Identification of Cellular Compositions in Different Microenvironments and Their Potential Impacts on Hematopoietic Stem Cells HSCs Using Single-Cell RNA Sequencing with Systematical Confirmation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Laboratory Animals
- C57BL/6 female and male mice were purchased from Shanghai Southern Biological Model Center;
- Mouse embryos at 13.5d gestation were obtained by breeding C57BL/6 female and male mice.
2.2. Experimental Methods
2.2.1. Preparation of FL-MNCs
- C57BL/6 pregnant mice at 13.5 days of gestation were sacrificed via cervical dislocation. Thirty-three embryos were taken and transferred to the anatomical liquid, and the fetal liver was taken. The added sample diluent was blown repeatedly into cell suspension, and after cell counting, it was slowly added into the supernatant of mononuclear cell separation solution at a ratio of 1:1 and centrifuged for 30 min at 25 °C and 450 g;
- After centrifugation through a density gradient, all the mononuclear cells in the middle layer were separated, and PBS was added for washing 4 to 5 times. The cells were counted and centrifuged at 1000× g for 10 min at 4 °C to obtain fetal liver mononuclear cells. The main materials used in the experiments were centrifuge tubes (Corning, Wujiang, China), dishes (Corning, Wujiang, China), and filters (MiltenyiBiotec, Bergisch Gladbach Cologne, Germany); anatomical liquid (7% FBS in PBS); FBS (Thermo Fisher, Shanghai, China); sample diluent (PBS, Yuanpei, Shanghai, China) and the supernatant of the mononuclear cell separation (Stemcell, Technologies, Vancouver, Canada).
2.2.2. Preparation of BM-MNCs
- Ten 4-week-old C57BL/6 female mice were sacrificed via cervical dislocation. The femur was taken, and the bone marrow cells were taken into a 1 mL syringe and collected in a 15 mL centrifuge tube. After repeated blowing and homogenization, the mixture was filtered through a 30 μm filter screen into a new 15 mL centrifuge tube. Cells were counted, added at a 1:1 ratio to the supernatant of the mononuclear cell separation, and centrifuged for 30 min at 25 °C and 450 g;
- After centrifuging through a density gradient, all the mononuclear cells in the middle layer were absorbed, and PBS was added 4–5 times for washing. After cell counting, BM-MNCs were obtained by centrifuging at 4 °C and 1000× g for 10 min.
2.2.3. Culture and Collection of FL Stromal Cells
2.2.4. Treatment of Cultured Cells
2.2.5. 10x Genomics Single-cell Transcription Set Sequencing
