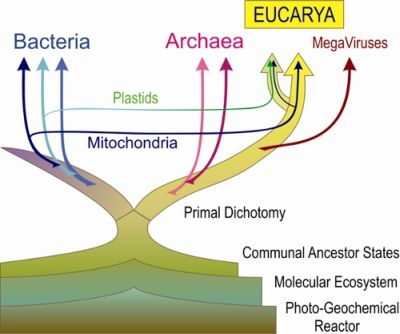
Primal Eukaryogenesis: On the Communal Nature of Precellular States, Ancestral to Modern Life
Abstract
:1. Preface

2. The Photoactive Metal Sulfide Scenario
3. The Phospho-Riboside Connection
4. Molecular Ecosystems in Biogenic Photochemical Reactors
4.1. Recasting the Plot
- (i)
- to couple energy from the environment into usable chemical forms;
- (ii)
- to carry out specific catalytic functions;
- (iii)
- to make and/or copy macromolecules;
- (iv)
- to give some of these informational significance.
4.2. A Porespace Setting in Shallow Sediments

4.3. Catching and Utilizing Photons by Photoactive Minerals

4.4. Membrane Compartmentalization of Charge Separation

4.5. Additional Potential of UV-Facilitated Biochemistry
5. Early Protogenes
- Was cellularization a very early precondition for Darwinian evolution as such?
- Or was it rather a relatively late manifestation of cellular escape, after a long period of subcellular evolution in indistinctly bounded assemblages of macromolecular hydrogels?
6. From Genes to Chromosomes
7. Cellular Emancipation and Escape
7.1. The Coenocytic Alternative to "Cell-like Vesicles Too Early"
7.2. The "Karyogenic Proto-Coenocyte Hypothesis"-Emergence of Multiple Protonuclei
7.3. Cellular Escape Events
8. Rethinking the Primal Dichotomy and the Primordial Trefoil in the Universal Tree of Life
8.1. Nesting Eukaryotes Between Two Phylodomains of Prokaryotic Lineages
8.2. Can Fossil Scarcity Constrain Eukaryogenic Time Scales?
8.3. Ambiguous Leads to the Emergence of Three Phylodomains
8.4. Ecological Considerations-Engulfment and Gamete Fusion
8.5. Continuity of Ancient RNA Functions
8.6. Significance of a Complex Protoeukaryotic Stemline
8.7. Multiple Escape Events at the Base of Prokaryotic Lineages
- How can there be room for shared archaeal/bacterial characters that are not automatically represented in eukaryotes as well?
8.8. Eukaryotes, Organelles and Megaviruses
9. Concluding Prospects
Note Added in Proof
Acknowledgements
References
- Follmann, H.; Brownson, C. Darwin's warm little pond revisited: from molecules to the origin of life. Naturwissenschaften 2009, 96, 1265–1292. [Google Scholar] [CrossRef]
- Peretó, J. Controversies on the origin of life. Int. Microbiol. 2005, 8, 23–31. [Google Scholar]
- Pascal, R.; Boiteau, L.; Forterre, P.; Gargaud, M.; Lazcano, A.; Lopez-Garcia, P.; Maurel, M.C.; Moreira, D.; Peretó, J.; Prieur, D.; Reisse, J. Prebiotic chemistry-biochemistry-emergence of life (4.4-2 Ga). Earth Moon Planets 2006, 98, 153–203. [Google Scholar] [CrossRef]
- Egel, R. Integrative perspectives: In quest of a coherent framework for origins of life on earth. In Origins of Life: The Primal Self-Organization; Egel, R., Lankenau, D.-H., Mulkidjanian, A.Y., Eds.; Springer-Verlag: Heidelberg, Germany, 2011; pp. 289–360. [Google Scholar]
- Mulkidjanian, A.Y. Origin of life in the Zinc World: 1. Photosynthetic, porous edifices built of hydrothermally precipitated zinc sulfide (ZnS) as cradles of life on Earth. Biol. Direct 2009, 4, 26–39. [Google Scholar] [CrossRef]
- Mulkidjanian, A.Y.; Galperin, M.Y. On the origin of life in the Zinc World: 2. Validation of the hypothesis on the photosynthesizing zinc sulfide edifices as cradles of life on Earth. Biol. Direct 2009, 4, 27–37. [Google Scholar] [CrossRef]
- Zhang, X.V.; Ellery, S.P.; Friend, C.M.; Holland, H.D.; Michel, F.M.; Schoonen, M.A.A.; Martin, S.T. Photodriven reduction and oxidation reactions on colloidal semiconductor particles: Implications for prebiotic synthesis. J. Photochem. Photobiol. A Chem. 2007, 185, 301–311. [Google Scholar] [CrossRef]
- Guzman, M.I.; Martin, S.T. Oxaloacetate-to-malate conversion by mineral photoelectrochemistry: Implications for the viability of the reductive tricarboxylic acid cycle in prebiotic chemistry. Int. J. Astrobiol. 2008, 7, 271–278. [Google Scholar] [CrossRef]
- Guzman, M.I.; Martin, S.T. Prebiotic metabolism: Production by mineral photoelectrochemistry of alpha-ketocarboxylic acids in the reductive tricarboxylic acid cycle. Astrobiology 2009, 9, 833–842. [Google Scholar] [CrossRef]
- Guzman, M.I.; Martin, S.T. Photo-production of lactate from glyoxylate: How minerals can facilitate energy storage in a prebiotic world. Chem. Commun. 2010, 46, 2265–2267. [Google Scholar] [CrossRef]
- Morowitz, H.J.; Kostelnik, J.D.; Yang, J.; Cody, G.D. The origin of intermediary metabolism. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 7704–7708. [Google Scholar]
- Smith, E.; Morowitz, H.J. Universality in intermediary metabolism. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 13168–13173. [Google Scholar] [CrossRef]
- Morowitz, H.J.; Smith, E. Energy flow and the organization of life. Complexity 2007, 13, 51–59. [Google Scholar] [CrossRef]
- Shapiro, R. Small molecule interactions were central to the origin of life. Q. Rev. Biol. 2006, 81, 105–125. [Google Scholar] [CrossRef]
