa subunit) Located inside the Quinone Binding Cavity of Bovine Mitochondrial Complex I. Biochemistry 2016, 55, 3189–3197. [Google Scholar] [CrossRef]
- Andrews, B.; Carroll, J.; Ding, S.; Fearnley, I.M.; Walker, J.E. Assembly factors for the membrane arm of human complex I. Proc. Natl. Acad. Sci. USA 2013, 110, 18934–18939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crofts, A.R.; Hong, S.; Wilson, C.; Burton, R.; Victoria, D.; Harrison, C.; Schulten, K. The mechanism of ubihydroquinone oxidation at the Qo-site of the cytochrome bc1 complex. Biochim. Biophys. Acta Bioenerg. 2013, 1827, 1362–1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, X.-R.; Zhou, W.-X.; Zhang, Y.-X. The effects of Liuwei Dihuang decoction on the gene expression in the hippocampus of senescence-accelerated mouse. Fitoterapia 2007, 78, 175–181. [Google Scholar] [CrossRef]
- Kadenbach, B. Complex IV—The regulatory center of mitochondrial oxidative phosphorylation. Mitochondrion 2020. [Google Scholar] [CrossRef]
- Collins, J.F. Molecular, Genetic, and Nutritional Aspects of Major and Trace Minerals; Academic Press: London, UK; Cambridge, MA, USA, 2016; ISBN 978-0-12-802376-1. [Google Scholar]
- Maurer, I. A selective defect of cytochrome c oxidase is present in brain of Alzheimer disease patients. Neurobiol. Aging 2000, 21, 455–462. [Google Scholar] [CrossRef]
- He, J.; Carroll, J.; Ding, S.; Fearnley, I.M.; Montgomery, M.G.; Walker, J.E. Assembly of the peripheral stalk of ATP synthase in human mitochondria. Proc. Natl. Acad. Sci. USA 2020, 117, 29602–29608. [Google Scholar] [CrossRef]
- Mukherjee, S.; Russell, J.C.; Carr, D.T.; Burgess, J.D.; Allen, M.; Serie, D.J.; Boehme, K.L.; Kauwe, J.S.K.; Naj, A.C.; Fardo, D.W.; et al. Systems biology approach to late-onset Alzheimer’s disease genome-wide association study identifies novel candidate genes validated using brain expression data and Caenorhabditis elegans experiments. Alzheimer’s Dement. 2017, 13, 1133–1142. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Hu, Y.; Wang, B.; Wang, S.; Zhang, X. Metabolic Dysregulation Contributes to the Progression of Alzheimer’s Disease. Front. Neurosci. 2020, 14, 1107. [Google Scholar] [CrossRef] [PubMed]
- Sonntag, K.-C.; Ryu, W.-I.; Amirault, K.M.; Healy, R.A.; Siegel, A.J.; McPHIE, N.L.; Forester, B.; Cohen, B.M. Late-onset Alzheimer’s disease is associated with inherent changes in bioenergetics profiles. Sci. Rep. 2017, 7, 14038. [Google Scholar] [CrossRef] [Green Version]
- Schlachter, C.R.; Klapper, V.; Radford, T.; Chruszcz, M. Comparative studies of Aspergillus fumigatus 2-methylcitrate synthase and human citrate synthase. Biol. Chem. 2019, 400, 1567–1581. [Google Scholar] [CrossRef]
- Lloyd, S.J.; Lauble, H.; Prasad, G.S.; Stout, C.D. The mechanism of aconitase: 1.8 Å resolution crystal structure of the S642A: Citrate complex. Protein Sci. 2008, 8, 2655–2662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eder, M.; Fritz-Wolf, K.; Kabsch, W.; Wallimann, T.; Schlattner, U. Crystal structure of human ubiquitous mitochondrial creatine kinase. Proteins Struct. Funct. Bioinform. 2000, 39, 216–225. [Google Scholar] [CrossRef]
- Schlattner, U.; Tokarska-Schlattner, M.; Wallimann, T. Mitochondrial creatine kinase in human health and disease. Biochim. Biophys. Acta Mol. Basis Dis. 2006, 1762, 164–180. [Google Scholar] [CrossRef] [PubMed]
- Khait, I.; Togliatti, A.; Benzecry, J.M.; Wieringa, B.; Holtzman, D. Altered brain phosphocreatine and ATP regulation when mitochondrial creatine kinase is absent. J. Neurosci. Res. 2001, 66, 866–872. [Google Scholar] [CrossRef]
- van der Bliek, A.M.; Sedensky, M.M.; Morgan, P.G. Cell Biology of the Mitochondrion. Genetics 2017, 207, 843–871. [Google Scholar] [CrossRef] [Green Version]
- Butterfield, D.A.; Halliwell, B. Oxidative stress, dysfunctional glucose metabolism and Alzheimer disease. Nat. Rev. Neurosci. 2019, 20, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Bresciani, G.; da Cruz, I.B.M.; González-Gallego, J. Manganese Superoxide Dismutase and Oxidative Stress Modulation. Adv. Appl. Microbiol. 2015, 68, 87–130. [Google Scholar] [CrossRef]