2.2.6. Sequencing Data Conversion and Quality Control
2.2.7. Data Processing and Clustering Analysis
2.2.8. Differentially Expressed Gene Analysis
2.2.9. Gene Ontology (GO) Enrichment Analysis of Differentially Expressed Genes
2.2.10. Quantitative Real-Time PCR (qRT-PCR)
2.2.11. Confocal Image Analysis of Cell Surface Labeling
2.2.12. Statistical Analysis
3. Results
3.1. Cellular Composition Analysis
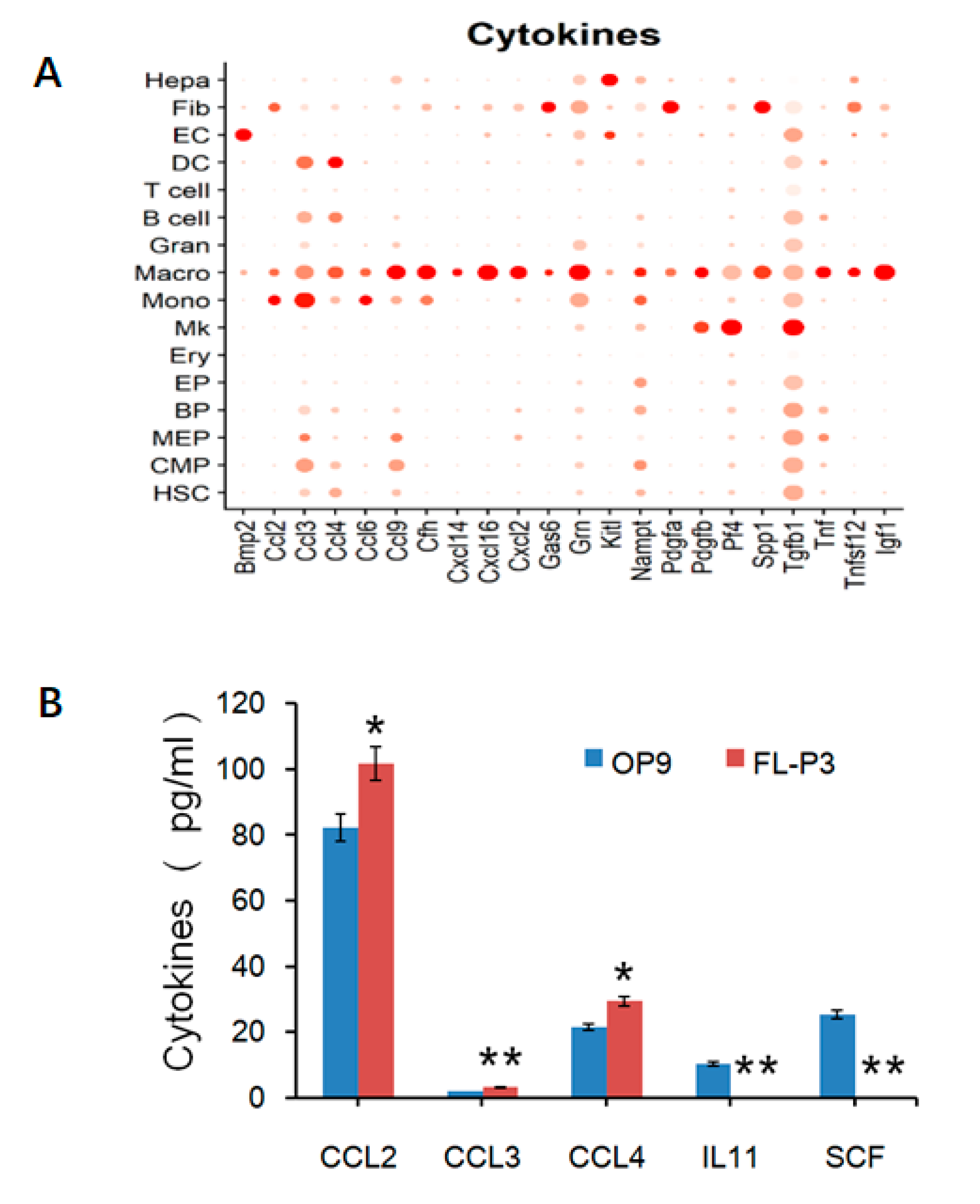
3.2. Differential Gene Expression Analysis
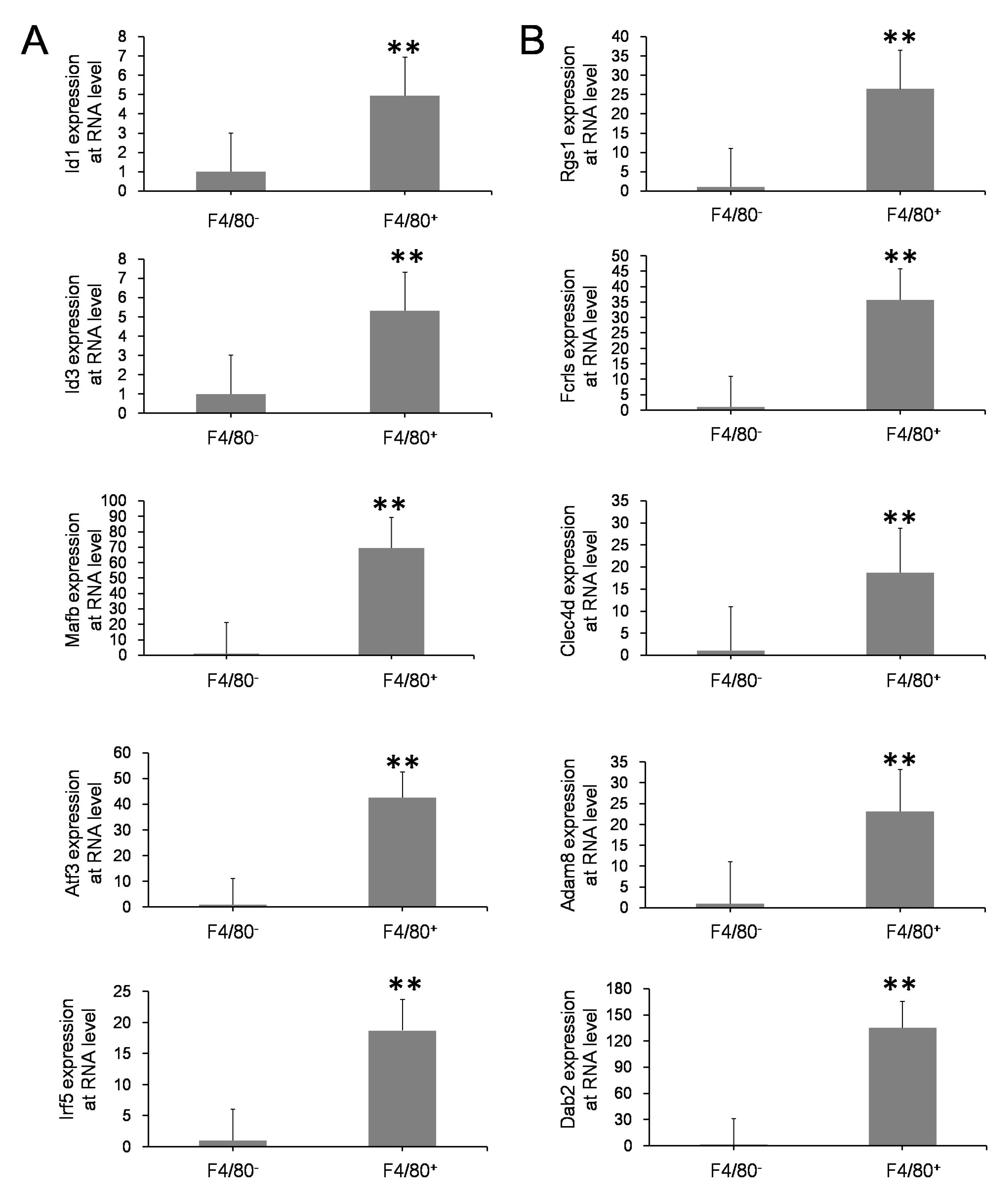
3.3. Validation of Esults with Various Methods
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seita, J.; Weissman, I.L. Hematopoietic stem cell: Self-renewal versus differentiation. Wiley Interdiscip Rev. Syst. Biol. Med. 2010, 2, 640–653. [Google Scholar] [CrossRef] [PubMed]
- Eaves, C.J. Hematopoietic stem cells: Concepts, definitions, and the new reality. Blood 2015, 125, 2605–2613. [Google Scholar] [CrossRef] [PubMed]
- Nazaraliyev, A.; Richard, E.; Sawai, C.M. In-vivo differentiation of adult hematopoietic stem cells from a single-cell point of view. Curr. Opin. Hematol. 2020, 27, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Chen, M.J.; Huang, W.Y.; Yan, J.B.; Xiao, Y.P.; Gong, Z.J.; Ren, Z.R.; Huang, S.Z. In utero transplantation of human hematopoetic stem cells into fetal goats under B-type ultrasonographic scan: An experimental model for the study of potential prenatal therapy. Eur. J. Obstet. Gynecol. Reprod. Biol. 2005, 118, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Sun, W.; Wang, J.; Ruan, H.; Zhang, X.; Ye, Y.; Shen, S.; Wang, C.; Lu, W.; Cheng, K.; et al. Conjugation of haematopoietic stem cells and platelets decorated with anti-PD-1 antibodies augments anti-leukaemia efficacy. Nat. Biomed Eng. 2018, 2, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Koschmieder, S.; Vetrie, D. Epigenetic dysregulation in chronic myeloid leukaemia: A myriad of mechanisms and therapeutic options. Semin. Cancer Biol. 2018, 51, 180–197. [Google Scholar] [CrossRef]
- Zhang, J.; Yan, J.; Zeng, F. Recent Progress on Genetic Diagnosis and Therapy for β-Thalassemia in China and Around the World. Hum. Gene Ther. 2018, 29, 197–203. [Google Scholar] [CrossRef]
- Palis, J.; Yoder, M.C. Yolk-sac hematopoiesis: The first blood cells of mouse and man. Exp. Hematol. 2001, 29, 927–936. [Google Scholar] [CrossRef]
- Yamane, T. Mouse Yolk Sac Hematopoiesis. Front. Cell Dev. Biol. 2018, 6, 80. [Google Scholar] [CrossRef]