- Shapiro, R. A simpler origin for life. Scient. Amer. 2007, 296, 46–53. [Google Scholar] [CrossRef]
- Hunding, A.; Kepes, F.; Lancet, D.; Minsky, A.; Norris, V.; Raine, D.; Sriram, K.; Root-Bernstein, R. Compositional complementarity and prebiotic ecology in the origin of life. BioEssays 2006, 28, 399–412. [Google Scholar] [CrossRef]
- Kurakin, A. The self-organizing fractal theory as a universal discovery method: the phenomenon of life. Theor. Biol. Med. Model. 2011, 8, 4:1–4:66. [Google Scholar]
- Haldane, J.B.S. The origin of life. The Rationalist Annual 1929. [Google Scholar] -Reproduced in: Haldane, J.B.S. Science and Life: Essays of a Rationalist; Pemberton Publishing in association with Barrie & Rockliff: London, UK, 1968; pp. 1–11. [Google Scholar]
- Russell, M.J.; Hall, A.J. The emergence of life from iron monosulphide bubbles at a submarine hydrothermal redox and pH front. J. Geol. Soc. Lond. 1997, 154, 377–402. [Google Scholar] [CrossRef]
- Martin, W.; Russell, M.J. On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2003, 358, 59–85. [Google Scholar] [CrossRef]
- Martin, W.; Baross, J.; Kelley, D.; Russell, M.J. Hydrothermal vents and the origin of life. Nat. Rev. Microbiol. 2008, 6, 805–814. [Google Scholar]
- Koonin, E.V. On the origin of cells and viruses: primordial virus world scenario. Ann. N.Y. Acad. Sci. 1178, 47–64. [Google Scholar]
- Blair, N.E.; Bonner, W.A. A model for the enantiomeric enrichment of polypeptides on the primitive earth. Orig. Life Evol. Biosph. 1981, 11, 331–335. [Google Scholar] [CrossRef]
- Cheng, C.M.; Fan, C.; Wan, R.; Tong, C.Y.; Miao, Z.W.; Chen, J.; Zhao, Y.F. Phosphorylation of adenosine with trimetaphosphate under simulated prebiotic conditions. Orig. Life. Evol. Biosph. 2002, 32, 219–224. [Google Scholar] [CrossRef]
- Commeyras, A.; Collet, H.; Boiteau, L.; Taillades, J.; Vandenabeele-Trambouze, O.; Cottet, H.; Biron, J.-P.; Plasson, R.; Mion, L.; Lagrille, O.; et al. Prebiotic synthesis of sequential peptides on the Hadean beach by a molecular engine working with nitrogen oxides as energy sources. Polym. Int. 2002, 51, 661–665. [Google Scholar]
- Lathe, R. Fast tidal cycling and the origin of life. Icarus 2004, 168, 18–22. [Google Scholar] [CrossRef]
- Bywater, R.P.; Conde-Frieboes, K. Did life begin on the beach? Astrobiology 2005, 5, 568–574. [Google Scholar] [CrossRef]
- Plankensteiner, K.; Reiner, H.; Rode, B.M. Prebiotic chemistry: The amino acid and peptide world. Curr. Org. Chem. 2005, 9, 1107–1114. [Google Scholar] [CrossRef]
- Brack, A. From interstellar amino acids to prebiotic catalytic peptides: A review. Chem. Biodiv. 2007, 4, 665–679. [Google Scholar] [CrossRef]
- Deamer, D.; Weber, A.L. Bioenergetics and life's origins. Cold Spring Harb. Perspect. Biol. 2010, 2, a004929:1–a004929:16. [Google Scholar]
- Trinks, H.; Schröder, W.; Biebricher, C.K. Ice and the origin of life. Orig. Life Evol. Biosph. 2005, 35, 429–445. [Google Scholar] [CrossRef]
- Price, P.B. Microbial life in glacial ice and implications for a cold origin of life. FEMS Microbiol. Ecol. 2007, 59, 217–231. [Google Scholar] [CrossRef]
- Gilbert, W. Origin of life: The RNA world. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- The RNA World: The Nature of Modern RNA Suggests a Prebiotic RNA World; Gesteland, R.F.; Cech, T.R.; Atkins, J.F. (Eds.) Cold Spring Harbor Lab Press: Cold Spring Harbor, NY, USA, 2006.
- Robertson, M.P.; Joyce, G.F. The origins of the RNA World. Cold Spring Harb. Perspect. Biol. 2010, 2, a003608:1–a003608:22. [Google Scholar]
- Lankenau, D.-H. Two RNA Worlds: toward the origin of replication, genes, recombination, and repair. In Origins of Life: The Primal Self-Organization; Egel, R., Lankenau, D.-H., Mulkidjanian, A.Y., Eds.; Springer-Verlag: Heidelberg, Germany, 2011; pp. 225–286. [Google Scholar]
- Kurland, C.G. The RNA dreamtime. BioEssays 2010, 32, 866–871. [Google Scholar] [CrossRef]
- Moulton, V.; Gardner, P.; Pointon, R.; Creamer, L.; Jameson, G.; Penny, D. RNA folding argues against a hot-start origin of life. J. Mol. Evol. 2000, 51, 416–421. [Google Scholar]
- Kritsky, M.; Telegina, T. Role of nucleotide-like coenzymes in primitive evolution. In Origins: Cellular Origin, Life in Extreme Habitats and Astrobiology; Seckbach, J., Ed.; Kluver Academic Publishers: Dordrecht, NL, 2005; pp. 215–231. [Google Scholar]
- Sharov, A.A. Coenzyme autocatalytic network on the surface of oil microspheres as a model for the origin of life. Int J. Mol. Sci. 2009, 10, 1838–1852. [Google Scholar] [CrossRef]
- Raffaelli, N. Nicotinamide coenzyme synthesis: A case of ribonucleotide emergence or a byproduct of the RNA world? In Origins of Life: The Primal Self-Organization; Egel, R., Lankenau, D.-H., Mulkidjanian, A.Y., Eds.; Springer-Verlag: Heidelberg, Germany, 2011; pp. 185–208. [Google Scholar]
- Rode, B.M. Peptides and the origin of life. Peptides 1999, 20, 773–786. [Google Scholar] [CrossRef]
