Current Nuclear Engineering Strategies in the Green Microalga Chlamydomonas reinhardtii
Abstract
:1. Introduction
2. Strain Domestication for Efficient Nuclear Transformation
3. Promoters and Terminators
4. Optimization of Transgene Sequences
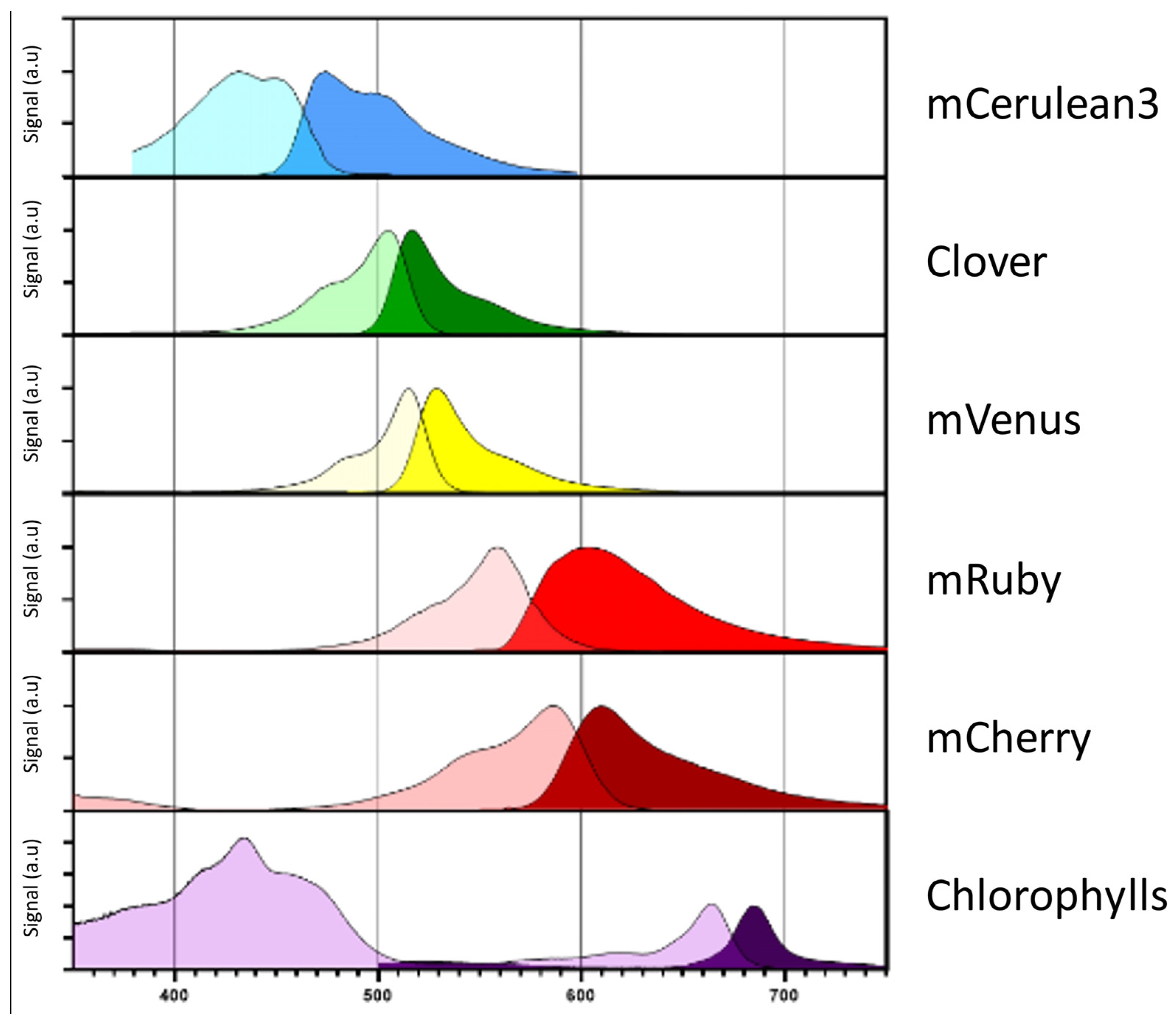
5. Selection Markers and Reporters for Gene Expression
| Type | Gene | Screening Mechanism | References |
|---|---|---|---|
| Auto | ARG7 | Growth in arginine-free medium | [72] |
| Auto | NIT1 | Growth in ammonium-free medium | [14] |
| Auto | SPD1 | Growth in spermidine-free medium | [21] |
| AB | aphVII | Resistance to hygromycin B | [76] |
| AB | aphVIII | Resistance to paromomycin, neomycin and kanamycin | [74,75] |
| AB | Shble | Resistance to bleomycin and sapromycin | [77,78] |
| AB | aadA | Resistance to spectinomycin and streptomycin | [79,80] |
| AB | NptII | Resistance to paromomycin, neomycin and kanamycin | [81] |
| AB | TetX | Resistance to tetracycline | [82] |
| AB | NAT | Resistance to nourseothricin | [83] |
| AB | CRY-1 | Resistance to cryptopleurine and emetine | [84] |
| AB | BSR | Resistance to blasticidin S | [85] |
| Herb | GAT | Resistance to glyphosate | [86] |
| Herb | PDS (R268T) | Resistance to norflurazon | [86] |
| Herb | protox rs-3 | Resistance to oxyfluorfen | [86] |
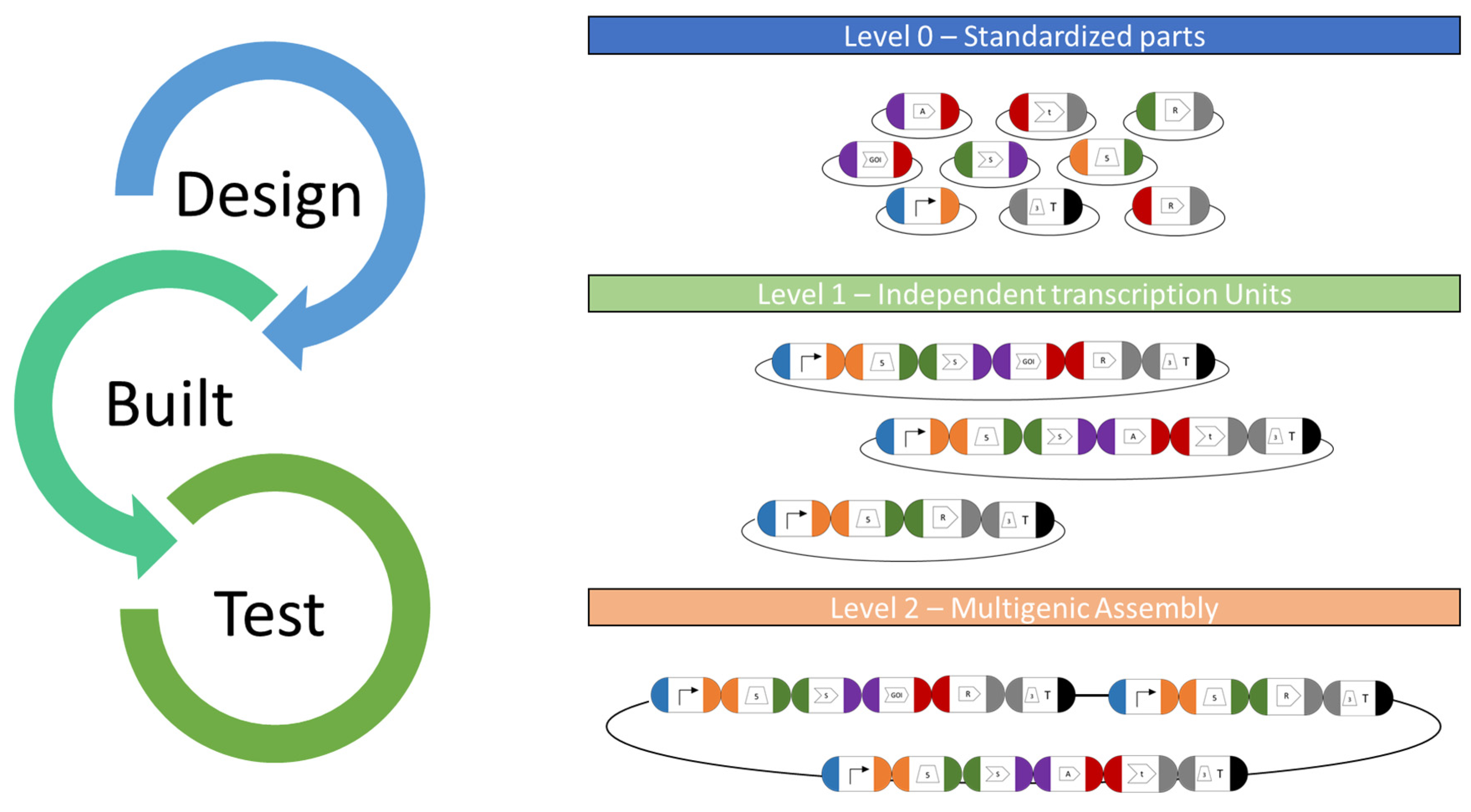
6. Vector System
7. Biotechnological Application
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mitra, D.; van Leeuwen, J.; Lamsal, B. Heterotrophic/mixotrophic cultivation of oleaginous Chlorella vulgaris on industrial co-products. Algal Res. 2012, 1, 40–48. [Google Scholar] [CrossRef]
- Bhola, V.; Swalaha, F.; Ranjith Kumar, R.; Singh, M.; Bux, F. Overview of the potential of microalgae for CO2 sequestration. Int. J. Environ. Sci. Technol. 2014, 11, 2103–2118. [Google Scholar] [CrossRef] [Green Version]
- Loera-Quezada, M.M.; Leyva-Gonzalez, M.A.; Velazquez-Juarez, G.; Sanchez-Calderon, L.; Do Nascimento, M.; Lopez-Arredondo, D.; Herrera-Estrella, L. A novel genetic engineering platform for the effective management of biological contaminants for the production of microalgae. Plant Biotechnol. J. 2016, 14, 2066–2076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, R.E.; Purton, S. Cytosine deaminase as a negative selectable marker for the microalgal chloroplast: A strategy for the isolation of nuclear mutations that affect chloroplast gene expression. Plant J. 2014, 80, 915–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franklin, S.E.; Mayfield, S.P. Prospects for molecular farming in the green alga Chlamydomonas. Curr. Opin. Plant Biol. 2004, 7, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Neupert, J.; Karcher, D.; Bock, R. Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. 2009, 57, 1140–1150. [Google Scholar] [CrossRef]
- Dementyeva, P.; Freudenberg, R.A.; Baier, T.; Rojek, K.; Wobbe, L.; Weisshaar, B.; Kruse, O. A novel, robust and mating-competent Chlamydomonas reinhardtii strain with an enhanced transgene expression capacity for algal biotechnology. Biotechnol. Rep. 2021, 31, e00644. [Google Scholar] [CrossRef]
- Abdallah, M.N.; Wellman, G.B.; Overmans, S.; Lauersen, K.J. Combinatorial Engineering Enables Photoautotrophic Growth in High Cell Density Phosphite-Buffered Media to Support Engineered Chlamydomonas reinhardtii Bio-Production Concepts. Front. Microbiol. 2022, 13, 1337. [Google Scholar] [CrossRef]
- Crozet, P.; Navarro, F.J.; Willmund, F.; Mehrshahi, P.; Bakowski, K.; Lauersen, K.J.; Pérez-Pérez, M.-E.; Auroy, P.; Gorchs Rovira, A.; Sauret-Gueto, S.; et al. Birth of a Photosynthetic Chassis: A MoClo Toolkit Enabling Synthetic Biology in the Microalga Chlamydomonas reinhardtii. ACS Synth. Biol. 2018, 7, 2074–2086. [Google Scholar] [CrossRef] [Green Version]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Marechal-Drouard, L.; et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef] [Green Version]