- Islam, T. Oxidative stress and mitochondrial dysfunction-linked neurodegenerative disorders. Neurol. Res. 2017, 39, 73–82. [Google Scholar] [CrossRef]
- Dai, D.-F.; Chiao, Y.-A.; Martin, G.; Marcinek, D.; Basisty, N.; Quarles, E.; Rabinovitch, P. Mitochondrial-Targeted Catalase. Prog. Mol. Biol. Transl. Sci. 2017, 146, 203–241. [Google Scholar] [CrossRef]
- Sies, H.; Berndt, C.; Jones, D.P. Oxidative Stress. Annu. Rev. Biochem. 2017, 86, 715–748. [Google Scholar] [CrossRef]
- Tramutola, A.; Lanzillotta, C.; Perluigi, M.; Butterfield, D.A. Oxidative stress, protein modification and Alzheimer disease. Brain Res. Bull. 2017, 133, 88–96. [Google Scholar] [CrossRef]
- Radi, R. Oxygen radicals, nitric oxide, and peroxynitrite: Redox pathways in molecular medicine. Proc. Natl. Acad. Sci. USA 2018, 115, 5839–5848. [Google Scholar] [CrossRef] [Green Version]
- Butterfield, D.A.; Gu, L.; Di Domenico, F.; Robinson, R.A. Mass spectrometry and redox proteomics: Applications in disease. Mass Spectrom. Rev. 2013, 33, 277–301. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, D.A.; Perluigi, M.; Reed, T.; Muharib, T.; Hughes, C.P.; Robinson, R.A.; Sultana, R. Redox Proteomics in Selected Neurodegenerative Disorders: From Its Infancy to Future Applications. Antioxid. Redox Signal. 2012, 17, 1610–1655. [Google Scholar] [CrossRef] [Green Version]
- Perluigi, M.; Swomley, A.M.; Butterfield, D.A. Redox proteomics and the dynamic molecular landscape of the aging brain. Ageing Res. Rev. 2014, 13, 75–89. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Boyd-Kimball, D. Mitochondrial Oxidative and Nitrosative Stress and Alzheimer Disease. Antioxidants 2020, 9, 818. [Google Scholar] [CrossRef]
- Ghezzi, P.; Bonetto, V. Redox proteomics: Identification of oxidatively modified proteins. Proteomics 2003, 3, 1145–1153. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, D.A.; Sultana, R. Redox proteomics: Understanding oxidative stress in the progression of age-related neurodegenerative disorders. Expert Rev. Proteom. 2008, 5, 157–160. [Google Scholar] [CrossRef] [Green Version]
- Shacter, E.; Williams, J.A.; Lim, M.; Levine, R.L. Differential susceptibility of plasma proteins to oxidative modification: Examination by western blot immunoassay. Free Radic. Biol. Med. 1994, 17, 429–437. [Google Scholar] [CrossRef]
- Berlett, B.S.; Stadtman, E.R. Protein Oxidation in Aging, Disease, and Oxidative Stress. J. Biol. Chem. 1997, 272, 20313–20316. [Google Scholar] [CrossRef] [Green Version]
- Stadtman, E.R.; Berlett, B.S. Reactive Oxygen-Mediated Protein Oxidation in Aging and Disease. Drug Metab. Rev. 1998, 30, 225–243. [Google Scholar] [CrossRef]
- Perluigi, M.; Butterfield, D.A. Oxidative Stress and Down Syndrome: A Route toward Alzheimer-Like Dementia. Curr. Gerontol. Geriatr. Res. 2011, 2012, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Yeo, W.-S.; Lee, S.J.; Lee, J.R.; Kim, K.P. Nitrosative Protein Tyrosine Modifications: Biochemistry and Functional Signifi-cance. BMB Rep. 2008, 41, 194–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramaniam, R.; Roediger, F.; Jordan, B.; Mattson, M.P.; Keller, J.N.; Waeg, G.; Butterfield, D.A. The Lipid Peroxidation Product, 4-Hydroxy-2-trans-Nonenal, Alters the Conformation of Cortical Synaptosomal Membrane Proteins. J. Neurochem. 2002, 69, 1161–1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sultana, R.; Butterfield, D.A. Proteomics Identification of Carbonylated and HNE-Bound Brain Proteins in Alzheimer’s Disease. Adv. Struct. Saf. Stud. 2009, 566, 123–135. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.; Creasy, D.M.; Cottrell, J.S. Probability-Based Protein Identification by Searching Sequence Data-bases Using Mass Spectrometry Data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Eng, J.K.; McCormack, A.L.; Yates, J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994, 5, 976–989. [Google Scholar] [CrossRef] [Green Version]