- Milner-White, E.J.; Russell, M.J. Sites for phosphates and iron-sulfur thiolates in the first membranes: 3 to 6 residue anion-binding motifs (nests). Orig. Life Evol. Biosph. 2005, 35, 19–27. [Google Scholar] [CrossRef]
- van der Gulik, P.; Massar, S.; Gilis, D.; Buhrman, H.; Rooman, M. The first peptides: The evolutionary transition between prebiotic amino acids and early proteins. J. Theor. Biol. 2009, 261, 531–539. [Google Scholar] [CrossRef]
- Vauthey, S.; Santoso, S.; Gong, H.; Watson, N.; Zhang, S. Molecular self-assembly of surfactant-like peptides to form nanotubes and nanovesicles. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 5355–5360. [Google Scholar] [CrossRef]
- Fishkis, M. Steps towards the formation of a protocell: the possible role of short peptides. Orig. Life Evol. Biosph. 2007, 37, 537–553. [Google Scholar] [CrossRef]
- Zhang, S. Plausible lipid-like peptides: prebiotic molecular self-assembly in water. In Fitness of the Cosmos for Life: Biochemistry and Fine-Tuning; Barrow, J.D., Morris, S.C., Freeland, S.J., Harper, C.L., Jr., Eds.; Cambridge University Press: Cambridge, UK, 2008; pp. 440–455. [Google Scholar]
- Egel, R. Peptide-dominated membranes preceding the genetic takeover by RNA: latest thinking on a classic controversy. BioEssays 2009, 31, 1100–1109. [Google Scholar] [CrossRef]
- Fishkis, M. Emergence of self-reproduction in cooperative chemical evolution of prebiological molecules. Orig. Life Evol. Biosph. 2011, 41, 261–275. [Google Scholar] [CrossRef]
- Dyson, F.J. A model for the origin of life. J. Mol. Evol. 1982, 18, 344–350. [Google Scholar] [CrossRef]
- Kauffman, S.A. The Origins of Order; Oxford University Press: Oxford, UK, 1993. [Google Scholar]
- Hordijk, W.; Kauffman, S.A.; Steel, M. Required levels of catalysis for emergence of autocatalytic sets in models of chemical reaction systems. Int. J. Mol. Sci. 2011, 12, 3085–3101. [Google Scholar] [CrossRef]
- de Duve, C. Selection by differential molecular survival: A possible mechanism of early chemical evolution. Proc. Natl. Acad. Sci. U. S. A. 1987, 84, 8253–8256. [Google Scholar] [CrossRef]
- Davidovich, C.; Belousoff, M.; Bashan, A.; Yonath, A. The evolving ribosome: from non-coded peptide bond formation to sophisticated translation machinery. Res. Microbiol. 2009, 160, 487–492. [Google Scholar] [CrossRef]
- Belousoff, M.J.; Davidovich, C.; Zimmerman, E.; Caspi, Y.; Wekselman, I.; Rozenszajn, L.; Shapira, T.; Sade-Falk, O.; Taha, L.; Bashan, A.; et al. Ancient machinery embedded in the contemporary ribosome. Biochem. Soc. Trans. 2010, 38, 422–427. [Google Scholar] [CrossRef]
- Weinger, J.S.; Parnell, K.M.; Dorner, S.; Green, R.; Strobel, S.A. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat. Struct. Mol. Biol. 2004, 11, 1101–1106. [Google Scholar] [CrossRef]
- Steitz, T.A. A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell Biol. 2008, 9, 242–253. [Google Scholar] [CrossRef]
- Woese, C. Molecular mechanics of translation: A reciprocating ratchet mechanism. Nature 1970, 226, 817–820. [Google Scholar] [CrossRef]
- Zhang, W.; Dunkle, J.A.; Cate, J.H.D. Structures of the ribosome in intermediate states of ratcheting. Science 2009, 325, 1014–1017. [Google Scholar] [CrossRef]
- Schmeing, T.M.; Huang, K.S.; Strobel, S.A.; Steitz, T.A. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature 2005, 438, 520–524. [Google Scholar] [CrossRef]
- Agmon, I.; Bashan, A.; Zarivach, R.; Yonath, A. Symmetry at the active site of the ribosome: structural and functional implications. Biol. Chem. 2005, 386, 833–844. [Google Scholar] [CrossRef]
- Luisi, P.L.; Walde, P.; Oberholzer, T. Lipid vesicles as possible intermediates in the origin of life. Curr. Opin. Colloid Interface Sci. 1999, 4, 33–39. [Google Scholar] [CrossRef]
- Segré, D.; Ben-Eli, D.; Deamer, D.W.; Lancet, D. The Lipid World. Orig. Life Evol. Biosph. 2001, 31, 119–145. [Google Scholar] [CrossRef]
- Budin, I.; Szostak, J.W. Expanding roles for diverse physical phenomena during the origin of life. Annu. Rev. Biophys. 2010, 39, 245–263. [Google Scholar] [CrossRef]
- Koch, A.L.; Silver, S. The first cell. Adv. Microb. Physiol. 2005, 50, 227–259. [Google Scholar] [CrossRef]
- Cairns-Smith, A.G. Genetic Takeover and the Mineral Origins of Life; Cambridge University Press: Cambridge, UK, 1982. [Google Scholar]
- Baaske, P.; Weinert, F.M.; Duhr, S.; Lemke, K.H.; Russell, M.J.; Braun, D. Extreme accumulation of nucleotides in simulated hydrothermal pore systems. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 9346–9351. [Google Scholar]
- Branciamore, S.; Gallori, E.; Szathmary, E.; Czaran, T. The origin of life: Chemical evolution of a metabolic system in a mineral honeycomb? J. Mol. Evol. 2009, 69, 458–469. [Google Scholar] [CrossRef] [Green Version]
- Hansma, H.G. Possible origin of life between mica sheets. J. Theor. Biol. 2010, 266, 175–188. [Google Scholar] [CrossRef]
- Maynard Smith, J.; Szathmáry, E. The Major Transitions in Evolution; Freeman: Oxford, UK, 1995. [Google Scholar]
