Is Life Unique?
Abstract
1. Introduction
- What are some of the criteria that allow us to distinguish life from non-life?
- Can life spontaneously generate from physicodynamic interactions alone?
- Will our intelligence ever be able artificially to construct life from inanimate chemical components?
- What exactly does it mean for life to die?
2. The Simplest Known Free-Living Organism
3. The Invariant Characteristics of Life
- (1)
- Delineate itself from its environment through the production and maintenance of a membrane equivalent. In theoretical early life, this membrane equivalent would most likely have been a rudimentary or quasi-active-transport membrane necessary for selective absorption of nutrients, excretion of wastes, and overcoming osmotic and toxic gradients,
- (2)
- Write, store, and pass along into progeny Prescriptive Information (PI) (instructions, both genetic and epigenetic/epigenomic) needed for organization; provide instructions for energy derivation and for needed metabolite production and function; symbolically encode and communicate functional message through a transmission channel to a receiver/decoder/destination/effector mechanism; integrate past, present and future time into its biological PI (instruction) content (PI instructions can be implemented now or any time in the future. In addition, according to evolution theory, these instructions embody a protracted history of derivation and former control. PI is thus largely time-independent, a feature that bespeaks its formal rather than physical essence.),
- (3)
- Bring to pass the above recipe instructions into the production or acquisition of actual catalysts, coenzymes, cofactors, etc.; physically orchestrate the biochemical processes/pathways of metabolic reality; manufacture and maintain physical cellular architecture; establish and operate a semiotic system using “signal molecules”,
- (4)
- Capture, transduce, store, and call up energy for utilization (intuitive, useful work),
- (5)
- Actively self-replicate and eventually reproduce, not just passively polymerize or crystallize; pass along the apparatus and “know-how” for homeostatic metabolism and reproduction into progeny,
- (6)
- Self-monitor and repair its constantly deteriorating physical matrix of bioinstruction retention/transmission, and of architecture,
- (7)
- Develop and grow from immaturity to reproductive maturity,
- (8)
- Productively react to environmental stimuli. Respond in an efficacious manner that is supportive of survival, development, growth, and reproduction, and
- (9)
- Possess enough relative genetic stability, yet sufficient mutability and diversity, to allow for adaptation and potential evolution.
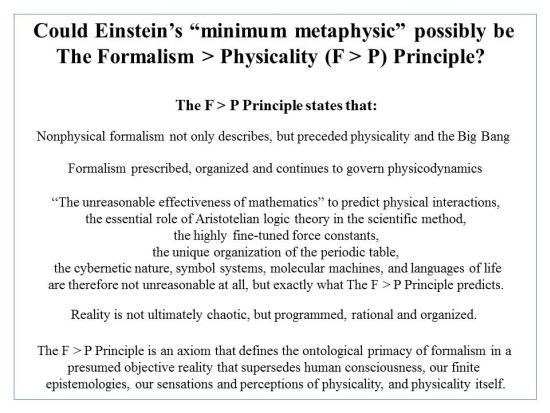
4. Life Is Formally Controlled, Not Just Physicodynamically Constrained
5. Life Is Largely “Computed” by Algorithmic Processing of Linear Digital Programming
Just as complex electronic circuits are built from simple Boolean gates, diverse biological functions, including signal transduction, differentiation, and stress response, frequently uses biochemical switches as a functional module [132].
6. Life Is Instructed and Controlled by Prescriptive Information (PI)
7. The Layers and Dimensions of Formal Biological PI Continue to Grow
8. Life Pursues Biofunctional Goals, and Succeeds
9. The Law of Organizational and Cybernetic Deterioration/Decline (The OCD Law)
10. Choice-Contingent Causation and Control (CCCC)
11. Life Organizes Its Components into Sustained Functional Systems (SFS)
12. Life Is Governed by the Genetic Selection (GS) Principle
13. Life Is Organized, Not Self-Ordered
14. Under No Circumstances Can Chaos Produce Life’s Ultra-Organization
- (1)
- Calculus
- (2)
- Algorithm
- (3)
- Program that achieves computational success
- (4)
- Organizer of formal function
- (5)
- Bona fide system
- (1)
- Highly ordered
- (2)
- Monotonous
- (3)
- Predictable
- (4)
- Regular (vortices, sand piles)
- (5)
- Low informational
- (6)
- Strings of momentary states
15. Life Accomplishes Intuitive (Functional) Work, Not Just the Physics’ Definition of Work
16. Conclusions
References and Notes
- Rizzotti, M. Defining Life: The Central Problem in Theoretical Biology; University of Padova Press: Padova, Italy, 1996; p. 208. [Google Scholar]