- Ross, P.L.; Huang, Y.N.; Marchese, J.N.; Williamson, B.; Parker, K.; Hattan, S.; Khainovski, N.; Pillai, S.; Dey, S.; Daniels, S.; et al. Multiplexed Protein Quantitation in Saccharomyces cerevisiae Using Amine-reactive Isobaric Tagging Reagents. Mol. Cell. Proteom. 2004, 3, 1154–1169. [Google Scholar] [CrossRef] [Green Version]
- Rauniyar, N.; Prokai, L. Isotope-coded dimethyl tagging for differential quantification of posttranslational protein carbonylation by 4-hydroxy-2-nonenal, an end-product of lipid peroxidation. J. Mass Spectrom. 2011, 46, 976–985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, V.-K.; Singh, T.G.; Mehta, V. Stressed Mitochondria: A Target to Intrude Alzheimer’s Disease. Mitochondrion 2021. [Google Scholar] [CrossRef] [PubMed]
- Perluigi, M.; Sultana, R.; Cenini, G.; di Domenico, F.; Memo, M.; Pierce, W.M.; Coccia, R.; Butterfield, D.A. Redox proteomics identification of 4-hydroxynonenal-modified brain proteins in Alzheimer’s disease: Role of lipid peroxidation in Alzheimer’s disease pathogenesis. Proteom. Clin. Appl. 2009, 3, 682–693. [Google Scholar] [CrossRef] [Green Version]
- Reed, T.T.; Pierce, W.M.; Markesbery, W.R.; Butterfield, D.A. Proteomic identification of HNE-bound proteins in early Alzheimer disease: Insights into the role of lipid peroxidation in the progression of AD. Brain Res. 2009, 1274, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Sultana, R.; Poon, H.F.; Cai, J.; Pierce, W.M.; Merchant, M.; Klein, J.B.; Markesbery, W.R.; Butterfield, D.A. Identification of nitrated proteins in Alzheimer’s disease brain using a redox proteomics approach. Neurobiol. Dis. 2006, 22, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Terni, B.; Boada, J.; Portero-Otin, M.; Pamplona, R.; Ferrer, I. Mitochondrial ATP-Synthase in the Entorhinal Cortex Is a Target of Oxidative Stress at Stages I/II of Alzheimer’s Disease Pathology. Brain Pathol. 2010, 20, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Sultana, R.; Robinson, R.A.S.; di Domenico, F.; Abdul, H.M.; Clair, D.K.S.; Markesbery, W.R.; Cai, J.; Pierce, W.M.; Butterfield, D.A. Proteomic identification of specifically carbonylated brain proteins in APPNLh/APPNLh×PS-1P264L/PS-1P264L human double mutant knock-in mice model of Alzheimer disease as a function of age. J. Proteom. 2011, 74, 2430–2440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoshan-Barmatz, V.; Nahon-Crystal, E.; Shteinfer-Kuzmine, A.; Gupta, R. VDAC1, mitochondrial dysfunction, and Alzheimer’s disease. Pharmacol. Res. 2018, 131, 87–101. [Google Scholar] [CrossRef]
- Shoshan-Barmatz, V.; Maldonado, E.N.; Krelin, Y. VDAC1 at the crossroads of cell metabolism, apoptosis and cell stress. Cell Stress 2017, 1, 11–36. [Google Scholar] [CrossRef]
- Geula, S.; Naveed, H.; Liang, J.; Shoshan-Barmatz, V. Structure-based Analysis of VDAC1 Protein. J. Biol. Chem. 2012, 287, 2179–2190. [Google Scholar] [CrossRef] [Green Version]
- Geula, S.; Ben-Hail, D.; Shoshan-Barmatz, V. Structure-based analysis of VDAC1: N-terminus location, translocation, channel gating and association with anti-apoptotic proteins. Biochem. J. 2012, 444, 475–485. [Google Scholar] [CrossRef] [Green Version]
- Abu-Hamad, S.; Arbel, N.; Calo, D.; Arzoine, L.; Israelson, A.; Keinan, N.; Ben-Romano, R.; Friedman, O.; Shoshan-Barmatz, V. The VDAC1 N-terminus is essential both for apoptosis and the protective effect of anti-apoptotic proteins. J. Cell Sci. 2009, 122, 1906–1916. [Google Scholar] [CrossRef] [Green Version]
- Arbel, N.; Shoshan-Barmatz, V. Voltage-dependent Anion Channel 1-based Peptides Interact with Bcl-2 to Prevent Antiapoptotic Activity. J. Biol. Chem. 2010, 285, 6053–6062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arbel, N.; Ben-Hail, D.; Shoshan-Barmatz, V. Mediation of the Antiapoptotic Activity of Bcl-xL Protein upon Interaction with VDAC1 Protein. J. Biol. Chem. 2012, 287, 23152–23161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varda, S.-B.; Yakov, K.; Quan, C. VDAC1 as a Player in Mitochondria-Mediated Apoptosis and Target for Modulating Apop-tosis. Curr. Med. Chem. 2017, 24, 4435–4446. [Google Scholar]