- Gallaher, S.D.; Fitz-Gibbon, S.T.; Strenkert, D.; Purvine, S.O.; Pellegrini, M.; Merchant, S.S. High-throughput sequencing of the chloroplast and mitochondrion of Chlamydomonas reinhardtii to generate improved de novo assemblies, analyze expression patterns and transcript speciation, and evaluate diversity among laboratory strains and wild isolates. Plant J. 2018, 93, 545–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Craig, R.J.; Gallaher, S.D.; Shu, S.; Salomé, P.A.; Jenkins, J.W.; Blaby-Haas, C.E.; Purvine, S.O.; O’Donnell, S.; Barry, K.; Grimwood, J.; et al. The Chlamydomonas Genome Project, version 6: Reference assemblies for mating-type plus and minus strains reveal extensive structural mutation in the laboratory. Plant Cell 2023, 35, 644–672. [Google Scholar] [CrossRef] [PubMed]
- Kindle, K.L. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 1990, 87, 1228–1232. [Google Scholar] [CrossRef]
- Kindle, K.L.; Schnell, R.A.; Fernández, E.; Lefebvre, P.A. Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J. Cell Biol. 1989, 109, 2589–2601. [Google Scholar] [CrossRef] [PubMed]
- El-Sheekh, M.M.; Almutairi, A.W.; Touliabah, H.E. Construction of a novel vector for the nuclear transformation of the unicellular green alga Chlamydomonas reinhardtii and its stable expression. J. Taibah Univ. Sci. 2019, 13, 529–535. [Google Scholar] [CrossRef]
- Shimogawara, K.; Fujiwara, S.; Grossman, A.; Usuda, H. High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 1998, 148, 1821–1828. [Google Scholar] [CrossRef]
- Kumar, S.V.; Misquitta, R.W.; Reddy, V.S.; Rao, B.J.; Rajam, M.V. Genetic transformation of the green alga—Chlamydomonas reinhardtii by Agrobacterium tumefaciens. Plant Sci. 2004, 166, 731–738. [Google Scholar] [CrossRef]
- Sodeinde, O.A.; Kindle, K.L. Homologous recombination in the nuclear genome of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 1993, 90, 9199–9203. [Google Scholar] [CrossRef]
- Angstenberger, M.; de Signori, F.; Vecchi, V.; Dall’Osto, L.; Bassi, R. Cell Synchronization Enhances Nuclear Transformation and Genome Editing via Cas9 Enabling Homologous Recombination in Chlamydomonas reinhardtii. ACS Synth. Biol. 2020, 9, 2840–2850. [Google Scholar] [CrossRef]
- Greiner, A.; Kelterborn, S.; Evers, H.; Kreimer, G.; Sizova, I.; Hegemann, P. Targeting of Photoreceptor Genes in Chlamydomonas reinhardtii via Zinc-Finger Nucleases and CRISPR/Cas9. Plant Cell 2017, 29, 2498–2518. [Google Scholar] [CrossRef] [Green Version]
- Freudenberg, R.A.; Wittemeier, L.; Einhaus, A.; Baier, T.; Kruse, O. The Spermidine Synthase Gene SPD1: A Novel Auxotrophic Marker for Chlamydomonas reinhardtii Designed by Enhanced CRISPR/Cas9 Gene Editing. Cells 2022, 11, 837. [Google Scholar] [CrossRef] [PubMed]
- Casas-Mollano, J.A.; Jeong, B.-r.; Xu, J.; Moriyama, H.; Cerutti, H. The MUT9p kinase phosphorylates histone H3 threonine 3 and is necessary for heritable epigenetic silencing in Chlamydomonas. Proc. Natl. Acad. Sci. USA 2008, 105, 6486–6491. [Google Scholar] [CrossRef] [PubMed]
- Shaver, S.; Casas-Mollano, J.A.; Cerny, R.L.; Cerutti, H. Origin of the polycomb repressive complex 2 and gene silencing by an E(z) homolog in the unicellular alga Chlamydomonas. Epigenetics 2010, 5, 301–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neupert, J.; Gallaher, S.D.; Lu, Y.; Strenkert, D.; Segal, N.a.; Barahimipour, R.; Fitz-Gibbon, S.T.; Schroda, M.; Merchant, S.S.; Bock, R. An epigenetic gene silencing pathway selectively acting on transgenic DNA in the green alga Chlamydomonas. Nat. Commun. 2020, 11, 6269. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Yamasaki, T.; Kurniasih, S.D.; Hou, L.; Li, X.; Ivanova, N.; Okada, S.; Ohama, T. Robust expression of heterologous genes by selection marker fusion system in improved Chlamydomonas strains. J. Biosci. Bioeng. 2015, 120, 239–245. [Google Scholar] [CrossRef] [Green Version]