- Ross, C.; Boroviak, T.E. Origin and function of the yolk sac in primate embryogenesis. Nat. Commun. 2020, 11, 3760. [Google Scholar] [CrossRef]
- Dzierzak, E.; Bigas, A. Blood Development: Hematopoietic Stem Cell Dependence and Independence. Cell Stem Cell 2018, 22, 639–651. [Google Scholar] [CrossRef]
- Taoudi, S.; Gonneau, C.; Moore, K.; Sheridan, J.M.; Blackburn, C.C.; Taylor, E.; Medvinsky, A. Extensive hematopoietic stem cell generation in the AGM region via maturation of VE-cadherin+CD45+ pre-definitive HSCs. Cell Stem Cell 2008, 3, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Rybtsov, S.A.; Lagarkova, M.A. Development of Hematopoietic Stem Cells in the Early Mammalian Embryo. Biochemistry 2019, 84, 190–204. [Google Scholar] [CrossRef]
- Hou, S.; Li, Z.; Zheng, X.; Gao, Y.; Dong, J.; Ni, Y.; Wang, X.; Li, Y.; Ding, X.; Chang, Z.; et al. Embryonic endothelial evolution towards first hematopoietic stem cells revealed by single-cell transcriptomic and functional analyses. Cell Res. 2020, 30, 376–392. [Google Scholar] [CrossRef] [PubMed]
- Ema, H.; Nakauchi, H. Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood 2000, 95, 2284–2288. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K.; Yoshimoto, M.; Takebe, T. Fetal liver hematopoiesis: From development to delivery. Stem Cell Res. Ther. 2021, 12, 139. [Google Scholar] [CrossRef]
- Ganuza, M.; Hall, T.; Myers, J.; Nevitt, C.; Sánchez-Lanzas, R.; Chabot, A.; Ding, J.; Kooienga, E.; Caprio, C.; Finkelstein, D.; et al. Murine foetal liver supports limited detectable expansion of life-long haematopoietic progenitors. Nat. Cell Biol. 2022, 24, 1475–1486. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, Y.S.; Li, H.; Kang, Y.L.; Chen, W.C.; Cheng, W.C.; Lai, D.M. Loss of Cxcl12/Sdf-1 in adult mice decreases the quiescent state of hematopoietic stem/progenitor cells and alters the pattern of hematopoietic regeneration after myelosuppression. Blood 2011, 117, 429–439. [Google Scholar] [CrossRef]
- Ya-nan, C.; Shu-yue, F.; Ji, M.; Xue, Y.; Zeng, F. Research progress on cell composition and function of hematopoietic microenvironment. Life Sci. 2020, 32, 325–334. [Google Scholar]
- Lyu, F.; Han, F.; Ge, C.; Mao, W.; Chen, L.; Hu, H.; Chen, G.; Lang, Q.; Fang, C. OmicStudio: A composable bioinformatics cloud platform with real-time feedback that can generate high-quality graphs for publication. iMeta 2023, 2, e85. [Google Scholar] [CrossRef]