- Manczak, M.; Reddy, P.H. Abnormal interaction of VDAC1 with amyloid beta and phosphorylated tau causes mitochondrial dysfunction in Alzheimer’s disease. Hum. Mol. Genet. 2012, 21, 5131–5146. [Google Scholar] [CrossRef] [PubMed]
- Abu-Hamad, S.; Zaid, H.; Israelson, A.; Nahon, E.; Shoshan-Barmatz, V. Hexokinase-I Protection against Apoptotic Cell Death Is Mediated via Interaction with the Voltage-dependent Anion Channel-1. J. Biol. Chem. 2008, 283, 13482–13490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smilansky, A.; Dangoor, L.; Nakdimon, I.; Ben-Hail, D.; Mizrachi, D.; Shoshan-Barmatz, V. The Voltage-dependent Anion Channel 1 Mediates Amyloid β Toxicity and Represents a Potential Target for Alzheimer Disease Therapy. J. Biol. Chem. 2015, 290, 30670–30683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quint, P.; Reutzel, R.; Mikulski, R.; McKenna, R.; Silverman, D.N. Crystal structure of nitrated human manganese superoxide dismutase: Mechanism of inactivation. Free Radic. Biol. Med. 2006, 40, 453–458. [Google Scholar] [CrossRef]
- Lushchak, O.V.; Piroddi, M.; Galli, F.; Lushchak, V.I. Aconitase post-translational modification as a key in linkage between Krebs cycle, iron homeostasis, redox signaling, and metabolism of reactive oxygen species. Redox Rep. 2013, 19, 8–15. [Google Scholar] [CrossRef] [Green Version]
- Shen, L.; Chen, C.; Yang, A.; Chen, Y.; Liu, Q.; Ni, J. Redox proteomics identification of specifically carbonylated proteins in the hippocampi of triple transgenic Alzheimer’s disease mice at its earliest pathological stage. J. Proteom. 2015, 123, 101–113. [Google Scholar] [CrossRef]
- Boyd-Kimball, D.; Poon, H.F.; Lynn, B.C.; Cai, J.; Pierce, W.M.; Klein, J.B.; Ferguson, J.; Link, C.D.; Butterfield, D.A. Proteomic identification of proteins specifically oxidized in Caenorhabditis elegans expressing human Aβ(1–42): Implications for Alzheimer’s disease. Neurobiol. Aging 2006, 27, 1239–1249. [Google Scholar] [CrossRef]
- Hsieh, J.-Y.; Shih, W.-T.; Kuo, Y.-H.; Liu, G.-Y.; Hung, H.-C. Functional Roles of Metabolic Intermediates in Regulating the Human Mitochondrial NAD(P)+-Dependent Malic Enzyme. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Minárik, P.; Tomásková, N.; Kollárová, M.; Antalík, M. Malate dehydrogenases--structure and function. Gen. Physiol. Biophys. 2002, 21, 257–265. [Google Scholar]
- Bubber, P.; Haroutunian, V.; Fisch, G.; Blass, J.P.; Gibson, G.E. Mitochondrial abnormalities in Alzheimer brain: Mechanistic implications. Ann. Neurol. 2005, 57, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Jagust, W. Imaging the evolution and pathophysiology of Alzheimer disease. Nat. Rev. Neurosci. 2018, 19, 687–700. [Google Scholar] [CrossRef]
- Veitch, D.P.; Weiner, M.W.; Aisen, P.S.; Beckett, L.A.; Cairns, N.J.; Green, R.C.; Harvey, D.; Jack, C.R.; Jagust, W.; Morris, J.C.; et al. Understanding disease progression and improving Alzheimer’s disease clinical trials: Recent highlights from the Alzheimer’s Disease Neuroimaging Initiative. Alzheimer’s Dement. 2019, 15, 106–152. [Google Scholar] [CrossRef]
- Leibovitz, M.B.E.; Siegel, B.V. Aspects of Free Radical Reactions in Biological Systems: Aging. J. Gerontol. 1980, 35, 45–56. [Google Scholar] [CrossRef]
- Flynn, J.M.; Melov, S. SOD2 in mitochondrial dysfunction and neurodegeneration. Free Radic. Biol. Med. 2013, 62, 4–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcus, D.L.; Strafaci, J.A.; Freedman, M.L. Differential Neuronal Expression of Manganese Superoxide Dismutase in Alz-heimer’s Disease. Med. Sci. Monit. 2006, 12, BR8–BR14. [Google Scholar]
- Anantharaman, M.; Tangpong, J.; Keller, J.N.; Murphy, M.P.; Markesbery, W.R.; Kiningham, K.K.; Clair, D.K.S. β-Amyloid Mediated Nitration of Manganese Superoxide Dismutase. Am. J. Pathol. 2006, 168, 1608–1618. [Google Scholar] [CrossRef] [Green Version]
- Sompol, P.; Ittarat, W.; Tangpong, J.; Chen, Y.; Doubinskaia, I.; Batinic-Haberle, I.; Abdul, H.; Butterfield, D.; Clair, D.S. A neuronal model of Alzheimer’s disease: An insight into the mechanisms of oxidative stress–mediated mitochondrial injury. Neuroscience 2008, 153, 120–130. [Google Scholar] [CrossRef] [Green Version]