- Joyce, G.F. Origins of Life: The Central Concepts See Forward; Jones and Bartlett: Boston, MA, USA, 1994. [Google Scholar]
- Trevors, J.T.; Psenner, R. From self-assembly of life to present-day bacteria: A possible role for nanocells. FEMS Microbiol. Rev. 2001, 25, 573–582. [Google Scholar] [CrossRef]
- Monnard, P.; Deamer, D. Membrane self-assembly processes: Steps toward the first cellular life. Anat. Rec. 2002, 268, 196–207. [Google Scholar] [CrossRef]
- Deamer, D.; Dworkin, J.P.; Sandford, S.A.; Bernstein, M.P.; Allamandola, L.J. The first cell membranes. Astrobiology 2002, 2, 371–381. [Google Scholar] [CrossRef]
- Szostak, J.W.; Bartel, D.P.; Luisi, P.L. Synthesizing life. Nature 2001, 409, 387–390. [Google Scholar] [CrossRef]
- Segre, D.; Ben-Eli, D.; Deamer, D.W.; Lancet, D. The lipid world. Orig. Life Evol. Biosph. 2001, 31, 119–145. [Google Scholar] [CrossRef]
- Fischer, A.; Oberholzer, T.; Luisi, P.L. Giant vesicles as models to study the interactions between membranes and proteins. Biochim. Biophys. Acta 2000, 1467, 177–188. [Google Scholar] [CrossRef]
- Luisi, P.L.; Walde, P.; Oberholzer, T. Lipid vesicles as possible intermediates in the origin of life. Curr. Opin. Colloid Interface Sci. 1999, 3, 33–39. [Google Scholar]
- Deamer, D.W. The first living systems: A bioenergetic perspective. Microbiol. Mol. Biol. Rev. 1997, 61, 239–261. [Google Scholar]
- Oberholzer, T.; Wick, R.; Luisi, P.L.; Biebricher, C.K. Enzymatic RNA replication in self-reproducing vesicles: An approach to a minimal cell. Biochem. Biophys. Res. Commun. 1995, 207, 250–257. [Google Scholar] [CrossRef]
- Chakrabarti, A.C.; Deamer, D.W. Permeation of membranes by the neutral form of amino acids and peptides: Relevance to the origin of peptide translocation. J. Mol. Evol. 1994, 39, 1–5. [Google Scholar]
- Chakrabarti, A.C.; Breaker, R.R.; Joyce, G.F.; Deamer, D.W. Production of RNA by a polymerase protein encapsulated within phospholipid vesicles. J. Mol. Evol. 1994, 39, 555–559. [Google Scholar] [CrossRef]
- Wick, R.; Luisi, P.L. Enzyme-containing liposomes can endogenously produce membrane-constituting lipids. Chem. Biol. 1996, 3, 277–285. [Google Scholar] [CrossRef]
- Luisi, P.L. Autopoiesis: A review and a reappraisal. Naturwissenschaften 2003, 90, 49–59. [Google Scholar]
- Dyson, F.J. A model for the origin of life. J. Mol. Evol. 1982, 18, 344–350. [Google Scholar] [CrossRef]
- Dyson, F.J. Origins of Life, 2nd ed.; Cambridge University Press: Cambridge, UK, 1998. [Google Scholar]
- Guimaraes, R.C. Linguistics of biomolecules and the protein-first hypothesis for the origins of cells. J. Biol. Phys. 1994, 20, 193–199. [Google Scholar] [CrossRef]
- De Duve, C. A research proposal on the origin of life. Orig. Life Evol. Biosph. 2003, 33, 559–574. [Google Scholar] [CrossRef]
- Lahav, N. The synthesis of primitive ‘living’ forms: Definitions, goals, strategies and evolution synthesizers. Orig. Life Evol. Biosph. 1985, 16, 129–149. [Google Scholar] [CrossRef]
- Shapiro, R. A replicator was not involved in the origin of life. IUBMB Life 2000, 49, 173–176. [Google Scholar]
- Pattee, H.H. Dynamic and linguistic modes of complex systems. Int. J. Gen. Syst. 1977, 3, 259–266. [Google Scholar] [CrossRef]
- Rocha, L.M. Syntactic autonomy: Or why there is no autonomy without symbols and how self-organizing systems might evolve them. Ann. N. Y. Acad. Sci. 2000, 207–223. [Google Scholar]
- Rocha, L.M. The physics and evolution of symbols and codes: Reflections on the work of howard pattee. Biosystems 2001, 60, 1–4. [Google Scholar] [CrossRef]
- Rocha, L.M. Evolution with material symbol systems. Biosystems 2001, 60, 95–121. [Google Scholar] [CrossRef]
- Rocha, L.M.; Hordijk, W. Material representations: From the genetic code to the evolution of cellular automata. Artif. Life 2005, 11, 189–214. [Google Scholar] [CrossRef]
- Rocha, L.M.; Joslyn, C. Simulations of Embodied Evolving Semiosis: Emergent Semantics in Artificial Environments. In Proceedings of the 1998 Conference on Virtual Worlds and Simulation, San Diego, CA, USA, 11–14 January 1998; Landauer, C., Bellman, K.L., Eds.; The Society for Computer Simulation International: San Diego, CA, USA; pp. 233–238.