- Ruecker, O.; Zillner, K.; Groebner-Ferreira, R.; Heitzer, M. Gaussia-luciferase as a sensitive reporter gene for monitoring promoter activity in the nucleus of the green alga Chlamydomonas reinhardtii. Mol. Genet. Genom. 2008, 280, 153–162. [Google Scholar] [CrossRef]
- Butanaev, A.M. [Use of the hygromycin phosphotransferase gene as the dominant selective marker for Chlamydomonas reinhardtii transformation]. Mol. Biol. 1994, 28, 1061–1068. [Google Scholar]
- El-Ayouty, Y.; El-Manawy, I.; Nasih, S.; Hamdy, E.; Kebeish, R. Engineering Chlamydomonas reinhardtii for Expression of Functionally Active Human Interferon-α. Mol. Biotechnol. 2019, 61, 134–144. [Google Scholar] [CrossRef]
- Díaz-Santos, E.; de la Vega, M.; Vila, M.; Vigara, J.; León, R. Efficiency of different heterologous promoters in the unicellular microalga Chlamydomonas reinhardtii. Biotechnol. Prog. 2013, 29, 319–328. [Google Scholar] [CrossRef]
- Walker, T.L.; Becker, D.K.; Collet, C. Characterisation of the Dunaliella tertiolecta RbcS genes and their promoter activity in Chlamydomonas reinhardtii. Plant Cell Rep. 2005, 23, 727–735. [Google Scholar] [CrossRef] [Green Version]
- Schmollinger, S.; Mühlhaus, T.; Boyle, N.R.; Blaby, I.K.; Casero, D.; Mettler, T.; Moseley, J.L.; Kropat, J.; Sommer, F.; Strenkert, D.; et al. Nitrogen-Sparing Mechanisms in Chlamydomonas Affect the Transcriptome, the Proteome, and Photosynthetic Metabolism. Plant Cell 2014, 26, 1410–1435. [Google Scholar] [CrossRef] [Green Version]
- Kozminski, K.G.; Diener, D.R.; Rosenbaum, J.L. High level expression of nonacetylatable alpha-tubulin in Chlamydomonas reinhardtii. Cell Motil. Cytoskelet. 1993, 25, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Schroda, M.; Blocker, D.; Beck, C.F. The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 2000, 21, 121–131. [Google Scholar] [CrossRef]
- Schroda, M.; Beck, C.F.; Vallon, O. Sequence elements within an HSP70 promoter counteract transcriptional transgene silencing in Chlamydomonas. Plant J. 2002, 31, 445–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Hu, Z.; Wang, C.; Li, S.; Lei, A. Efficient expression of green fluorescent protein (GFP) mediated by a chimeric promoter in Chlamydomonas reinhardtii. Chin. J. Oceanol. Limnol. 2008, 26, 242–247. [Google Scholar] [CrossRef]
- Amendola, S.; Kneip, J.S.; Meyer, F.; Perozeni, F.; Cazzaniga, S.; Lauersen, K.J.; Ballottari, M.; Baier, T. Metabolic Engineering for Efficient Ketocarotenoid Accumulation in the Green Microalga Chlamydomonas reinhardtii. ACS Synth. Biol. 2023, 12, 820–831. [Google Scholar] [CrossRef]
- Perozeni, F.; Cazzaniga, S.; Baier, T.; Zanoni, F.; Zoccatelli, G.; Lauersen, K.J.; Wobbe, L.; Ballottari, M. Turning a green alga red: Engineering astaxanthin biosynthesis by intragenic pseudogene revival in Chlamydomonas reinhardtii. Plant Biotechnol. J. 2020, 18, 2053–2067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pivato, M.; Perozeni, F.; Licausi, F.; Cazzaniga, S.; Ballottari, M. Heterologous expression of cyanobacterial Orange Carotenoid Protein (OCP2) as a soluble carrier of ketocarotenoids in Chlamydomonas reinhardtii. Algal Res. 2021, 55, 102255. [Google Scholar] [CrossRef]
- Perozeni, F.; Stella, G.R.; Ballottari, M. LHCSR Expression under HSP70/RBCS2 Promoter as a Strategy to Increase Productivity in Microalgae. Int. J. Mol. Sci. 2018, 19, 155. [Google Scholar] [CrossRef] [Green Version]