- Trevors, J.T.; Pollack, G.H. Hypothesis: The origin of life in a hydrogel environment. Prog. Biophys. Mol. Biol. 2005, 89, 1–8. [Google Scholar] [CrossRef]
- Spitzer, J.; Poolman, B. The role of biomacromolecular crowding, ionic strength, and physicochemical gradients in the complexities of life’s emergence. Microbiol. Mol. Biol. Rev. 2009, 73, 371–388. [Google Scholar] [CrossRef]
- Trevors, J.T. Hypothesized origin of microbial life in a prebiotic gel and the transition to a living biofilm and microbial mats. C. R. Biologies 2011, 334, 269–272. [Google Scholar] [CrossRef]
- Pollack, G.H.; Figueroa, X.; Zhao, Q. The minimal cell and life’s origin: Role of water and aqueous interfaces. In The Minimal Cell: The Biophysics of Cell Compartment and the Origin of Cell Functionality; Luisi, P.L., Stano, P., Eds.; Springer: Dordrecht, The Netherlands, 2011. [Google Scholar]
- Griffiths, G. Cell evolution and the problem of membrane topology. Nat. Rev. Mol. Cell Biol. 2007, 8, 1018–1024. [Google Scholar] [CrossRef]
- Sobolewski, A.L.; Domcke, W. On the mechanism of rapid non-radiative decay in intramolecularly hydrogen-bonded π systems. Chem. Phys. Lett. 1999, 300, 533–539. [Google Scholar] [CrossRef]
- Sobolewski, A.L.; Domcke, W. The chemical physics of the photostability of life. Europhys. News 2006, 37, 20–23. [Google Scholar] [CrossRef]
- Shemesh, D.; Hattig, C.; Domcke, W. Photophysics of the Trp-Gly dipeptide: Role of electron and proton transfer processes for efficient excited-state deactivation. Chem. Phys. Lett. 2009, 482, 38–43. [Google Scholar] [CrossRef]
- Mulkidjanian, A.Y.; Cherepanov, D.A.; Galperin, M.Y. Survival of the fittest before the beginning of life: Selection of the first oligonucleotide-like polymers by UV light. BMC Evol. Biol. 2003, 3, 12:1–12:7. [Google Scholar]
- Brack, A.; Barbier, B. Chemical activity of simple basic peptides. Orig. Life. Evol. Biosph. 1990, 20, 139–144. [Google Scholar] [CrossRef]
- Braun, S.; Humphreys, C.; Fraser, E.; Brancale, A.; Bochtler, M.; Dale, T.C. Amyloid-associated nucleic acid hybridisation. PLoS One 2011, 6, e19125:1–e19125:11. [Google Scholar]
- Hoffmann, M.R.; Martin, S.T.; Choi, W.; Bahnemann, D.W. Environmental applications of semiconductor photocatalysis. Chem. Rev. 1995, 95, 69–96. [Google Scholar]
- Mulkidjanian, A.Y.; Galperin, M.Y. Evolutionary origins of membrane proteins. In Structural Bioinformatics of Membrane Proteins; Frishman, D., Ed.; Springer: Wien, Austria, 2010; pp. 1–28. [Google Scholar]
- Takagi, M.; Goto, S.; Matsuda, T. Photo-reaction of lipoic acid and related organic disulphides: reductive acylation with aldehydes. J. Chem. Soc. Chem. Commun. 1976, 92–93. [Google Scholar]
- Weber, A.L. Formation of the thioester, N,S-diacetylcysteine, from acetaldehyde and N,N'- diacetylcystine in aqueous solution with ultraviolet light. J. Mol. Evol. 1981, 17, 103–107. [Google Scholar] [CrossRef]
- Di Sabato, G.; Jencks, W.P. Mechanism and catalysis of acyl phosphates II. Hydrolysis. J. Am. Chem. Soc. 1961, 83, 4400–4405. [Google Scholar] [CrossRef]
- Weber, A.L. Formation of pyrophosphate, tripolyphosphate, and phosphorylimidazole with the thioester, N,S-diacetylcysteamine, as the condensing agent. J. Mol. Evol. 1981, 18, 24–29. [Google Scholar] [CrossRef]
- Weber, A.L. Formation of pyrophosphate on hydroxyapatite with thioesters as condensing agents. BioSystems 1982, 15, 183–189. [Google Scholar]
- Wächtershäuser, G. Evolution of the first metabolic cycles. Proc. Natl. Acad. Sci. U. S. A. 1990, 87, 200–204. [Google Scholar] [CrossRef]
- de Duve, C. Clues from present-day biology: the thioester world. In The Molecular Origins of Life; Brack, A., Ed.; Cambridge Univ Press: Cambridge, UK, 1998; pp. 219–236. [Google Scholar]
- Maheen, G.; Tian, G.; Wang, Y.; He, C.; Shi, Z.; Yuan, H.; Feng, S. Resolving the enigma of prebiotic C-O-P bond formation: Prebiotic hydrothermal synthesis of important biological phosphate esters. Heteroatom Chem. 2010, 21, 161–167. [Google Scholar]
- Maheen, G.; Wang, Y.; Wang, Y.; Shi, Z.; Tian, G.; Feng, S. Mimicking the prebiotic acidic hydrothermal environment: One-pot prebiotic hydrothermal synthesis of glucose phosphates. Heteroatom Chem. 2011, 22, 186–191. [Google Scholar] [CrossRef]
- Powner, M.W.; Gerland, B.; Sutherland, J.D. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature 2009, 459, 239–242. [Google Scholar] [CrossRef]
- Szostak, J.W. Origins of life: systems chemistry on early Earth. Nature 2009, 459, 171–172. [Google Scholar] [CrossRef]
- Hagan, W.J., Jr. Uracil-catalyzed synthesis of acetyl phosphate: a photochemical driver for protometabolism. ChemBioChem 2010, 11, 383–387. [Google Scholar] [CrossRef]
- Conrad, M. The geometry of evolution. BioSystems 1990, 24, 61–81. [Google Scholar] [CrossRef]
- Woese, C.R. Evolution of the genetic code. Proc. Natl. Acad. Sci. U. S. A. 1973, 54, 1546–1552. [Google Scholar] [CrossRef]