- Pereto, J. Controversies on the origin of life. Int. Microbiol. 2005, 8, 23–31. [Google Scholar]
- Ruiz-Mirazo, K.; Pereto, J.; Moreno, A. A universal definition of life: Autonomy and open-ended evolution. Orig. Life Evol. Biosph. 2004, 34, 323–346. [Google Scholar] [CrossRef]
- Ruiz-Mirazo, K.; Umerez, J.; Moreno, A. Enabling conditions for ‘open-ended evolution’. Biol. Philos. 2008, 23, 67–85. [Google Scholar]
- Smith, J.M. The 1999 crafoord prize lectures. The idea of information in biology. Q. Rev. Biol. 1999, 74, 395–400. [Google Scholar]
- Smith, J.M. The concept of information in biology. Philos. Sci. 2000, 67, 177–194, (entire issue is an excellent discussion).. [Google Scholar]
- Szathmary, E. From RNA to language. Curr. Biol. 1996, 6. [Google Scholar]
- Szathmary, E. Biological information, kin selection, and evolutionary transitions. Theor. Popul. Biol. 2001, 59, 11–14. [Google Scholar] [CrossRef]
- Francois, J. The Logic of Living System: A History of Heredity; Allen Lane: London, UK, 1974. [Google Scholar]
- Alberts, B.; Bray, D.; Lewis, J.; Raff, M.; Roberts, K.; Watson, J.D. Molecular Biology of the Cell; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Davidson, E.H.; Rast, J.P.; Oliveri, P.; Ransick, A.; Calestani, C.; Yuh, C.H.; Minokawa, T.; Amore, G.; Hinman, V.; Arenas-Mena, C.; et al. A genomic regulatory network for development. Science 2002, 295, 1669–1678. [Google Scholar]
- Wolpert, L.; Smith, J.; Jessell, T.; Lawrence, P. Principles of Development; Oxford University Press: Oxford, UK, 2002. [Google Scholar]
- Stegmann, U.E. Genetic information as instructional content. Phil. Sci. 2005, 72, 425–443. [Google Scholar] [CrossRef]
- Barbieri, M. Biology with information and meaning. Hist. Philos. Life Sci. 2004, 25, 243–254. [Google Scholar] [CrossRef]
- Abel, D.L. Is Life Reducible to Complexity? In Fundamentals of Life; Palyi, G., Zucchi, C., Caglioti, L., Eds.; Elsevier: Paris, France, 2002; pp. 57–72. [Google Scholar]
- Abel, D.L.; Trevors, J.T. Three subsets of sequence complexity and their relevance to biopolymeric information. Theor. Biol. Med. Model. 2005, 2. Available online: http://www.tbiomed.com/content/2/1/29 (accessed on 26 December 2011).
- Chen, S.; Zhang, Y.E.; Long, M. New genes in drosophila quickly become essential. Science 2010, 330, 1682–1685. [Google Scholar]
- Freiberg, C.; Wieland, B.; Spaltmann, F.; Ehlert, K.; Brotz, H.; Labischinski, H. Identification of novel essential Escherichia coli genes conserved among pathogenic bacteria. J. Mol. Microbiol. Biotechnol. 2001, 3, 483–489. [Google Scholar]
- Razin, S.; Yogev, D.; Naot, Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 1998, 62, 1094–1156. [Google Scholar]
- Pattee, H.H. Clues from Molecular Symbol Systems. In Dahlem Workshop Reports: Life Sciences Research Report 19: Signed and Spoken Language: Biological Constraints on Linguistic Form; Bellugi, U., Studdert-Kennedy, M., Eds.; Verlag Chemie GmbH: Dahlem Konferenzen, Weinheim, Germany, 1980; pp. 261–274. [Google Scholar]
- Gánti, T. The Principles of Life; Oxford University Press: Oxford, UK, 2003; p. 200. [Google Scholar]
- Jablonka, E.; Lamb, M. Epigenetic Inheritance and Evolution; Oxford University Press: Oxford, UK, 1995. [Google Scholar]
- Jablonka, E.; Szathmary, E. The evolution of information storage and heredity. Trends Ecol. Evol. 1995, 10, 206–211. [Google Scholar] [CrossRef]
- Palyi, G.; Zucchi, C.; Caglioti, L. Proceedings of the Workshop on Life: A Satellite Meeting before the Millennial World Meeting of University Professors. Modena, Italy, 3–8 September 2000.
- Palyi, G.; Zucchi, C.; Caglioti, L. Fundamentals of Life; Elsevier: Paris, France, 2002. [Google Scholar]
- Yockey, H.P. Origin of life on earth and shannon’s theory of communication. Comput. Chem. 2000, 24, 105–123. [Google Scholar]
- Yockey, H.P. Information Theory, Evolution, and the Origin of Life. In Fundamentals of Life; Palyi, G., Zucchi, C., Caglioti, L., Eds.; Elsevier: Paris, France, 2002; pp. 335–348. [Google Scholar]
- Yockey, H.P. On the Role of Information Theory in Mathematical Biology, Radiation on Biology and Medicine, Geneva Presentation Volume; Addison Wesley Publication Company: Boston, MA, USA, 1958. [Google Scholar]
- Yockey, H.P. Information Theory with Applications to Biogenesis and Evolution. In Biogenesis Evolution Homeostasis; Locker, A., Ed.; Springer-Verlag: New York, NY, USA, 1973; Heidelberg and Berlin, Germany. [Google Scholar]
- Yockey, H.P. An application of information theory to the central dogma and the sequence hypothesis. J. Theor. Biol. 1974, 46, 369–406. [Google Scholar] [CrossRef]
- Yockey, H.P. A calculation of the probability of spontaneous biogenesis by information theory. J. Theor. Biol. 1977, 67, 377–398. [Google Scholar] [CrossRef]
- Yockey, H.P. On the information content of cytochrome c. J. Theor. Biol. 1977, 67, 345–398. [Google Scholar] [CrossRef]