- Baier, T.; Kros, D.; Feiner, R.C.; Lauersen, K.J.; Muller, K.M.; Kruse, O. Engineered Fusion Proteins for Efficient Protein Secretion and Purification of a Human Growth Factor from the Green Microalga Chlamydomonas reinhardtii. ACS Synth. Biol. 2018, 7, 2547–2557. [Google Scholar] [CrossRef]
- Baier, T.; Wichmann, J.; Kruse, O.; Lauersen, K.J. Intron-containing algal transgenes mediate efficient recombinant gene expression in the green microalga Chlamydomonas reinhardtii. Nucleic Acids Res. 2018, 46, 6909–6919. [Google Scholar] [CrossRef] [PubMed]
- Einhaus, A.; Steube, J.; Freudenberg, R.A.; Barczyk, J.; Baier, T.; Kruse, O. Engineering a powerful green cell factory for robust photoautotrophic diterpenoid production. Metab. Eng. 2022, 73, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Lauersen, K.J.; Baier, T.; Wichmann, J.; Wordenweber, R.; Mussgnug, J.H.; Hubner, W.; Huser, T.; Kruse, O. Efficient phototrophic production of a high-value sesquiterpenoid from the eukaryotic microalga Chlamydomonas reinhardtii. Metab. Eng. 2016, 38, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Lauersen, K.J.; Wichmann, J.; Baier, T.; Kampranis, S.C.; Pateraki, I.; Moller, B.L.; Kruse, O. Phototrophic production of heterologous diterpenoids and a hydroxy-functionalized derivative from Chlamydomonas reinhardtii. Metab. Eng. 2018, 49, 116–127. [Google Scholar] [CrossRef]
- Einhaus, A.; Baier, T.; Rosenstengel, M.; Freudenberg, R.A.; Kruse, O. Rational Promoter Engineering Enables Robust Terpene Production in Microalgae. ACS Synth. Biol. 2021, 10, 847–856. [Google Scholar] [CrossRef]
- Fischer, N.; Rochaix, J.D. The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol. Genet. Genom. 2001, 265, 888–894. [Google Scholar] [CrossRef]
- Weiner, I.; Atar, S.; Schweitzer, S.; Eilenberg, H.; Feldman, Y.; Avitan, M.; Blau, M.; Danon, A.; Tuller, T.; Yacoby, I. Enhancing heterologous expression in Chlamydomonas reinhardtii by transcript sequence optimization. Plant J. 2018, 94, 22–31. [Google Scholar] [CrossRef] [Green Version]
- Barjona do Nascimento Coutinho, P.; Friedl, C.; Buchholz, R.; Stute, S.C. Chemical regulation of Fea1 driven transgene expression in Chlamydomonas reinhardtii. Algal Res. 2017, 26, 323–329. [Google Scholar] [CrossRef]
- Ohresser, M.; Matagne, R.F.; Loppes, R. Expression of the arylsulphatase reporter gene under the control of the nit1 promoter in Chlamydomonas reinhardtii. Curr. Genet. 1997, 31, 264–271. [Google Scholar] [CrossRef]
- Lee, S.; Lee, Y.J.; Choi, S.; Park, S.-B.; Tran, Q.-G.; Heo, J.; Kim, H.-S. Development of an alcohol-inducible gene expression system for recombinant protein expression in Chlamydomonas reinhardtii. J. Appl. Phycol. 2018, 30, 2297–2304. [Google Scholar] [CrossRef] [Green Version]
- Beltran-Aguilar, A.G.; Peraza-Echeverria, S.; López-Ochoa, L.A.; Borges-Argáez, I.C.; Herrera-Valencia, V.A. A novel salt-inducible CrGPDH3 promoter of the microalga Chlamydomonas reinhardtii for transgene overexpression. Appl. Microbiol. Biotechnol. 2019, 103, 3487–3499. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.M.; Merchant, S. Two copper-responsive elements associated with the Chlamydomonas Cyc6 gene function as targets for transcriptional activators. Plant Cell 1995, 7, 623–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehrshahi, P.; Nguyen, G.T.D.T.; Gorchs Rovira, A.; Sayer, A.; Llavero-Pasquina, M.; Lim Huei Sin, M.; Medcalf, E.J.; Mendoza-Ochoa, G.I.; Scaife, M.A.; Smith, A.G. Development of Novel Riboswitches for Synthetic Biology in the Green Alga Chlamydomonas. ACS Synth. Biol. 2020, 9, 1406–1417. [Google Scholar] [CrossRef] [PubMed]