- Popescu, D.M.; Botting, R.A.; Stephenson, E.; Green, K.; Webb, S.; Jardine, L.; Calderbank, E.F.; Polanski, K.; Goh, I.; Efremova, M.; et al. Decoding human fetal liver haematopoiesis. Nature 2019, 574, 365–371. [Google Scholar] [CrossRef]
- Mansoori, B.; Mohammadi, A.; Ditzel, H.J.; Duijf, P.H.G.; Khaze, V.; Gjerstorff, M.F.; Baradaran, B. HMGA2 as a Critical Regulator in Cancer Development. Genes 2021, 12, 269. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Kubota, S.; Iimori, M.; Hamashima, A.; Murakami, H.; Bai, J.; Morii, M.; Yokomizo-Nakano, T.; Osato, M.; Araki, K.; et al. The acidic domain of Hmga2 and the domain’s linker region are critical for driving self-renewal of hematopoietic stem cell. Int. J. Hematol. 2022, 115, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Karimi Dermani, F.; Gholamzadeh Khoei, S.; Afshar, S.; Amini, R. The potential role of nucleophosmin (NPM1) in the development of cancer. J. Cell. Physiol. 2021, 236, 7832–7852. [Google Scholar] [CrossRef]
- Jia, W.; Yao, Z.; Zhao, J.; Guan, Q.; Gao, L. New perspectives of physiological and pathological functions of nucleolin (NCL). Life Sci. 2017, 186, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Jakubison, B.; Keller, J.R. Protection of hematopoietic stem cells from stress-induced exhaustion and aging. Curr. Opin. Hematol. 2020, 27, 225–231. [Google Scholar] [CrossRef]
- Sarrazin, S.; Mossadegh-Keller, N.; Fukao, T.; Aziz, A.; Mourcin, F.; Vanhille, L.; Kelly Modis, L.; Kastner, P.; Chan, S.; Duprez, E.; et al. MafB restricts M-CSF-dependent myeloid commitment divisions of hematopoietic stem cells. Cell 2009, 138, 300–313. [Google Scholar] [CrossRef]
- Huang, X.; Ferris, S.T.; Kim, S.; Choudhary, M.N.K.; Belk, J.A.; Fan, C.; Qi, Y.; Sudan, R.; Xia, Y.; Desai, P.; et al. Differential usage of transcriptional repressor Zeb2 enhancers distinguishes adult and embryonic hematopoiesis. Immunity 2021, 54, 1417–1432.e1417. [Google Scholar] [CrossRef]
- Eto, K.; Leavitt, A.L.; Nakano, T.; Shattil, S.J. Development and analysis of megakaryocytes from murine embryonic stem cells. Methods Enzym. 2003, 365, 142–158. [Google Scholar]
- Gaur, M.; Kamata, T.; Wang, S.; Moran, B.; Shattil, S.J.; Leavitt, A.D. Megakaryocytes derived from human embryonic stem cells: A genetically tractable system to study megakaryocytopoiesis and integrin function. J. Thromb. Haemost. 2006, 4, 436–442. [Google Scholar] [CrossRef]