- Yockey, H.P. Information Theory and Molecular Biology; Cambridge University Press: Cambridge, UK, 1992; p. 408. [Google Scholar]
- Mayr, E. The Place of Biology in the Sciences and Its Conceptional Structure. In The Growth of Biological Thought: Diversity, Evolution, and Inheritance; Mayr, E., Ed.; Harvard University Press: Cambridge, MA, USA, 1982; pp. 21–82. [Google Scholar]
- Mayr, E. Introduction, pp. 1-7; Is Biology an Autonomous Science? pp. 8-23. In Toward a New Philosophy of Biology, Part 1; Mayr, E., Ed.; Harvard University Press: Cambridge, MA, USA, 1988. [Google Scholar]
- Monod, J. Chance and Necessity; Knopf: New York, NY, USA, 1972. [Google Scholar]
- Bohr, N. Light and life. Nature 1933, 131, 421–423. [Google Scholar] [CrossRef]
- Küppers, B.-O. Information and the Origin of Life; MIT Press: Cambridge, MA, USA, 1990; p. 215. [Google Scholar]
- Bedau, M.A. An aristotelian account of minimal chemical life. Astrobiology 2010, 10, 1011–1020. [Google Scholar] [CrossRef]
- Barbieri, M. The Organic Codes: An Introduction to Semantic Biology; Cambridge University Press: Cambridge, UK, 2003. [Google Scholar]
- Barbieri, M. The definitions of information and meaning: Two possible boundaries between physics and biology (Translated from Italian). Rev. Biol. Biol. Forum 2004, 97, 91–110. [Google Scholar]
- Barbieri, M. Life is ‘artifact-making’. J. Biosemiotics 2005, 1, 113–142. [Google Scholar]
- Barbieri, M. Is the Cell a Semiotic System? In Introduction to Biosemiotics: The New Biological Synthesis; Barbieri, M., Ed.; Springer-Verlag New York, Inc.: Secaucus, NJ, USA, 2006. [Google Scholar]
- Barbieri, M. Introduction to Biosemiotics: The New Biological Synthesis; Springer-Verlag: Dordrecht, The Netherlands, 2006. [Google Scholar]
- Barbieri, M. Semantic biology and the mind-body problem: The theory of the conventional mind. Biological Theory 2006, 1, 352–356. [Google Scholar] [CrossRef]
- Barbieri, M. Has Biosemiotics Come of Age? In Introduction to Biosemiotics: The New Biological Synthesis; Barbieri, M., Ed.; Springer: Dorcrecht, The Netherlands, 2007; pp. 101–114. [Google Scholar]
- Barbieri, M. Is the Cell a Semiotics System? In Introduction to Biosemiotics: The New Biological Synthesis; Barbieri, M., Ed.; Springer: Dordrecht, The Netherlands, 2007; pp. 179–208. [Google Scholar]
- Barbieri, M. Biosemiotic Research Trends; Nova Science Publishers, Inc.: New York, NY, USA, 2007. [Google Scholar]
- Barbieri, M. The Codes of Life: The Rules of Macroevolution (Biosemiotics); Springer: Dordrecht, The Netherlands, 2007. [Google Scholar]
- Barbieri, M. Biosemiotics: A new understanding of life. Naturwissenschaften 2008, 95, 577–599. [Google Scholar] [CrossRef]
- Barbieri, M. Cosmos and history: Life is semiosis; the biosemiotic view of nature. J. Nat. Soc. Philos. 2008, 4, 29–51. [Google Scholar]
- Johnson, D.E. Chapter 10: What Might be a Protocell’s Minimal “Genome”? In The First Gene: The Birth of Programming, Messaging and Formal Control; Abel, D.L., Ed.; LongView Press-Academic: Biol. Res. Div.: New York, NY, USA, 2011; pp. 287–303. [Google Scholar]
- Abel, D.L. What is Protobiocybernetics? In The First Gene: The Birth of Programming, Messaging and Formal Control; Abel, D.L., Ed.; LongView Press-Academic: Biolog. Res. Div.: New York, NY, USA, 2011; pp. 1–18, Chapter 1. [Google Scholar]
- Abel, D.L. The Three Fundamental Categories of Reality. In The First Gene: The Birth of Programming, Messaging and Formal Control; Abel, D.L., Ed.; LongView Press-Academic: Biolog. Res. Div.: New York, NY, USA, 2011; pp. 19–54, Chapter 2. [Google Scholar]
- Abel, D.L. The Cybernetic Cut and Configurable Switch (Cs) Bridge. In The First Gene: The Birth of Programming, Messaging and Formal Control; Abel, D.L., Ed.; LongView Press-Academic: Biol. Res. Div.: New York, NY, USA, 2011; pp. 55–74, Chapter 3. [Google Scholar]
- Abel, D.L. What Utility does Order, Pattern or Complexity Prescribe? In The First Gene: The Birth of Programming, Messaging and Formal Control; Abel, D.L., Ed.; LongView Press-Academic: Biol. Res. Div.: New York, NY, USA, 2011; pp. 75–116, Chapter 4. [Google Scholar]
- Abel, D.L. Linear Digital Material Symbol Systems (Mss). In The First Gene: The Birth of Programming, Messaging and Formal Control; Abel, D.L., Ed.; LongView Press-Academic: Biol. Res. Div.: New York, NY, USA, 2011; pp. 135–160, Chapter 6. [Google Scholar]
- Abel, D.L. The Formalism > Physicality (F > P) Principle. In The First Gene: The Birth of Programming, Messaging and Formal Control; Abel, D.L., Ed.; LongView Press-Academic: New York, NY, USA, 2011; pp. 325–356, Chapter 12. [Google Scholar]
- Abel, D.L. The biosemiosis of prescriptive information. Semiotica 2009, 2009, 1–19. [Google Scholar] [CrossRef]
- Abel, D.L. Prescriptive Information (PI). Available online: http://www.scitopics.com/Prescriptive_Information_PI.html (accessed on 26 December 2011).