- Wong, J.T.-F. Evolution of the genetic code. Microbiol. Sci. 1988, 5, 164–181. [Google Scholar]
- Davis, B.K. Evolution of the genetic code. Progr. Biophys. Mol. Biol. 1999, 72, 157–243. [Google Scholar] [CrossRef]
- Koonin, E.V.; Novozhilov, A.S. Origin and evolution of the genetic code: the universal enigma. IUBMB Life 2009, 61, 99–111. [Google Scholar] [CrossRef]
- Rodin, A.S.; Szathmáry, E.; Rodin, S.N. One ancestor for two codes viewed from the perspective of two complementary modes of tRNA aminoacylation. Biol. Direct 2009, 4, 4–1. [Google Scholar]
- Johnson, D.B.F.; Wang, L. Imprints of the genetic code in the ribosome. Proc. Natl. Acad. Sci.U. S. A. 2010, 107, 8298–8303. [Google Scholar] [CrossRef]
- Freeland, S.J.; Knight, R.D.; Landweber, L.F.; Hurst, L.D. Early fixation of an optimal genetic code. Mol. Biol. Evol. 2000, 17, 511–518. [Google Scholar] [CrossRef]
- Itzkovitz, S.; Alon, U. The genetic code is nearly optimal for allowing additional information within protein-coding sequences. Genome Res. 2007, 17, 405–412. [Google Scholar] [CrossRef]
- Butler, T.; Goldenfeld, N.; Mathew, D.; Luthey-Schulten, Z. Extreme genetic code optimality from a molecular dynamics calculation of amino acid polar requirement. Phys. Rev. E 2009, 79, 060901:1–060901:4. [Google Scholar]
- Vetsigian, K.; Woese, C.R.; Goldenfeld, N. Collective evolution and the genetic code. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 10696–10701. [Google Scholar] [CrossRef]
- Woese, C. The universal ancestor. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 6854–6859. [Google Scholar] [CrossRef]
- Calabretta, R.; Nolfi, S.; Parisi, D.; Wagner, P. A case study of the evolution of modularity: towards a bridge between evolutionary biology, artificial life, neuro- and cognitive science. In Artificial Life VI: Proceedings of the Sixth International Conference on Artificial Life; Adami, C., Belew, R.K., Kitano, H., Taylor, C.E., Eds.; MIT Press: Cambridge, MA, USA, 1998; pp. 275–284. [Google Scholar]
- Dauscher, P.; Uthmann, T. Self-organized modularization in evolutionary algorithms. Evolut. Comput. 2005, 13, 303–328. [Google Scholar] [CrossRef]
- Polani, D.; Dauscher, P.; Uthmann, T. On a quantitative measure for modularity based on information theory. In Advances in Artificial Life. ECAL 2005; Capcarrère, M.S., Freitas, A.A., Bentley, P.J., Johnson, C.G., Timmis, J., Eds.; Springer-Verlag: Heidelberg, Germany, 2005; pp. 393–402. [Google Scholar]
- Schimmel, P. Origin of genetic code: A needle in the haystack of tRNA sequences. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 4521–4522. [Google Scholar] [CrossRef]
- Rodin, A.S.; Szathmáry, E.; Rodin, S.N. On origin of genetic code and tRNA before translation. Biol. Direct 2011, 6, 14–24. [Google Scholar] [CrossRef]
- Modularity: Understanding the Development and Evolution of Natural Complex Systems; Callebaut, W.; Rasskin-Gutman, D. (Eds.) MIT Press: Cambridge, MA, USA, 2005.
- Pereira-Leal, J.B.; Levy, E.D.; Teichmann, S.A. The origins and evolution of functional modules: lessons from protein complexes. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2006, 361, 507–517. [Google Scholar] [CrossRef]
- Bonabeau, E.; Dorigo, M.; Theraulaz, G. Swarm intelligence: From natural to artificial systems; Oxford University Press: Oxford, UK, 1999. [Google Scholar]
- Chen, H.; Zhu, Y.; Hu, K.; He, X. Hierarchical swarm model: A new approach to optimization. Discrete Dyn. Nat. Soc. 2010, 2010, 379649:1–379649:30. [Google Scholar]
- Sun, J.; Deem, M. Spontaneous emergence of modularity in a model of evolving individuals. Phys. Rev. Lett. 2007, 99, 228107–1. [Google Scholar]
- Goldenfeld, N.; Woese, C. Biology's next revolution. Nature 2007, 445, 369. [Google Scholar] [CrossRef]
- Cohen, E.; Kessler, D.A.; Levine, H. Recombination dramatically speeds up evolution of finite populations. Phys. Rev. Lett. 2005, 94, 098102:1–098102:5. [Google Scholar]
- Reanney, D.C. Genetic error and genome design. Trends Genet. 1986, 2, 41–46. [Google Scholar] [CrossRef]
- Lehman, N. A case for the extreme antiquity of recombination. J. Mol. Evol. 2003, 56, 770–777. [Google Scholar] [CrossRef]
- Vogan, A.A.; Higgs, P.G. The advantages and disadvantages of horizontal gene transfer and the emergence of the first species. Biology Direct 2011, 6, 1–14. [Google Scholar] [CrossRef]
- Lehman, N.; Unrau, P.J. Recombination during in vitro evolution. J. Mol. Evol. 2005, 61, 245–252. [Google Scholar] [CrossRef]
- Lehman, N. A recombination-based model for the origin and early evolution of genetic information. Chem. Biodivers. 2008, 5, 1707–1717. [Google Scholar] [CrossRef]
- Lankenau, D.H. The legacy of the germ line-maintaining sex and life in metazoans: Cognitive roots of the concept of hierarchical selection. In Recombination and Meiosis-Models, Means and Evolution; Egel, R., Lankenau, D.H., Eds.; Springer-Verlag: Heidelberg, Germany, Genome Dyn. Stab. 2007, 3, 289-339.