- Dhar, S.K.; Tangpong, J.; Chaiswing, L.; Oberley, T.D.; Clair, D.K.S. Manganese Superoxide Dismutase Is a p53-Regulated Gene That Switches Cancers between Early and Advanced Stages. Cancer Res. 2011, 71, 6684–6695. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Chaiswing, L.; Velez, J.M.; Batinic-Haberle, I.; Colburn, N.H.; Oberley, T.D.; Clair, D.K.S. p53 Translocation to Mitochondria Precedes Its Nuclear Translocation and Targets Mitochondrial Oxidative Defense Protein-Manganese Superoxide Dismutase. Cancer Res. 2005, 65, 3745–3750. [Google Scholar] [CrossRef] [Green Version]
- Dai, C.-Q.; Luo, T.-T.; Luo, S.-C.; Wang, J.-Q.; Wang, S.-M.; Bai, Y.-H.; Yang, Y.-L.; Wang, Y.-Y. p53 and mitochondrial dysfunction: Novel insight of neurodegenerative diseases. J. Bioenerg. Biomembr. 2016, 48, 337–347. [Google Scholar] [CrossRef] [Green Version]
- Cenini, G.; Sultana, R.; Memo, M.; Butterfield, D.A. Effects of oxidative and nitrosative stress in brain on p53 proapoptotic protein in amnestic mild cognitive impairment and Alzheimer disease. Free Radic. Biol. Med. 2008, 45, 81–85. [Google Scholar] [CrossRef] [Green Version]
- Cenini, G.; Sultana, R.; Memo, M.; Butterfield, D.A. Elevated levels of pro-apoptotic p53 and its oxidative modification by the lipid peroxidation product, HNE, in brain from subjects with amnestic mild cognitive impairment and Alzheimer’s disease. J. Cell. Mol. Med. 2008, 12, 987–994. [Google Scholar] [CrossRef]
- Sultana, R.; Mecocci, P.; Mangialasche, F.; Cecchetti, R.; Baglioni, M.; Butterfield, D.A. Increased Protein and Lipid Oxidative Damage in Mitochondria Isolated from Lymphocytes from Patients with Alzheimer’s Disease: Insights into the Role of Oxidative Stress in Alzheimer’s Disease and Initial Investigations into a Potential Biomarker for this Dementing Disorder. J. Alzheimer’s Dis. 2011, 24, 77–84. [Google Scholar] [CrossRef] [Green Version]
- Sultana, R.; Baglioni, M.; Cecchetti, R.; Cai, J.; Klein, J.B.; Bastiani, P.; Ruggiero, C.; Mecocci, P.; Butterfield, D.A. Lymphocyte mitochondria: Toward identification of peripheral biomarkers in the progression of Alzheimer disease. Free Radic. Biol. Med. 2013, 65, 595–606. [Google Scholar] [CrossRef] [Green Version]
- Feldhaus, P.; Fraga, D.B.; Ghedim, F.V.; de Luca, R.D.; Bruna, T.D.; Heluany, M.; Matos, M.P.; Ferreira, G.K.; Jeremias, I.C.; Heluany, C.; et al. Evaluation of respiratory chain activity in lymphocytes of patients with Alzheimer disease. Metab. Brain Dis. 2011, 26, 229–236. [Google Scholar] [CrossRef]
- Cardoso, S.M.; Proença, M.; Santos, S.; Santana, I.; Oliveira, C.R. Cytochrome c oxidase is decreased in Alzheimer’s disease platelets. Neurobiol. Aging 2004, 25, 105–110. [Google Scholar] [CrossRef]
- di Domenico, F.; Barone, E.; Perluigi, M.; Butterfield, D.A. Strategy to reduce free radical species in Alzheimer’s disease: An update of selected antioxidants. Expert Rev. Neurother. 2014, 15, 19–40. [Google Scholar] [CrossRef]
- Reddy, P.H. Mitochondrial Oxidative Damage in Aging and Alzheimer’s Disease: Implications for Mitochondrially Targeted Antioxidant Therapeutics. J. Biomed. Biotechnol. 2006, 2006, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Maczurek, A.; Hager, K.; Kenklies, M.; Sharman, M.; Martins, R.; Engel, J.; Carlson, D.A.; Münch, G. Lipoic acid as an anti-inflammatory and neuroprotective treatment for Alzheimer’s disease. Adv. Drug Deliv. Rev. 2008, 60, 1463–1470. [Google Scholar] [CrossRef] [PubMed]
- Joshi, Y.B.; Praticò, D. Vitamin E in aging, dementia, and Alzheimer’s disease. Biofactors 2012, 38, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Sung, S.; Yao, Y.; Uryu, K.; Yang, H.; Lee, V.M.; Trojanowski, J.Q.; Praticò, D. Early Vitamin E supplementation in young but not aged mice reduces Aβ levels and amyloid deposition in a transgenic model of Alzheimer’s disease. FASEB J. 2003, 18, 323–325. [Google Scholar] [CrossRef]
- Praticò, D.; Clark, C.M.; Liun, F.; Rokach, J.; Lee, V.Y.-M.; Trojanowski, J.Q. Increase of Brain Oxidative Stress in Mild Cognitive Impairment. Arch. Neurol. 2002, 59, 972–976. [Google Scholar] [CrossRef] [Green Version]