- Abel, D.L. The First Gene: The Birth of Programming, Messaging and Formal Control; LongView Press-Academic: Biolog. Res. Div.: New York, NY, USA, 2011; p. 389. [Google Scholar]
- Abel, D.L. Moving ‘Far from Equilibrium’ in a Prebitoic Environment: The Role of Maxwell’s Demon in Life Origin. In Genesis—In the Beginning: Precursors of Life, Chemical Models and Early Biological Evolution; Seckbach, J., Gordon, R., Eds.; Springer: Dordrecht, The Netherlands, 2011. [Google Scholar]
- Cleland, C.E.; Chyba, C.F. Defining ‘Life’. Orig. Life Evol. Biosph. 2002, 32, 387–393. [Google Scholar] [CrossRef]
- Abel, D.L. To What Degree can We Reduce “Life” Without “Loss of Life”? In Workshop on Life: A Satellite Meeting before the Millenial World Meeting of University Professor; Palyi, G., Caglioti, L., Zucchi, C., Eds.; University of Modena: Modena, Italy, 2000. [Google Scholar]
- Abel, D.L. Life Origin: The Role of Complexity at the Edge of Chaos. In Washington Science 2006; Chandler, J., Kay, P., Eds.; Headquarters of the National Science Foundation: Arlington, VA, USA, 2006. [Google Scholar]
- Abel, D.L. Complexity, self-organization, and emergence at the edge of chaos in life-origin models. J. Wash. Acad. Sci. 2007, 93, 1–20. [Google Scholar]
- Abel, D.L. The Capabilities of Chaos and Complexity. In Society for Chaos Theory: Society for Complexity in Psychology and the Life Sciences; International Conference at Virginia Commonwealth University: Richmond, VA, USA, 2008. [Google Scholar] [Green Version]
- Henry, C.; Overbeek, R.; Stevens, R.L. Building the blueprint of life. Biotechnol. J. 2010, 5, 695–704. [Google Scholar] [CrossRef]
- Dorman, C.J. Regulation of transcription by DNA supercoiling in Mycoplasma genitalium: Global control in the smallest known self-replicating genome. Mol. Microbiol. 2011, 81, 302–304. [Google Scholar] [CrossRef]
- Butt, A.M.; Tahir, S.; Nasrullah, I.; Idrees, M.; Lu, J.; Tong, Y. Mycoplasma genitalium: A comparative genomics study of metabolic pathways for the identification of drug and vaccine targets. Infect. Genet. Evol. 2011, 12, 53–62. [Google Scholar]
- Pennisi, E. Genomics. Synthetic genome brings new life to bacterium. Science 2010, 328, 958–959. [Google Scholar] [CrossRef]
- McCutcheon, J.P. The bacterial essence of tiny symbiont genomes. Curr. Opin. Microbiol. 2010, 13, 73–78. [Google Scholar] [CrossRef]
- Kwok, R. Genomics: DNA’s master craftsmen. Nature 2010, 468, 22–25. [Google Scholar] [CrossRef]
- Benders, G.A.; Noskov, V.N.; Denisova, E.A.; Lartigue, C.; Gibson, D.G.; Assad-Garcia, N.; Chuang, R.Y.; Carrera, W.; Moodie, M.; Algire, M.A.; et al. Cloning whole bacterial genomes in yeast. Nucleic Acids Res. 2010, 38, 2558–2569. [Google Scholar] [CrossRef]
- Suthers, P.F.; Dasika, M.S.; Kumar, V.S.; Denisov, G.; Glass, J.I.; Maranas, C.D. A genome-scale metabolic reconstruction of Mycoplasma genitalium, Ips189. PLoS Comput. Biol. 2009, 5. [Google Scholar]
- Zhang, W.; Baseman, J.B. Transcriptional regulation of Mg_149, an osmoinducible lipoprotein gene from Mycoplasma genitalium. Mol. Microbiol. 2011, 81, 327–339. [Google Scholar] [CrossRef]
- Zhang, W.; Baseman, J.B. Transcriptional response of Mycoplasma genitalium to osmotic stress. Microbiology 2011, 157, 548–556. [Google Scholar] [CrossRef]
- Abel, D.L.; Trevors, J.T. More than Metaphor: Genomes are Objective Sign Systems. In Biosemiotic Research Trends; Barbieri, M., Ed.; Nova Science Publishers: New York, NY, USA, 2007; pp. 1–15. [Google Scholar]
- Abel, D.L. What is life? (under Definitions). Available online: http://www.lifeorigin.info (accessed on 26 December 2011).
- Abel, D.L. Constraints vs. controls. Open Cybern. Syst. J. 2010, 4, pp. 14–27. Available online: http://www.benthamscience.com/open/tocsj/articles/V004/14TOCSJ.pdf (accessed on 26 December 2011).