- Fedorov, A.; Fedorova, L. Introns: mighty elements from the RNA world. J. Mol. Evol. 2004, 59, 718–721. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Poole, A.M.; Penny, D. Relics from the RNA world. J. Mol. Evol. 1998, 46, 18–36. [Google Scholar] [CrossRef]
- Penny, D.; Hoeppner, M.P.; Poole, A.M.; Jeffares, D.C. An overview of the introns-first theory. J. Mol. Evol. 2009, 69, 527–540. [Google Scholar] [CrossRef]
- Kooter, J.M.; de Lange, T.; Borst, P. Discontinuous synthesis of mRNA in trypanosomes. EMBO J. 1984, 3, 2387–2392. [Google Scholar]
- Günzl, A. The pre-mRNA splicing machinery of trypanosomes: complex or simplified? Eukaryot. Cell 2010, 9, 1159–1170. [Google Scholar] [CrossRef]
- Dorit, R.L.; Schoenbacher, L.; Gilbert, W. How big is the universe of exons? Science 1990, 250, 1377–1382. [Google Scholar]
- Gilbert, W.; de Souza, S.J.; Long, M. Origin of genes. Proc. Natl. Acad. Sci. U. S. A. 1997, 94, 7698–7703. [Google Scholar]
- Patthy, L. Genome evolution and the evolution of exon-shuffling-a review. Gene 1999, 238, 103–114. [Google Scholar] [CrossRef]
- Roy, S.W.; Gilbert, W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat. Rev. Genet. 2006, 7, 211–221. [Google Scholar]
- Siefert, J.L.; Martin, K.A.; Abdi, F.; Widger, WR.; Fox, G.E. Conserved gene clusters in bacterial genomes provide further support for the primacy of RNA. J. Mol. Evol. 1997, 45, 467–472. [Google Scholar] [CrossRef]
- Dandekar, T.; Snel, B.; Huynen, M.; Bork, P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci. 1998, 23, 324–328. [Google Scholar] [CrossRef]
- Glansdorff, N. On the origin of operons and their possible role in evolution toward thermophily. J. Mol. Evol. 1999, 49, 432–438. [Google Scholar] [CrossRef]
- Fondi, M.; Emiliani, G.; Fani, R. Origin and evolution of operons and metabolic pathways. Res. Microbiol. 2009, 160, 502–512. [Google Scholar] [CrossRef]
- Emiliani, G.; Fondi, M.; Liò, P.; Fani, R. Evolution of metabolic pathways and evolution of genomes. In Geomicrobiology, Molecular and Environmental Perspective.; Barton, L.L., Mandl, M., Loy, A., Eds.; Springer: Dordrecht, NL, 2010; pp. 37–68. [Google Scholar]
- Møller-Jensen, J.; Jensen, R.B.; Gerdes, K. Plasmid and chromosome segregation in prokaryotes. Trends Microbiol. 2000, 8, 313–320. [Google Scholar] [CrossRef]
- Yanagida, M. Basic mechanism of eukaryotic chromosome segregation. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2005, 360, 609–621. [Google Scholar] [CrossRef]
- Dye, N.A.; Shapiro, L. The push and pull of the bacterial cytoskeleton. Trends Cell Biol. 2007, 17, 239–245. [Google Scholar] [CrossRef]
- Blackburn, E.H. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 2005, 579, 859–862. [Google Scholar] [CrossRef]
- Blackburn, E.H.; Collins, K. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb. Perspect. Biol. 2010, 2, a003558:1–a003558:9. [Google Scholar]
- Khan, S.A. Rolling-circle replication of bacterial plasmids. Microbiol. Mol. Biol. Rev. 1997, 61, 442–455. [Google Scholar]
- Fiore-Donno, A.-M.; Berney, C.; Pawlowski, J.; Baldauf, S.L. Higher-order phylogeny of plasmodial slime molds (Myxogastria) based on elongation factor-1 a and small subunit rRNA gene sequences. J. Eukaryot. Microbiol. 2005, 52, 1–10. [Google Scholar] [CrossRef]
- Hoppe, T.; Kutschera, U. In the shadow of Darwin: Anton deBary's origin of myxomycetology and a molecular phylogeny of the plasmodial slime molds. Theory Biosci. 2010, 129, 15–23. [Google Scholar] [CrossRef]
- Egel, R.; Penny, D. On the origin of meiosis in eukaryotic evolution: Coevolution of meiosis and mitosis from feeble beginnings. In Recombination and Meiosis-Models, Means and Evolution; Egel, R., Lankenau, D.H., Eds.; Springer-Verlag: Heidelberg, Germany, Genome Dyn. Stab. 2007, 3, 249-388.
- Woese, C.R.; Fox, G. The concept of cellular evolution. J. Mol. Evol. 1977, 10, 1–6. [Google Scholar] [CrossRef]
- Kandler, O. The early diversification of life. In Early Life on Earth: Nobel Symposium 84; Bengtson, S., Ed.; Columbia University Press: New York, NY, USA, 1994; pp. 152–160. [Google Scholar]
- Kandler, O. Cell wall biochemistry in Archaea and its phylogenetic implications. J. Biol. Phys. 1994, 20, 165–169. [Google Scholar] [CrossRef]
- Kandler, O. Cell wall biochemistry and three-domain concept of life. System. Appl. Microbiol. 1994, 16, 501–509. [Google Scholar] [CrossRef]
- Moreira, D.; López-García, P. The last common ancestor of modern cells. In Lectures in astrobiology; Gargaud, M., Martin, H., Claeys, P., Eds.; Springer-Verlag: Berlin, Germany, 2007; Volume II, Adv. Astrobiol. Biogeophys. pp. 305-317. [Google Scholar]
- Blobel, G. Intracellular protein topogenesis. Proc. Natl. Acad. Sci. U. S. A. 1980, 77, 1496–1500. [Google Scholar] [CrossRef]
- Cavalier-Smith, T. Obcells as proto-organisms: Membrane heredity, lithophosphorylation, and the origins of the genetic code, the first cells, and photosynthesis. J. Mol. Evol. 2001, 53, 555–595. [Google Scholar] [CrossRef]
- Collins, O.R.; Haskins, E.F. Genetics of somatic fusion in Physarum polycephalum: the PpII strain. Genetics 1972, 71, 63–71. [Google Scholar]
- Woldringh, C.L.; Nanninga, N. Structural and physical aspects of bacterial chromosome segregation. J. Struct. Biol. 2006, 156, 273–283. [Google Scholar] [CrossRef]
- Iborra, F.J. Can visco-elastic phase separation, macromolecular crowding and colloidal physics explain nuclear organisation? Theor. Biol. Med. Model. 2007, 12, 4–15. [Google Scholar]
- Rippe, K. Dynamic organization of the cell nucleus. Curr. Opin. Genet. Dev. 2007, 17, 373–380. [Google Scholar] [CrossRef]
- Lake, J.A.; Rivera, M.C. Was the nucleus the first endosymbiont? Proc. Natl. Acad. Sci. U. S. A. 1994, 91, 2880–2881. [Google Scholar] [CrossRef]