- Mangialasche, F.; Xu, W.; Kivipelto, M.; Costanzi, E.; Ercolani, S.; Pigliautile, M.; Cecchetti, R.; Baglioni, M.; Simmons, A.; Soininen, H.; et al. Tocopherols and tocotrienols plasma levels are associated with cognitive impairment. Neurobiol. Aging 2012, 33, 2282–2290. [Google Scholar] [CrossRef] [PubMed]
- Fillenbaum, G.G.; Kuchibhatla, M.N.; Hanlon, J.T.; Artz, M.B.; Pieper, C.F.; Schmader, K.E.; Dysken, M.W.; Gray, S.L. Dementia and Alzheimer’s Disease in Community-Dwelling Elders Taking Vitamin C and/or Vitamin E. Ann. Pharmacother. 2005, 39, 2009–2014. [Google Scholar] [CrossRef]
- Dysken, M.W.; Sano, M.; Asthana, S.; Vertrees, J.E.; Pallaki, M.; Llorente, M.; Love, S.; Schellenberg, G.D.; McCarten, J.R.; Malphurs, J.; et al. Effect of Vitamin E and Memantine on Functional Decline in Alzheimer Disease: The TEAM-AD VA cooperative randomized trial. JAMA 2014, 311, 33–44. [Google Scholar] [CrossRef]
- Shetty, R.A.; Ikonne, U.S.; Forster, M.J.; Sumien, N. Coenzyme Q10 and α-tocopherol reversed age-associated functional impairments in mice. Exp. Gerontol. 2014, 58, 208–218. [Google Scholar] [CrossRef] [Green Version]
- Kontush, A.; Mann, U.; Arlt, S.; Ujeyl, A.; Lührs, C.; Müller-Thomsen, T.; Beisiegel, U. Influence of vitamin E and C supplementation on lipoprotein oxidation in patients with Alzheimer’s disease. Free Radic. Biol. Med. 2001, 31, 345–354. [Google Scholar] [CrossRef]
- Bonda, D.J.; Wang, X.; Perry, G.; Nunomura, A.; Tabaton, M.; Zhu, X.; Smith, M.A. Oxidative stress in Alzheimer disease: A possibility for prevention. Neuropharmacology 2010, 59, 290–294. [Google Scholar] [CrossRef]
- Moreira, P.I.; Harris, P.L.; Zhu, X.; Santos, M.S.; Oliveira, C.R.; Smith, M.A.; Perry, G. Lipoic Acid and N-acetyl Cysteine Decrease Mitochondrial-Related Oxidative Stress in Alzheimer Disease Patient Fibroblasts. J. Alzheimer’s Dis. 2007, 12, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Poon, H.F.; Farr, S.A.; Thongboonkerd, V.; Lynn, B.C.; Banks, W.A.; Morley, J.E.; Klein, J.B.; Butterfield, D.A. Proteomic analysis of specific brain proteins in aged SAMP8 mice treated with alpha-lipoic acid: Implications for aging and age-related neurodegenerative disorders. Neurochem. Int. 2005, 46, 159–168. [Google Scholar] [CrossRef]
- Abdul, H.M.; Butterfield, D.A. Involvement of PI3K/PKG/ERK1/2 signaling pathways in cortical neurons to trigger protection by cotreatment of acetyl-L-carnitine and α-lipoic acid against HNE-mediated oxidative stress and neurotoxicity: Implications for Alzheimer’s disease. Free Radic. Biol. Med. 2007, 42, 371–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shenk, J.C.; Liu, J.; Fischbach, K.; Xu, K.; Puchowicz, M.; Obrenovich, M.E.; Gasimov, E.; Alvarez, L.M.; Ames, B.N.; Lamanna, J.C.; et al. The effect of acetyl-L-carnitine and R-α-lipoic acid treatment in ApoE4 mouse as a model of human Alzheimer’s disease. J. Neurol. Sci. 2009, 283, 199–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turunen, M.; Olsson, J.; Dallner, G. Metabolism and function of coenzyme Q. Biochim. Biophys. Acta Biomembr. 2004, 1660, 171–199. [Google Scholar] [CrossRef] [Green Version]
- Dallner, G.; Sindelar, P.J. Regulation of ubiquinone metabolism. Free Radic. Biol. Med. 2000, 29, 285–294. [Google Scholar] [CrossRef]
- Beyer, R.E. An analysis of the role of coenzyme Q in free radical generation and as an antioxidant. Biochem. Cell Biol. 1992, 70, 390–403. [Google Scholar] [CrossRef]
- Formigli, L.; Zecchi-Orlandini, S.; Orlandini, G.; Carella, G.; Brancato, R.; Papucci, L.; Schiavone, N.; Witort, E.; Donnini, M.; Lapucci, A.; et al. Coenzyme Q10 Prevents Apoptosis by Inhibiting Mitochondrial Depolarization Independently of Its Free Radical Scavenging Property. J. Biol. Chem. 2003, 278, 28220–28228. [Google Scholar] [CrossRef] [Green Version]
- Beal, M.F. Mitochondrial Dysfunction and Oxidative Damage in Alzheimer’s and Parkinson’s Diseases and Coenzyme Q10as a Potential Treatment. J. Bioenerg. Biomembr. 2004, 36, 381–386. [Google Scholar] [CrossRef]