- Abel, D.L.; Trevors, J.T. Self-organization vs. self-ordering events in life-origin models. Phys. Life Rev. 2006, 3, 211–228. [Google Scholar] [CrossRef]
- Johnson, D.E. Programming of Life; Big Mac Publishers: Sylacauga, AL, USA, 2010; p. 127. [Google Scholar]
- Rocha, L.M. Evidence Sets and Contextual Genetic Algorithms: Exploring Uncertainty, Context, and Embodiment in Cognitive and Biological Systems; State University of New York: Binghamton, NY, USA, 1997. [Google Scholar]
- Whitehead, A.N. Symbolism: Its Meaning and Effect; Macmillan: New York, NY, USA, 1927. [Google Scholar]
- Cassirer, E. The Philosophy of Symbolic Forms, Vol 3: The Phenomena of Knowledge; Yale University Press: New Haven, CT, USA, 1957. [Google Scholar]
- Harnad, S. The symbol grounding problem. Phys. D 1990, 42, 335–346. [Google Scholar] [CrossRef]
- Pattee, H.H. Evolving self-reference: Matter, symbols, and semantic closure. Commun. Cogn. Artif. Intell. 1995, 12, 9–28. [Google Scholar]
- Pattee, H.H.; Kull, K. A biosemiotic conversation: Between physics and semiotics. Sign Sys. Stud. 2009, 37, 311–331. [Google Scholar]
- Cairns-Smith, A.G. Seven Clues to the Origin of Life; Cambridge University Press: Cambridge, UK, 1990; p. 130, Canto ed. [Google Scholar]
- Segre, D.; Ben-Eli, D.; Lancet, D. Compositional genomes: Prebiotic information transfer in mutually catalytic noncovalent assemblies. Proc. Natl. Acad. Sci. USA 2000, 97, 4112–4117. [Google Scholar]
- Segre, D.; Lancet, D. Composing life. EMBO Rep. 2000, 1, 217–222. [Google Scholar] [CrossRef]
- Segre, D.; Lancet, D.; Kedem, O.; Pilpel, Y. Graded autocatalysis replication domain (gard): Kinetic analysis of self-replication in mutually catalytic sets. Orig. Life Evol. Biosph. 1998, 28, 501–514. [Google Scholar] [CrossRef]
- Durston, K.K.; Chiu, D.K.Y. Functional Sequence Complexity in Biopolymers. In The First Gene: The Birth of Programming, Messaging and Formal Control; Abel, D.L., Ed.; LongView Press-Academic: Biol. Res. Div.: New York, NY, USA, 2011; pp. 117–133, Chapter 5. [Google Scholar]
- Durston, K.K.; Chiu, D.K.; Abel, D.L.; Trevors, J.T. Measuring the functional sequence complexity of proteins. Theor. Biol. Med. Model. 2007, 4, p. 47. Available online: http://www.tbiomed.com/content/4/1/47 (accessed on 26 December 2011).
- Durston, K.K.; Chiu, D.K.Y. A functional entropy model for biological sequences. Dyn. Contin. Discret. Impuls. Syst. Ser. B. 2005. Available online: http://www.newscholars.com/papers/Durston&Chiu%20paper.pdf (accessed on 26 December 2011).
- Turing, A.M. On computable numbers, with an application to the entscheidungs problem. Proc. Roy. Soc. Lond. Math. Soc. 1936, 42, 230–265, correction in 243, 544-546.. [Google Scholar] [CrossRef]
- Von Neumann, J. Functional Operators; Princeton University Press: Princeton, NJ, USA, 1950. [Google Scholar]
- Von Neumann, J. The General and Logical Theory of Automata. In The World of Mathematics Vol 4; Newman, J.R., Ed.; Simon and Schuster: New York, NY, USA, 1956. [Google Scholar]
- Von Neumann, J.; Aspray, W.; Burks, A.W. Papers of John Von Neumann on Computing and Computer Theory; MIT Press: Cambridge, MA, USA, 1987; p. xviii. [Google Scholar]
- Von Neumann, J.; Churchland, P.M.; Churchland, P.S. The Computer and the Brain, 2nd ed.; Yale University Press: New Haven, CT, USA, 2000; p. xxviii. [Google Scholar]
- Wiener, N. Cybernetics; Wiley: New York, NY, USA, 1948. [Google Scholar]
- Wiener, N. Cybernetics, Its Control and Communication in the Animal and the Machine, 2nd ed.; MIT Press: Cambridge, MA, USA, 1961. [Google Scholar]
- D’Onofrio, D.J.; An, G. A comparative approach for the investigation of biological information processing: An examination of the structure and function of computer hard drives and DNA. Theor. Biol. Med. Model. 2010, 7. [Google Scholar]
- Conrad, M. Microscopic-macroscopic interface in biological information processing. Biosystems 1983, 16, 345–363. [Google Scholar] [CrossRef]
- Wang, D. Discrepancy between mRNA and protein abundance: Insight from information retrieval process in computers. Comput. Biol. Chem. 2008, 32, 462–468. [Google Scholar] [CrossRef]