- Geisler, K.; Scaife, M.A.; Mordaka, P.M.; Holzer, A.; Tomsett, E.V.; Mehrshahi, P.; Mendoza Ochoa, G.I.; Smith, A.G. Exploring the Impact of Terminators on Transgene Expression in Chlamydomonas reinhardtii with a Synthetic Biology Approach. Life 2021, 11, 964. [Google Scholar] [CrossRef] [PubMed]
- López-Paz, C.; Liu, D.; Geng, S.; Umen, J.G. Identification of Chlamydomonas reinhardtii endogenous genic flanking sequences for improved transgene expression. Plant J. 2017, 92, 1232–1244. [Google Scholar] [CrossRef] [Green Version]
- Naya, H.; Romero, H.; Carels, N.; Zavala, A.; Musto, H. Translational selection shapes codon usage in the GC-rich genome of Chlamydomonas reinhardtii. FEBS Lett. 2001, 501, 127–130. [Google Scholar] [CrossRef] [Green Version]
- Barahimipour, R.; Strenkert, D.; Neupert, J.; Schroda, M.; Merchant, S.S.; Bock, R. Dissecting the contributions of GC content and codon usage to gene expression in the model alga Chlamydomonas reinhardtii. Plant J. 2015, 84, 704–717. [Google Scholar] [CrossRef] [Green Version]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- Rose, A.B. Introns as Gene Regulators: A Brick on the Accelerator. Front. Genet. 2018, 9, 672. [Google Scholar] [CrossRef] [Green Version]
- Laxa, M. Intron-Mediated Enhancement: A Tool for Heterologous Gene Expression in Plants? Front. Plant Sci. 2016, 7, 1977. [Google Scholar] [CrossRef] [Green Version]
- Shaul, O. How introns enhance gene expression. Int. J. Biochem. Cell Biol. 2017, 91, 145–155. [Google Scholar] [CrossRef]
- Ott, C.J.; Suszko, M.; Blackledge, N.P.; Wright, J.E.; Crawford, G.E.; Harris, A. A complex intronic enhancer regulates expression of the CFTR gene by direct interaction with the promoter. J. Cell Mol. Med. 2009, 13, 680–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, C.L.; Wu, Q.; Vega, V.B.; Chiu, K.P.; Ng, P.; Zhang, T.; Shahab, A.; Yong, H.C.; Fu, Y.; Weng, Z.; et al. A global map of p53 transcription-factor binding sites in the human genome. Cell 2006, 124, 207–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morello, L.; Bardini, M.; Sala, F.; Breviario, D. A long leader intron of the Ostub16 rice beta-tubulin gene is required for high-level gene expression and can autonomously promote transcription both in vivo and in vitro. Plant J. 2002, 29, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Gallegos, J.E.; Rose, A.B. Intron DNA Sequences Can Be More Important Than the Proximal Promoter in Determining the Site of Transcript Initiation. Plant Cell 2017, 29, 843–853. [Google Scholar] [CrossRef] [Green Version]
- Le Hir, H.; Gatfield, D.; Izaurralde, E.; Moore, M.J. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001, 20, 4987–4997. [Google Scholar] [CrossRef] [Green Version]
- Baier, T.; Jacobebbinghaus, N.; Einhaus, A.; Lauersen, K.J.; Kruse, O. Introns mediate post-transcriptional enhancement of nuclear gene expression in the green microalga Chlamydomonas reinhardtii. PLoS Genet. 2020, 16, e1008944. [Google Scholar] [CrossRef]
- Eichler-Stahlberg, A.; Weisheit, W.; Ruecker, O.; Heitzer, M. Strategies to facilitate transgene expression in Chlamydomonas reinhardtii. Planta 2009, 229, 873–883. [Google Scholar] [CrossRef]
- Lumbreras, V.; Stevens, D.R.; Purton, S. Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 1998, 14, 441–447. [Google Scholar] [CrossRef]
- Wichmann, J.; Baier, T.; Wentnagel, E.; Lauersen, K.J.; Kruse, O. Tailored carbon partitioning for phototrophic production of (E)-alpha-bisabolene from the green microalga Chlamydomonas reinhardtii. Metab. Eng. 2018, 45, 211–222. [Google Scholar] [CrossRef]
- Jaeger, D.; Baier, T.; Lauersen, K.J. Intronserter, an advanced online tool for design of intron containing transgenes. Algal Res. 2019, 42, 101588. [Google Scholar] [CrossRef]