- Moreira, P.I.; Zhu, X.; Wang, X.; Lee, H.-G.; Nunomura, A.; Petersen, R.B.; Perry, G.; Smith, M.A. Mitochondria: A therapeutic target in neurodegeneration. Biochim. Biophys. Acta Mol. Basis Dis. 2010, 1802, 212–220. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Yang, Y.; Li, G.; Wang, J.; Yang, E.S. Coenzyme Q10 Attenuates β-Amyloid Pathology in the Aged Transgenic Mice with Alzheimer Presenilin 1 Mutation. J. Mol. Neurosci. 2008, 34, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Young, M.L.; Franklin, J.L. The mitochondria-targeted antioxidant MitoQ inhibits memory loss, neuropathology, and extends lifespan in aged 3xTg-AD mice. Mol. Cell. Neurosci. 2019, 101, 103409. [Google Scholar] [CrossRef]
- James, A.M.; Sharpley, M.S.; Manas, A.-R.B.; Frerman, F.E.; Hirst, J.; Smith, R.A.J.; Murphy, M.P. Interaction of the Mitochondria-targeted Antioxidant MitoQ with Phospholipid Bilayers and Ubiquinone Oxidoreductases. J. Biol. Chem. 2007, 282, 14708–14718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, B.; Wang, X.; Bonda, D.; Perry, G.; Smith, M.; Zhu, X. Abnormal Mitochondrial Dynamics—A Novel Therapeutic Target for Alzheimer’s Disease? Mol. Neurobiol. 2010, 41, 87–96. [Google Scholar] [CrossRef] [Green Version]
- Dhanasekaran, A.; Kotamraju, S.; Kalivendi, S.V.; Matsunaga, T.; Shang, T.; Keszler, A.; Joseph, J.; Kalyanaraman, B. Supplementation of Endothelial Cells with Mitochondria-targeted Antioxidants Inhibit Peroxide-induced Mitochondrial Iron Uptake, Oxidative Damage, and Apoptosis. J. Biol. Chem. 2004, 279, 37575–37587. [Google Scholar] [CrossRef] [Green Version]
- McManus, M.J.; Murphy, M.P.; Franklin, J.L. The Mitochondria-Targeted Antioxidant MitoQ Prevents Loss of Spatial Memory Retention and Early Neuropathology in a Transgenic Mouse Model of Alzheimer’s Disease. J. Neurosci. 2011, 31, 15703–15715. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Prior, M.; Dargusch, R.; Roberts, A.; Riek, R.; Eichmann, C.; Chiruta, C.; Akaishi, T.; Abe, K.; Maher, P.; et al. A Novel Neurotrophic Drug for Cognitive Enhancement and Alzheimer’s Disease. PLoS ONE 2011, 6, e27865. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, J.; Currais, A.; Prior, M.; Fischer, W.; Chiruta, C.; Ratliff, E.; Daugherty, D.; Dargusch, R.; Finley, K.; Esparza-Moltó, P.B.; et al. The mitochondrial ATP synthase is a shared drug target for aging and dementia. Aging Cell 2018, 17, e12715. [Google Scholar] [CrossRef]
- Currais, A.; Goldberg, J.; Farrokhi, C.; Chang, M.; Prior, M.; Dargusch, R.; Daugherty, D.; Armando, A.; Quehenberger, O.; Maher, P.; et al. A comprehensive multiomics approach toward understanding the relationship between aging and dementia. Aging 2015, 7, 937–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prior, M.; Chiruta, C.; Currais, A.; Goldberg, J.; Ramsey, J.; Dargusch, R.; Maher, P.A.; Schubert, D. Back to the Future with Phenotypic Screening. ACS Chem. Neurosci. 2014, 5, 503–513. [Google Scholar] [CrossRef]
- Zhou, G.; Myers, R.; Li, Y.; Chen, Y.; Shen, X.; Fenyk-Melody, J.; Wu, M.; Ventre, J.; Doebber, T.; Fujii, N.; et al. Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Investig. 2001, 108, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.Q.; Folstein, M.F. Insulin, insulin-degrading enzyme and amyloid-β peptide in Alzheimer’s disease: Review and hypothesis. Neurobiol. Aging 2006, 27, 190–198. [Google Scholar] [CrossRef]
- Beisswenger, P.; Ruggiero-Lopez, D. Metformin inhibition of glycation processes. Diabetes Metab. 2003, 29, 6S95–6S103. [Google Scholar] [CrossRef]
- Cameron, A.R.; Morrison, V.L.; Levin, D.; Mohan, M.; Forteath, C.; Beall, C.; McNeilly, A.D.; Balfour, D.J.K.; Savinko, T.; Wong, A.K.F.; et al. Anti-Inflammatory Effects of Metformin Irrespective of Diabetes Status. Circ. Res. 2016, 119, 652–665. [Google Scholar] [CrossRef] [Green Version]