- Güttinger, S.; Laurell, E.; Kutay, U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 2009, 10, 178–191. [Google Scholar] [CrossRef]
- de Roos, A.D.G. The origin of the eukaryotic cell based on conservation of existing interfaces. Artif. Life 2006, 12, 513–523. [Google Scholar] [CrossRef]
- Martin, W. Archaebacteria (Archaea) and the origin of the eukaryotic nucleus. Curr. Opin. Microbiol. 2005, 8, 630–637. [Google Scholar] [CrossRef]
- Sanders, I.R.; Croll, D. Arbuscular mycorrhiza: The challenge to understand the genetics of the fungal partner. Annu. Rev. Genet. 2010, 44, 271–292. [Google Scholar] [CrossRef]
- Jany, J.L.; Pawlowska, T.E. Multinucleate spores contribute to evolutionary longevity of asexual Glomeromycota. Amer. Naturalist 2010, 175, 424–435. [Google Scholar] [CrossRef]
- Koonin, E.; Martin, W. On the origin of genomes and cells within inorganic compartments. Trends Genet. 2005, 21, 647–654. [Google Scholar] [CrossRef]
- Lane, N.; Allen, J.F.; Martin, W. How did LUCA make a living? Chemiosmosis in the origin of life. BioEssays 2010, 32, 271–280. [Google Scholar] [CrossRef]
- Fischer, H.M. Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 1994, 58, 352–386. [Google Scholar]
- Poole, A.M.; Jeffares, D.C.; Penny, D. Prokaryotes, the new kids on the block. BioEssays 1999, 21, 880–889. [Google Scholar] [CrossRef]
- Bapteste, E.; O'Malley, M.A.; Beiko, R.G.; Ereshefsky, M.; Gogarten, J.P.; Franklin-Hall, L.; Lapointe, F.J.; Dupré, J.; Dagan, T.; Boucher, Y.; Martin, W. Prokaryotic evolution and the tree of life are two different things. Biol. Direct 2009, 4, 34:1–34:20. [Google Scholar]
- Cavalier-Smith, T. Origin of the cell nucleus, mitosis and sex: roles of intracellular coevolution. Biol. Direct 2010, 5, 7–78. [Google Scholar] [CrossRef]
- Schopf, J.W.; Packer, B.M. Early Archean (3.3 billion to 3.5 billion-year-old) microfossils from Warrawoona group, Australia. Science 1987, 237, 70–73. [Google Scholar]
- Forterre, P. Thermoreduction, a hypothesis for the origin of prokaryotes. C. R. Acad. Sci. III. 1995, 318, 415–422. [Google Scholar]
- Embley, M.T.; Martin, W. Eukaryotic evolution, changes and challenges. Nature 2006, 440, 623–630. [Google Scholar] [CrossRef]
- Martin, W.; Dagan, T.; Koonin, E.V.; Dipippo, J.L.; Gogarten, J.P.; Lake, J.A. The evolution of eukaryotes. Science 2007, 316, 542–543, author reply 543. [Google Scholar]
- Gribaldo, S.; Poole, A.M.; Daubin, V.; Forterre, P.; Brochier-Armanet, C. The origin of eukaryotes and their relationship with the Archaea: are we at a phylogenomic impasse? Nat. Rev. Microbiol. 2010, 8, 743–752. [Google Scholar] [CrossRef]
- Dagan, T.; Roettger, M.; Bryant, D.; Martin, W. Genome Networks Root the Tree of Life between Prokaryotic Domains. Genome Biol. Evol. 2010, 2, 379–392. [Google Scholar] [CrossRef]
- Poole, A.; Penny, D. Eukaryote evolution: engulfed by speculation. Nature 2007, 447, 913. [Google Scholar] [CrossRef]
- Javaux, E.J.; Marshall, C.P.; Bekker, A. Organic-walled microfossils in 3.2-billion-year-old shallow-marine siliciclastic deposits. Nature 2010, 463, 934–938. [Google Scholar] [CrossRef]
- David, L.A.; Alm, E.J. Rapid evolutionary innovation during an Archaean Genetic Expansion. Nature 2010, 469, 93–96. [Google Scholar] [CrossRef]
- Fuerst, J.A. Intracellular compartmentation in planctomycetes. Annu. Rev. Microbiol. 2005, 59, 299–328. [Google Scholar] [CrossRef]
- Forterre, P.; Gribaldo, S. Bacteria with a eukaryotic touch: a glimpse of ancient evolution? Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 12739–12740. [Google Scholar] [CrossRef]
- McInerney, J.O.; Martin, W.F.; Koonin, E.V.; Allen, J.F.; Galperin, M.Y.; Lane, N.; Archibald, J.M.; Embley, T.M. Planctomycetes and eukaryotes: A case of analogy not homology. BioEssays 2011, 33, 810–817. [Google Scholar] [CrossRef]
- Fuerst, J.A.; Sagulenko, E. Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat. Rev. Microbiol. 2011, 9, 403–413. [Google Scholar] [CrossRef]
- Pace, N.R. Time for a change. Nature 2006, 441, 289. [Google Scholar] [CrossRef]
- Harris, J.K.; Kelley, S.T.; Spiegelman, G.B.; Pace, N.R. The genetic core of the universal ancestor. Genome Res. 2003, 13, 407–412. [Google Scholar] [CrossRef]
- Puigbò, P.; Wolf, Y.I.; Koonin, E.V. Search for a 'Tree of Life' in the thicket of the phylogenetic forest. J. Biol. 2009, 8, 59:1–59:17. [Google Scholar]
- Goldman, A.D.; Samudrala, R.; Baross, J.A. The evolution and functional repertoire of translation proteins following the origin of life. Biol. Direct 2010, 5, 15:1–15:12. [Google Scholar]
- Dagan, T.; Martin, W. The tree of one percent. Genome Biol. 2006, 7, 118:1–118:7. [Google Scholar]
- Bapteste, E.; Boucher, Y. Lateral gene transfer challenges principles of microbial systematics. Trends Microbiol. 2008, 16, 200–207. [Google Scholar] [CrossRef]
- Puigbò, P.; Wolf, Y.I.; Koonin, E.V. The tree and net components of prokaryote evolution. Genome Biol. Evol. 2010, 2, 745–756. [Google Scholar] [CrossRef]
- Raoult, D. The post-Darwinist rhizome of life. Lancet 2010, 375, 104–105. [Google Scholar] [CrossRef]
- Beauregard-Racine, J.; Bicep, C.; Schliep, K.; Lopez, P.; Lapointe, F.J.; Bapteste, E. Of Woods and Webs: Possible alternatives to the tree of life for studying genomic fluidity in E. coli. Biol. Direct 2011, 6, 39:1–39:21. [Google Scholar]
- Marcet-Houben, M.; Gabaldon, T. Acquisition of prokaryotic genes by fungal genomes. Trends Genet. 2010, 26, 5–8. [Google Scholar] [CrossRef]
- Doolittle, W.F. You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 1998, 14, 307–311. [Google Scholar] [CrossRef]