- Nishimura, T.; Hatoya, S.; Kanegi, R.; Sugiura, K.; Wijewardana, V.; Kuwamura, M.; Tanaka, M.; Yamate, J.; Izawa, T.; Takahashi, M.; et al. Generation of functional platelets from canine induced pluripotent stem cells. Stem Cells Dev. 2013, 22, 2026–2035. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Kodama, H.; Honjo, T. Generation of lymphohematopoietic cells from embryonic stem cells in culture. Science 1994, 265, 1098–1101. [Google Scholar] [CrossRef] [PubMed]
- Charan, M.; Das, S.; Mishra, S.; Chatterjee, N.; Varikuti, S.; Kaul, K.; Misri, S.; Ahirwar, D.K.; Satoskar, A.R.; Ganju, R.K. Macrophage migration inhibitory factor inhibition as a novel therapeutic approach against triple-negative breast cancer. Cell Death Dis. 2020, 11, 774. [Google Scholar] [CrossRef]
- Nobre, C.C.; de Araújo, J.M.; Fernandes, T.A.; Cobucci, R.N.; Lanza, D.C.; Andrade, V.S.; Fernandes, J.V. Macrophage Migration Inhibitory Factor (MIF): Biological Activities and Relation with Cancer. Pathol. Oncol. Res. 2017, 23, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Toubai, T.; Shono, Y.; Nishihira, J.; Ibata, M.; Suigita, J.; Kato, N.; Ohkawara, T.; Tone, S.; Lowler, K.P.; Ota, S.; et al. Serum macrophage migration inhibitory factor (MIF) levels after allogeneic hematopoietic stem cell transplantation. Int. J. Lab. Hematol. 2009, 31, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Becker-Herman, S.; Rozenberg, M.; Hillel-Karniel, C.; Gil-Yarom, N.; Kramer, M.P.; Barak, A.; Sever, L.; David, K.; Radomir, L.; Lewinsky, H.; et al. CD74 is a regulator of hematopoietic stem cell maintenance. PLoS Biol. 2021, 19, e3001121. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Shi, Q.; Zhang, Y.; Liang, G.; Kang, Z.; Huang, B.; Ma, D.; Wang, L.; Jiao, J.; Fang, X.; et al. Identification of HSC/MPP expansion units in fetal liver by single-cell spatiotemporal transcriptomics. Cell Res. 2022, 32, 38–53. [Google Scholar] [CrossRef]
- Wattrus, S.J.; Smith, M.L.; Rodrigues, C.P.; Hagedorn, E.J.; Kim, J.W.; Budnik, B.; Zon, L.I. Quality assurance of hematopoietic stem cells by macrophages determines stem cell clonality. Science 2022, 377, 1413–1419. [Google Scholar] [CrossRef]
- Winkler, I.G.; Sims, N.A.; Pettit, A.R.; Barbier, V.; Nowlan, B.; Helwani, F.; Poulton, I.J.; van Rooijen, N.; Alexander, K.A.; Raggatt, L.J.; et al. Bone marrow macrophages maintain hematopoietic stem cell (HSC) niches and their depletion mobilizes HSCs. Blood 2010, 116, 4815–4828. [Google Scholar] [CrossRef]
- Mariani, S.A.; Li, Z.; Rice, S.; Krieg, C.; Fragkogianni, S.; Robinson, M.; Vink, C.S.; Pollard, J.W.; Dzierzak, E. Pro-inflammatory Aorta-Associated Macrophages Are Involved in Embryonic Development of Hematopoietic Stem Cells. Immunity 2019, 50, 1439–1452.e1435. [Google Scholar] [CrossRef]