- Debuchy, R.; Purton, S.; Rochaix, J.D. The argininosuccinate lyase gene of Chlamydomonas reinhardtii: An important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 1989, 8, 2803–2809. [Google Scholar] [CrossRef] [PubMed]
- Remacle, C.; Cline, S.; Boutaffala, L.; Gabilly, S.; Larosa, V.; Barbieri, M.R.; Coosemans, N.; Hamel, P.P. The ARG9 gene encodes the plastid-resident N-acetyl ornithine aminotransferase in the green alga Chlamydomonas reinhardtii. Eukaryot. Cell 2009, 8, 1460–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sizova, I.; Greiner, A.; Awasthi, M.; Kateriya, S.; Hegemann, P. Nuclear gene targeting in Chlamydomonas using engineered zinc-finger nucleases. Plant J. 2013, 73, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Wright, G.D. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol. 1997, 5, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Berthold, P.; Schmitt, R.; Mages, W. An Engineered Streptomyces hygroscopicus aph 7″ Gene Mediates Dominant Resistance against Hygromycin B in Chlamydomonas reinhardtii. Protist 2002, 153, 401–412. [Google Scholar] [CrossRef]
- Stevens, D.R.; Purton, S.; Rochaix, J.D. The bacterial phleomycin resistance geneble as a dominant selectable marker inChlamydomonas. Mol. Gen. Genet. MGG 1996, 251, 23–30. [Google Scholar] [CrossRef] [Green Version]
- Chang, M.; Li, F.; Odom, O.W.; Lee, J.; Herrin, D.L. A cosmid vector containing a dominant selectable marker for cloning Chlamydomonas genes by complementation. Plasmid 2003, 49, 75–78. [Google Scholar] [CrossRef]
- Goldschmidt-Clermont, M. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: A selectable marker for site-directed transformation of chlamydomonas. Nucleic Acids Res. 1991, 19, 4083–4089. [Google Scholar] [CrossRef] [Green Version]
- Meslet-Cladière, L.; Vallon, O. Novel Shuttle Markers for Nuclear Transformation of the Green Alga Chlamydomonas reinhardtii. Eukaryot. Cell 2011, 10, 1670–1678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bingham, S.E.; Cox, J.C.; Strem, M.D. Expression of foreign DNA in Chlamydomonas reinhardtii. FEMS Microbiol. Lett. 1989, 65, 77–81. [Google Scholar] [CrossRef]
- Garcia-Echauri, S.A.; Cardineau, G.A. TETX: A novel nuclear selection marker for Chlamydomonas reinhardtii transformation. Plant Methods 2015, 11, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Peng, J.; Pan, J. Nourseothricin N-acetyl transferase (NAT), a new selectable marker for nuclear gene expression in Chlamydomonas. Plant Methods 2019, 15, 140. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.A.; Savereide, P.B.; Lefebvre, P.A. The CRY1 gene in Chlamydomonas reinhardtii: Structure and use as a dominant selectable marker for nuclear transformation. Mol. Cell. Biol. 1994, 14, 4011–4019. [Google Scholar] [CrossRef]
- de Carpentier, F.; Le Peillet, J.; Boisset, N.D.; Crozet, P.; Lemaire, S.D.; Danon, A. Blasticidin S Deaminase: A New Efficient Selectable Marker for Chlamydomonas reinhardtii. Front. Plant Sci. 2020, 11, 242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruggeman, A.J.; Kuehler, D.; Weeks, D.P. Evaluation of three herbicide resistance genes for use in genetic transformations and for potential crop protection in algae production. Plant Biotechnol. J. 2014, 12, 894–902. [Google Scholar] [CrossRef] [PubMed]
- Tsien, R.Y. THE GREEN FLUORESCENT PROTEIN. Annu. Rev. Biochem. 1998, 67, 509–544. [Google Scholar] [CrossRef]
- Shaner, N.C.; Patterson, G.H.; Davidson, M.W. Advances in fluorescent protein technology. J. Cell Sci. 2007, 120, 4247–4260. [Google Scholar] [CrossRef] [Green Version]
- Markwardt, M.L.; Kremers, G.J.; Kraft, C.A.; Ray, K.; Cranfill, P.J.; Wilson, K.A.; Day, R.N.; Wachter, R.M.; Davidson, M.W.; Rizzo, M.A. An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS ONE 2011, 6, e17896. [Google Scholar] [CrossRef]