- Repiščák, P.; Erhardt, S.; Rena, G.; Paterson, M.J. Biomolecular Mode of Action of Metformin in Relation to Its Copper Binding Properties. Biochemistry 2014, 53, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Sanders, M.J.; Grondin, P.O.; Hegarty, B.D.; Snowden, M.A.; Carling, D. Investigating the mechanism for AMP activation of the AMP-activated protein kinase cascade. Biochem. J. 2007, 403, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gallagher, D.; DeVito, L.M.; Cancino, G.I.; Tsui, D.; He, L.; Keller, G.M.; Frankland, P.W.; Kaplan, D.R.; Miller, F.D. Metformin Activates an Atypical PKC-CBP Pathway to Promote Neurogenesis and Enhance Spatial Memory Formation. Cell Stem Cell 2012, 11, 23–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, F.; Dong, R.R.; Zhong, K.L.; Ghosh, A.; Tang, S.S.; Long, Y.; Hu, M.; Miao, M.X.; Liao, J.M.; Sun, H.B.; et al. Antidiabetic drugs restore abnormal transport of amyloid-β across the blood–brain barrier and memory impairment in db/db mice. Neuropharmacology 2016, 101, 123–136. [Google Scholar] [CrossRef]
- Chiang, M.-C.; Cheng, Y.-C.; Chen, S.-J.; Yen, C.-H.; Huang, R.-N. Metformin activation of AMPK-dependent pathways is neuroprotective in human neural stem cells against Amyloid-beta-induced mitochondrial dysfunction. Exp. Cell Res. 2016, 347, 322–331. [Google Scholar] [CrossRef] [PubMed]




| Protein Name | UniProtKB Code (Mus musculus) | Gene Name | Expression Level (Cu-Treatment Versus Control) | Reference | |
|---|---|---|---|---|---|
| Mice Cortex | 3xTg-AD Mice Hippocampus | ||||
| NADH dehydrogenase [ubiquinone] flavoprotein 1 (CI-51kD) | Q91YT0 | NDUFV1 | ⇑ | [33] | |
| Cytochrome b-c1 complex subunit 2 (CIII-s2) | Q9DB77 | UQCRC2 | ⇑ | [33] | |
| ATP synthase subunit d (ATPase-d) | Q9DCX2 | ATP5PD | ⇓ | ⇓ | [33,34] |
| 75 kDa glucose-regulated protein (GRP75) | P38647 | HSPA9 | ⇓ | [33] | |
| 78 kDa glucose-regulated protein (GRP78) | P20029 | HSPA5 | ⇓ | [33] | |
| NADH dehydrogenase [ubiquinone] 1 α subcomplex subunit 1 (CI-α1) | O35683 | NDUFA1 | ⇓ | [34] | |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 2 (CI-49kD) | Q91WD5 | NDUFS2 | ⇑ | [34] | |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 8 (CI-23kD) | Q8K3J1 | NDUFS8 | ⇓ | [34] | |
| Creatine kinase U-type (Mia-CK) | P30275 | CKMT1 | ⇑ | [34] | |
| ATP-Citrate synthase (ATP-CS) | Q91V92 | ACLY | ⇑ | [34] | |
| Malate dehydrogenase (MDH) | P08249 | MDH2 | ⇑ | [34] | |
| Pyruvate dehydrogenase E1 component subunit α (PDHE1-A1) | P35486 | PDHA1 | ⇓ | [34] | |
| Pyruvate dehydrogenase (acetyl-transferring) kinase isozyme 2 (PDKII) | Q9JK42 | PDK2 | ⇑ | [34] | |
| Cytochrome b-c1 complex subunit Rieske (CIII-RISP) | Q9CR68 | UQCRFS1 | ⇓ | [34] | |
| Cytochrome c oxidase subunit 5A (CIV-COX5A) | P12787 | COX5A | ⇓ | [34] | |
| Cytochrome c oxidase subunit 5B (CIV-COX5B) | P19536 | COX5B | ⇓ | [34] | |
| Voltage-dependent anion-selective channel protein 1 (VDAC1) | Q60932 | VDAC1 | ⇓ | [34] | |
| Voltage-dependent anion-selective channel protein 2 (VDAC2) | Q60930 | VDAC2 | ⇓ | [34] | |
| Protein | Oxidative Modification 1 | Effect on Protein Activity | Expression Level (AD Versus Control) | AD Stage | Brain Region | Reference |
|---|---|---|---|---|---|---|
| Aconitase | HNE | ⇓ | ⇔ | Late | Hippocampus | [94] |
| MDH | HNE | ⇑ | Not reported | Early | IPL | [95] |
| ATPase-α | HNE | ⇓ | Not reported | Early | IPL | [95] |
| HNE | ⇓ | ⇓ | Late | IPL | [94] | |
| 3-NT | Not reported | ⇑ | Late | Hippocampus | [96] | |
| MnSOD (or SOD2) | HNE | ⇓ | Not reported | Early | IPL | [95] |
| HNE | ⇔ | ⇑ | Late | IPL | [94] | |
| VDAC1 | 3-NT | Not reported | ⇔ | Late | Hippocampus | [96] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tassone, G.; Kola, A.; Valensin, D.; Pozzi, C. Dynamic Interplay between Copper Toxicity and Mitochondrial Dysfunction in Alzheimer’s Disease. Life 2021, 11, 386. https://doi.org/10.3390/life11050386
Tassone G, Kola A, Valensin D, Pozzi C. Dynamic Interplay between Copper Toxicity and Mitochondrial Dysfunction in Alzheimer’s Disease. Life. 2021; 11(5):386. https://doi.org/10.3390/life11050386
Chicago/Turabian StyleTassone, Giusy, Arian Kola, Daniela Valensin, and Cecilia Pozzi. 2021. "Dynamic Interplay between Copper Toxicity and Mitochondrial Dysfunction in Alzheimer’s Disease" Life 11, no. 5: 386. https://doi.org/10.3390/life11050386