- Ramakrishnan, N.; Bhalla, U.S. Memory switches in chemical reaction space. PLoS Comput. Biol. 2008, 4. [Google Scholar]
- Benenson, Y.; Adar, R.; Paz-Elizur, T.; Livneh, Z.; Shapiro, E. DNA molecule provides a computing machine with both data and fuel. Proc. Natl. Acad. Sci. USA 2003, 100, 2191–2196. [Google Scholar]
- Benenson, Y.; Paz-Elizur, T.; Adar, R.; Keinan, E.; Livneh, Z.; Shapiro, E. Programmable and autonomous computing machine made of biomolecules. Nature 2001, 414, 430–434. [Google Scholar] [CrossRef]
- Shannon, C. Part I and II: A mathematical theory of communication. Bell Syst. Tech. J. 1948, XXVII, 379–423. [Google Scholar]
- Shannon, C. Part III: A mathematical theory of communication. Bell Syst. Tech. J. 1948, XXVII, 623–656. [Google Scholar]
- Adami, C. Introduction to Artificial Life; Springer/Telos: New York, NY, USA, 1998; p. 374. [Google Scholar]
- Bruza, P.D.; Song, D.W.; Wong, K.F. Aboutness from a common sense perspective. J. Am. Soc. Inf. Sci. 2000, 51, 1090–1105. [Google Scholar] [CrossRef]
- Hjorland, B. Towards a theory of aboutness, subject, topicallity, theme, domain, field, content . . . and relevance. J. Am. Soc. Inf. Syst. Technol. 2001, 52, 774–778. [Google Scholar] [CrossRef]
- Johnson, D.E. Probability’s Nature and Nature’s Probability (a Call to Scientific Integrity); Booksurge Publishing: Charleston, SC, USA, 2010. [Google Scholar]
- Oyama, S. The Ontogeny of Information: Developmenal Systems and Evolution (Science and Cultural Theory); Duke University Press: Durham, NC, USA, 2000. [Google Scholar]
- Sarkar, S. Information in genetics and developmental biology: Comments on Maynard Smith. Philos. Sci. 2000, 67, 208–213. [Google Scholar]
- Sarkar, S. Biological Information: A Skeptical Look at Some Central Dogmas of Molecular Biology. In The Philosophy and History of Molecular Biology: New Perspectives; Sarkar, S., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1996; pp. 187–231. [Google Scholar]
- Boniolo, G. Biology without information. Hist. Philos. Life Sci. 2003, 25, 255–273. [Google Scholar] [CrossRef]
- Salthe, S.N. Meaning in nature: Placing biosemitotics within pansemiotics. J. Biosemiotics 2005, 1, 287–301. [Google Scholar]
- Salthe, S.N. What is the Scope of Biosemiotics? Information in Living Systems. In Introduction to Biosemiotics: The New Biological Synthesis; Barbieri, M., Ed.; Springer-Verlag New York, Inc.: Dordrecht, The Netherlands and Secaucus, NJ, USA, 2006. [Google Scholar]
- Kurakin, A. Self-organization versus watchmaker: Molecular motors and protein translocation. Biosystems 2006, 84, 15–23. [Google Scholar] [CrossRef]
- Mahner, M.; Bunge, M.A. Foundations of Biophilosophy; Springer Verlag: Berlin, Germany, 1997. [Google Scholar]
- Kitcher, P. Battling the Undead; How (and How Not) to Resist Genetic Determinism. In Thinking About Evolution: Historical Philosophical and Political Perspectives; Singh, R.S., Krimbas, C.B., Paul, D.B., Beattie, J., Eds.; Cambridge University Press: Cambridge, UK, 2001; pp. 396–414. [Google Scholar]
- Szostak, J.W. Functional information: Molecular messages. Nature 2003, 423. [Google Scholar]
- Hazen, R.M.; Griffin, P.L.; Carothers, J.M.; Szostak, J.W. Functional information and the emergence of biocomplexity. Proc. Natl. Acad. Sci. USA 2007, 104, 8574–8581. [Google Scholar] [CrossRef]
- Sharov, A. Role of utility and inference in the evolution of functional information. Biosemiotics 2009, 2, 101–115. [Google Scholar] [CrossRef]
- Davis, M. Computability and Unsolvability; McGraw-Hill: New York, NY, USA, 1958. [Google Scholar]
- Abel, D.L. The capabilities of chaos and complexity. Int. J. Mol. Sci. 2009, 10, pp. 247–291. Available online: https://www.mdpi.com/1422-0067/10/1/247 (accessed on 26 December 2011).
- Trevors, J.T.; Abel, D.L. Chance and necessity do not explain the origin of life. Cell Biol. Int. 2004, 28, 729–739. [Google Scholar] [CrossRef]
- Abel, D.L. ‘The cybernetic cut’: Progressing from description to prescription in systems theory. Open Cybern. Syst. J. 2008, 2, pp. 234–244. Available online: www.bentham.org/open/tocsj/articles/V002/252TOCSJ.pdf (accessed on 26 December 2011).