- Travnickova, J.; Tran Chau, V.; Julien, E.; Mateos-Langerak, J.; Gonzalez, C.; Lelièvre, E.; Lutfalla, G.; Tavian, M.; Kissa, K. Primitive macrophages control HSPC mobilization and definitive haematopoiesis. Nat. Commun. 2015, 6, 6227. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Xue, W.; Li, M.; Dong, M.; Wang, J.; Wang, X.; Li, X.; Chen, K.; Zhang, W.; Wu, S.; et al. VCAM-1(+) macrophages guide the homing of HSPCs to a vascular niche. Nature 2018, 564, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Cossarizza, A.; Chang, H.D.; Radbruch, A.; Acs, A.; Adam, D.; Adam-Klages, S.; Agace, W.W.; Aghaeepour, N.; Akdis, M.; Allez, M.; et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies (second edition). Eur. J. Immunol. 2019, 49, 1457–1973. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Shao, L.; Chang, J.; Feng, W.; Liu, Y.L.; Cottler-Fox, M.H.; Emanuel, P.D.; Hauer-Jensen, M.; Bernstein, I.D.; Liu, L.; et al. M1 and M2 macrophages differentially regulate hematopoietic stem cell self-renewal and ex vivo expansion. Blood Adv. 2018, 2, 859–870. [Google Scholar] [CrossRef]
- Badham, H.J.; Winn, L.M. In utero exposure to benzene disrupts fetal hematopoietic progenitor cell growth via reactive oxygen species. Toxicol. Sci. 2010, 113, 207–215. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chi, Y.; Yang, G.; Guo, C.; Zhang, S.; Hong, L.; Tang, H.; Sang, X.; Wang, J.; Ma, J.; Xue, Y.; et al. Identification of Cellular Compositions in Different Microenvironments and Their Potential Impacts on Hematopoietic Stem Cells HSCs Using Single-Cell RNA Sequencing with Systematical Confirmation. Life 2023, 13, 2157. https://doi.org/10.3390/life13112157
Chi Y, Yang G, Guo C, Zhang S, Hong L, Tang H, Sang X, Wang J, Ma J, Xue Y, et al. Identification of Cellular Compositions in Different Microenvironments and Their Potential Impacts on Hematopoietic Stem Cells HSCs Using Single-Cell RNA Sequencing with Systematical Confirmation. Life. 2023; 13(11):2157. https://doi.org/10.3390/life13112157
Chicago/Turabian StyleChi, Yanan, Guanheng Yang, Chuanliang Guo, Shaoqing Zhang, Lei Hong, Huixiang Tang, Xiao Sang, Jie Wang, Ji Ma, Yan Xue, and et al. 2023. "Identification of Cellular Compositions in Different Microenvironments and Their Potential Impacts on Hematopoietic Stem Cells HSCs Using Single-Cell RNA Sequencing with Systematical Confirmation" Life 13, no. 11: 2157. https://doi.org/10.3390/life13112157