- Kremers, G.-J.; Goedhart, J.; van Munster, E.B.; Gadella, T.W.J. Cyan and Yellow Super Fluorescent Proteins with Improved Brightness, Protein Folding, and FRET Förster Radius. Biochemistry 2006, 45, 6570–6580. [Google Scholar] [CrossRef]
- Nelson, B.K.; Cai, X.; Nebenführ, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef]
- Lauersen, K.J.; Kruse, O.; Mussgnug, J.H. Targeted expression of nuclear transgenes in Chlamydomonas reinhardtii with a versatile, modular vector toolkit. Appl. Microbiol. Biotechnol. 2015, 99, 3491–3503. [Google Scholar] [CrossRef] [PubMed]
- Wiedenmann, J.; Schenk, A.; Röcker, C.; Girod, A.; Spindler, K.-D.; Nienhaus, G.U. A far-red fluorescent protein with fast maturation and reduced oligomerization tendency from Entacmaea quadricolor (Anthozoa, Actinaria). Proc. Natl. Acad. Sci. USA 2002, 99, 11646–11651. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Yamasaki, T.; Ohama, T. Expression levels of domestic cDNA cassettes integrated in the nuclear genomes of various Chlamydomonas reinhardtii strains. J. Biosci. Bioeng. 2014, 117, 613–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutiérrez, S.; Wellman, G.B.; Lauersen, K.J. Teaching an old ‘doc’ new tricks for algal biotechnology: Strategic filter use enables multi-scale fluorescent protein signal detection. Front. Bioeng. Biotechnol. 2022, 10, 979607. [Google Scholar] [CrossRef]
- Tannous, B.A.; Kim, D.E.; Fernandez, J.L.; Weissleder, R.; Breakefield, X.O. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol. Ther. 2005, 11, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.P.; Unch, J.; Binkowski, B.F.; Valley, M.P.; Butler, B.L.; Wood, M.G.; Otto, P.; Zimmerman, K.; Vidugiris, G.; Machleidt, T.; et al. Engineered Luciferase Reporter from a Deep Sea Shrimp Utilizing a Novel Imidazopyrazinone Substrate. ACS Chem. Biol. 2012, 7, 1848–1857. [Google Scholar] [CrossRef]
- Moore, S.J.; Lai, H.-E.; Kelwick, R.J.R.; Chee, S.M.; Bell, D.J.; Polizzi, K.M.; Freemont, P.S. EcoFlex: A Multifunctional MoClo Kit for E. coli Synthetic Biology. ACS Synth. Biol. 2016, 5, 1059–1069. [Google Scholar] [CrossRef] [Green Version]
- Iverson, S.V.; Haddock, T.L.; Beal, J.; Densmore, D.M. CIDAR MoClo: Improved MoClo Assembly Standard and New E. coli Part Library Enable Rapid Combinatorial Design for Synthetic and Traditional Biology. ACS Synth. Biol. 2016, 5, 99–103. [Google Scholar] [CrossRef]
- Lee, M.E.; DeLoache, W.C.; Cervantes, B.; Dueber, J.E. A Highly Characterized Yeast Toolkit for Modular, Multipart Assembly. ACS Synth. Biol. 2015, 4, 975–986. [Google Scholar] [CrossRef]
- Fonseca, J.P.; Bonny, A.R.; Kumar, G.R.; Ng, A.H.; Town, J.; Wu, Q.C.; Aslankoohi, E.; Chen, S.Y.; Dods, G.; Harrigan, P.; et al. A Toolkit for Rapid Modular Construction of Biological Circuits in Mammalian Cells. ACS Synth. Biol. 2019, 8, 2593–2606. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, R.; Gale, G.A.R.; Schiavon, A.A.; Puzorjov, A.; Malin, J.; Gillespie, M.D.; Vavitsas, K.; Zulkower, V.; Wang, B.; Howe, C.J.; et al. CyanoGate: A Modular Cloning Suite for Engineering Cyanobacteria Based on the Plant MoClo Syntax. Plant Physiol. 2019, 180, 39–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engler, C.; Youles, M.; Gruetzner, R.; Ehnert, T.M.; Werner, S.; Jones, J.D.; Patron, N.J.; Marillonnet, S. A golden gate modular cloning toolbox for plants. ACS Synth. Biol. 2014, 3, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Niemeyer, J.; Schroda, M. New destination vectors facilitate Modular Cloning for Chlamydomonas. Curr. Genet. 2022, 68, 531–536. [Google Scholar] [CrossRef]
- Niemeyer, J.; Scheuring, D.; Oestreicher, J.; Morgan, B.; Schroda, M. Real-time monitoring of subcellular H2O2 distribution in Chlamydomonas reinhardtii. Plant Cell 2021, 33, 2935–2949. [Google Scholar] [CrossRef]