- Beiter, T.; Reich, E.; Williams, R.; Simon, P. Antisense transcription: A critical look in both directions. Cell. Mol. Life Sci. (CMLS) 2009, 66, 94–112. [Google Scholar] [CrossRef]
- Chen, J.; Sun, M.; Kent, W.J.; Huang, X.; Xie, H.; Wang, W.; Zhou, G.; Shi, R.Z.; Rowley, J.D. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res. 2004, 32, 4812–4820. [Google Scholar]
- Dornenburg, J.E.; DeVita, A.M.; Palumbo, M.J.; Wade, J.T. Widespread antisense transcription in Escherichia coli. mBio 2010, 1. [Google Scholar]
- Lluch-Senar, M.; Vallmitjana, M.; Querol, E.; Pinol, J. A new promoterless reporter vector reveals antisense transcription in Mycoplasma genitalium. Microbiology 2007, 153, 2743–2752. [Google Scholar] [CrossRef]
- Slonczewski, J.L. Concerns about recently identified widespread antisense transcription in Escherichia coli. mBio 2010, 1. [Google Scholar]
- Wade, J.T.; Dornenburg, J.E.; DeVita, A.M.; Palumbo, M.J. Reply to “concerns about recently identified widespread antisense transcription in Escherichia coli”. mBio 2010, 1. [Google Scholar]
- Yelin, R.; Dahary, D.; Sorek, R.; Levanon, E.Y.; Goldstein, O.; Shoshan, A.; Diber, A.; Biton, S.; Tamir, Y.; Khosravi, R.; et al. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 2003, 21, 379–386. [Google Scholar] [CrossRef]
- Dinger, M.E.; Pang, K.C.; Mercer, T.R.; Mattick, J.S. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities. PLoS Comput. Biol. 2008, 4. [Google Scholar]
- He, Y.; Vogelstein, B.; Velculescu, V.E.; Papadopoulos, N.; Kinzler, K.W. The antisense transcriptomes of human cells. Science 2008, 322, 1855–1857. [Google Scholar]
- Shintani, S.; O’hUigin, C.; Toyosawa, S.; Michalová, V.; Klein, J. Origin of gene overlap: The case of tcp1 and acat2. Genetics 1999, 152, 743–754. [Google Scholar]
- Sanna, C.; Li, W.-H.; Zhang, L. Overlapping genes in the human and mouse genomes. BMC Genomics 2008, 9. [Google Scholar]
- Sabath, N.; Landan, G.; Graur, D. A method for the simultaneous estimation of selection intensities in overlapping genes. PLoS ONE 2008, 3. [Google Scholar]
- Herzel, H.; Weiss, O.; Trifonov, E.N. Sequence periodicity in complete genomes of archaea suggests positive supercoiling. J. Biomol. Struct. Dyn. 1998, 16, 341–345. [Google Scholar] [CrossRef]
- Ohyama, T. DNA Conformation and Transcription; Landes Bioscience: Georgetown, TX, and New York, NY, USA, 2005. [Google Scholar]
- Nurse, P. Life, logic and information. Nature 2008, 454, 424–426. [Google Scholar] [CrossRef]
- De Silva, A.P.; Uchiyama, S. Molecular logic and computing. Nat. Nano 2007, 2, 399–410. [Google Scholar] [CrossRef]
- Korzeniewski, B. Cybernetic formulation of the definition of life. J. Theor. Biol. 2001, 209, 275–286. [Google Scholar] [CrossRef]
- Korzeniewski, B. Confrontation of the cybernetic definition of a living individual with the real world. Acta Biotheor. 2005, 53, 1–28. [Google Scholar] [CrossRef]
- Abel, D.L. The universal plausibility metric (Upm) & principle (Upp). Theor. Biol. Med. Model. 2009, 6. Available online: http://www.tbiomed.com/content/6/1/27 (accessed on 26 December 2011).
- Abel, D.L. The Universal Plausibility Metric and Principle. In The First Gene: The Birth of Programming, Messaging and Formal Control; Abel, D.L., Ed.; LongView Press-Academic: New York, NY, USA, 2011; pp. 305–324, Chapter 11. [Google Scholar]
- Dembski, W. The Design Inference: Eliminating Chance Through Small Probabilities; Cambridge University Press: Cambridge, UK, 1998. [Google Scholar]
- Abel, D.L. The GS (Genetic Selection) Principle. Front. Biosci. 2009, 14, pp. 2959–2969. Available online: http://www.bioscience.org/2009/v14/af/3426/fulltext.htm (accessed on 26 December 2011).
- Abel, D.L. The Genetic Selection (Gs) Principle. Available online: http://www.scitopics.com/The_Genetic_Selection_GS_Principle.html (accessed on 26 December 2011).
- Kaplan, M. Decision Theory as Philosophy; Cambridge University Press: Cambridge, UK, 1996; p. 227. [Google Scholar]
- Eigen, M.; Gardiner, W.C., Jr.; Schuster, P. Hypercycles and compartments. Compartments assists—But do not replace—Hypercyclic organization of early genetic information. J. Theor. Biol. 1980, 85, 407–411. [Google Scholar] [CrossRef]
- Eigen, M.; Schuster, P. Comments on “growth of a hypercycle” by King (1981). Biosystems 1981, 13. [Google Scholar]
- Eigen, M.; Schuster, P.; Sigmund, K.; Wolff, R. Elementary step dynamics of catalytic hypercycles. Biosystems 1980, 13, 1–22. [Google Scholar] [CrossRef]
- Smith, J.M. Hypercycles and the origin of life. Nature 1979, 280, 445–446. [Google Scholar] [CrossRef]
- Ycas, M. Codons and hypercycles. Orig. Life Evol. Biosph. 1999, 29, 95–108. [Google Scholar] [CrossRef]
- Melendez-Hevia, E.; Montero-Gomez, N.; Montero, F. From prebiotic chemistry to cellular metabolism—The chemical evolution of metabolism before darwinian natural selection. J. Theor. Biol. 2008, 252, 505–519. [Google Scholar] [CrossRef]
- Munteanu, A.; Sole, R.V. Phenotypic diversity and chaos in a minimal cell model. J. Theor. Biol. 2006, 240, 434–442. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Abel, D.L. Is Life Unique? Life 2012, 2, 106-134. https://doi.org/10.3390/life2010106
Abel DL. Is Life Unique? Life. 2012; 2(1):106-134. https://doi.org/10.3390/life2010106
Chicago/Turabian StyleAbel, David L. 2012. "Is Life Unique?" Life 2, no. 1: 106-134. https://doi.org/10.3390/life2010106
APA StyleAbel, D. L. (2012). Is Life Unique? Life, 2(1), 106-134. https://doi.org/10.3390/life2010106