- Gershenzon, J.; Dudareva, N. The function of terpene natural products in the natural world. Nat. Chem. Biol. 2007, 3, 408–414. [Google Scholar] [CrossRef]
- Cox-Georgian, D.; Ramadoss, N.; Dona, C.; Basu, C. Therapeutic and Medicinal Uses of Terpenes. In Medicinal Plants; Springer: Cham, Switzerland, 2019; pp. 333–359. [Google Scholar] [CrossRef]
- Masyita, A.; Mustika Sari, R.; Dwi Astuti, A.; Yasir, B.; Rahma Rumata, N.; Emran, T.B.; Nainu, F.; Simal-Gandara, J. Terpenes and terpenoids as main bioactive compounds of essential oils, their roles in human health and potential application as natural food preservatives. Food Chem. X 2022, 13, 100217. [Google Scholar] [CrossRef]
- Vavitsas, K.; Fabris, M.; Vickers, C.E. Terpenoid Metabolic Engineering in Photosynthetic Microorganisms. Genes 2018, 9, 520. [Google Scholar] [CrossRef] [Green Version]
- Jaeger, R.; Cuny, E. Terpenoids with Special Pharmacological Significance: A Review. Nat. Prod. Commun. 2016, 11, 1934578X1601100946. [Google Scholar] [CrossRef] [Green Version]
- Wichmann, J.; Lauersen, K.J.; Kruse, O. Green algal hydrocarbon metabolism is an exceptional source of sustainable chemicals. Curr. Opin. Biotechnol. 2020, 61, 28–37. [Google Scholar] [CrossRef]
- Kirby, J.; Keasling, J.D. Biosynthesis of Plant Isoprenoids: Perspectives for Microbial Engineering. Annu. Rev. Plant Biol. 2009, 60, 335–355. [Google Scholar] [CrossRef] [PubMed]
- Papaefthimiou, D.; Diretto, G.; Demurtas, O.C.; Mini, P.; Ferrante, P.; Giuliano, G.; Kanellis, A.K. Heterologous production of labdane-type diterpenes in the green alga Chlamydomonas reinhardtii. Phytochemistry 2019, 167, 112082. [Google Scholar] [CrossRef] [PubMed]
- Yahya, R.Z.; Wellman, G.B.; Overmans, S.; Lauersen, K.J. Engineered production of isoprene from the model green microalga Chlamydomonas reinhardtii. Metab. Eng. Commun. 2023, 16, e00221. [Google Scholar] [CrossRef] [PubMed]
- Freudenberg, R.A.; Baier, T.; Einhaus, A.; Wobbe, L.; Kruse, O. High cell density cultivation enables efficient and sustainable recombinant polyamine production in the microalga Chlamydomonas reinhardtii. Bioresour. Technol. 2021, 323, 124542. [Google Scholar] [CrossRef] [PubMed]
- Freudenberg, R.A.; Wittemeier, L.; Einhaus, A.; Baier, T.; Kruse, O. Advanced pathway engineering for phototrophic putrescine production. Plant Biotechnol. J. 2022, 20, 1968–1982. [Google Scholar] [CrossRef]
- Kiefer, A.M.; Niemeyer, J.; Probst, A.; Erkel, G.; Schroda, M. Production and secretion of functional SARS-CoV-2 spike protein in Chlamydomonas reinhardtii. Front. Plant Sci. 2022, 13, 988870. [Google Scholar] [CrossRef]
- Ramos-Martinez, E.M.; Fimognari, L.; Sakuragi, Y. High-yield secretion of recombinant proteins from the microalga Chlamydomonas reinhardtii. Plant Biotechnol. J. 2017, 15, 1214–1224. [Google Scholar] [CrossRef] [Green Version]
- Cazzaniga, S.; Perozeni, F.; Baier, T.; Ballottari, M. Engineering astaxanthin accumulation reduces photoinhibition and increases biomass productivity under high light in Chlamydomonas reinhardtii. Biotechnol. Biofuels Bioprod. 2022, 15, 77. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perozeni, F.; Baier, T. Current Nuclear Engineering Strategies in the Green Microalga Chlamydomonas reinhardtii. Life 2023, 13, 1566. https://doi.org/10.3390/life13071566
Perozeni F, Baier T. Current Nuclear Engineering Strategies in the Green Microalga Chlamydomonas reinhardtii. Life. 2023; 13(7):1566. https://doi.org/10.3390/life13071566
Chicago/Turabian StylePerozeni, Federico, and Thomas Baier. 2023. "Current Nuclear Engineering Strategies in the Green Microalga Chlamydomonas reinhardtii" Life 13, no. 7: 1566. https://doi.org/10.3390/life13071566