- Martin, W.; Rujan, T.; Richly, E.; Hansen, A.; Cornelsen, S.; Lins, T.; Leister, D.; Stoebe, B.; Hasegawa, M.; Penny, D. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Nat. Acad. Sci. U. S. A. 2002, 99, 12246–12251. [Google Scholar]
- Dacks, J.; Roger, A.J. The first sexual lineage and the relevance of facultative sex. J. Mol. Evol. 1999, 48, 779–783. [Google Scholar] [CrossRef]
- Logsdon, J.M.J. Evolutionary genetics: sex happens in Giardia. Curr. Biol. 2008, 18, R66–68. [Google Scholar] [CrossRef]
- Wilkins, A.S.; Holliday, R. The evolution of meiosis from mitosis. Genetics 2009, 181, 3–12. [Google Scholar] [CrossRef]
- Carlile, M. Prokaryotes and eukaryotes: strategies and successes. Trends Biochem. Sci. 1982, 7, 128–130. [Google Scholar] [CrossRef]
- Cavalier-Smith, T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 2002, 52, 297–354. [Google Scholar]
- Hartman, H.; Fedorov, A. The origin of the eukaryotic cell: a genomic investigation. Proc. Natl. Acad. Sci. USA 2002, 99, 1420–1425. [Google Scholar] [CrossRef]
- Poole, A. Eukaryote evolution: the importance of the stem group. In Evolutionary Genomics and Systems Biology; Caetano-Anollés, G., Ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2010; pp. 63–80. [Google Scholar]
- Poole, A.M.; Neumann, N. Reconciling an archaeal origin of eukaryotes with engulfment: A biologically plausible update of the Eocyte hypothesis. Res. Microbiol. 2011, 162, 71–76. [Google Scholar] [CrossRef]
- de Nooijer, S.; Holland, B.R.; Penny, D. The emergence of predators in early life: there was no Garden of Eden. PLoS One 2009, 4, e5507:1–e5507:10. [Google Scholar]
- Lonhienne, T.G.; Sagulenko, E.; Webb, R.I.; Lee, K.C.; Franke, J.; Devos, D.P.; Nouwens, A.; Carroll, B.J.; Fuerst, J.A. Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 12883–12888. [Google Scholar]
- Jékely, G. Origin of phagotrophic eukaryotes as social cheaters in microbial biofilms. Biol. Direct 2007, 2, 3:1–3:15. [Google Scholar]
- Gross, J.; Bhattacharya, D. Uniting sex and eukaryote origins in an emerging oxygenic world. Biol. Direct 2010, 5, 53:1–53:20. [Google Scholar]
- Poole, A.M.; Phillips, M.J.; Penny, D. Prokaryote and eukaryote evolvability. BioSystems 2003, 69, 163–185. [Google Scholar] [CrossRef]
- Kurland, C.G.; Collins, L.J.; Penny, D. Genomics and the irreducible nature of eukaryote cells. Science 2006, 312, 1011–1014. [Google Scholar]
- Poole, A.M. Did group II intron proliferation in an endosymbiont-bearing archaeon create eukaryotes? Biol. Direct 2006, 1, 36:1–36:6. [Google Scholar]
- Martin, W.; Koonin, E.V. Introns and the origin of nucleus-cytosol compartmentation. Nature 2006, 440, 41–45. [Google Scholar] [CrossRef]
- Koonin, E.V. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 2010, 11, 209–1. [Google Scholar]
- Stetter, K.O. The lesson of archaebacteria. In Early Life on Earth: Nobel Symposium 84; Bengtson, S., Ed.; Columbia University Press: New York, NY, USA, 1994; pp. 143–151. [Google Scholar]
- Stetter, K.O. Hyperthermophiles in the history of life. Phil. Trans. R. Soc. Lond. B 2006, 361, 1837–1843. [Google Scholar] [CrossRef]
- Levy, M.; Miller, S.L. The stability of the RNA bases: implications for the origin of life. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 7933–7938. [Google Scholar] [CrossRef]
- Moulton, V.; Gardner, P.P.; Pointon, R.F.; Creamer, L.K.; Jameson, G.B.; Penny, D. RNA folding argues against a hot-start origin of life. J. Mol. Evol. 2000, 51, 416–421. [Google Scholar]
- Galtier, N.; Tourasse, N.; Gouy, M. A nonhyperthermophilic common ancestor to extant life forms. Science 1999, 283, 220–221. [Google Scholar]
- Forterre, P.; Confalonieri, F.; Charbonnier, F.; Duguet, M. Speculations on the origin of life and thermophily: review of available information on reverse gyrase suggests that hyperthermophilic procaryotes are not so primitive. Orig. Life Evol. Biosph. 1995, 25, 235–249. [Google Scholar] [CrossRef]
- Forterre, P. A hot topic: the origin of hyperthermophiles. Cell 1996, 85, 789–792. [Google Scholar] [CrossRef]
- Boussau, B.; Blanquart, S.; Necsulea, A.; Lartillot, N.; Gouy, M. Parallel adaptations to high temperatures in the Archaean eon. Nature 2008, 456, 942–945. [Google Scholar] [CrossRef]
- Poole, A.M.; Jeffares, D.C.; Penny, D. The path from the RNA world. J. Mol. Evol. 1998, 46, 1–17. [Google Scholar] [CrossRef]
- Penny, D.; Poole, A. The nature of the last universal common ancestor. Curr. Opin. Genet. Dev. 1999, 9, 672–677. [Google Scholar] [CrossRef]
- Poole, A.M.; Penny, D. Evaluating hypotheses for the origin of eukaryotes. BioEssays 2007, 29, 74–84. [Google Scholar] [CrossRef]
- Collins, L.; Penny, D. Complex spliceosomal organization ancestral to extant eukaryotes. Mol. Biol. Evol. 2005, 22, 1053–1066. [Google Scholar] [CrossRef]
- Collins, L.J.; Penny, D. The RNA infrastructure: dark matter of the eukaryotic cell? Trends Genet. 2009, 25, 120–128. [Google Scholar]
- Collins, L.J.; Kurland, C.G.; Biggs, P.; Penny, D. The modern RNP world of eukaryotes. J. Hered. 2009, 100, 597–604. [Google Scholar] [CrossRef]
- Cech, T.R. The generality of self-splicing RNA: Relationship to nuclear mRNA splicing. Cell 1986, 44, 207–210. [Google Scholar] [CrossRef]
- Copertino, D.W.; Hallick, R.B. Group II and group III introns of twintrons: potential relationships with nuclear pre-mRNA introns. Trends Biochem. Sci. 1993, 18, 467–471. [Google Scholar] [CrossRef]
- Bonen, L.; Vogel, J. The ins and outs of group II introns. Trends Genet. 2001, 6, 322–331. [Google Scholar] [CrossRef]
- Wang, M.; Yafremava, L.S.; Caetano-Anollés, D.; Mittenthal, J.E.; Caetano-Anollés, G. Reductive evolution of architectural repertoires in proteomes and the birth of the tripartite world. Genome Res. 2007, 17, 1572–1585. [Google Scholar] [CrossRef]
- Wang, M.; Caetano-Anollés, G. The evolutionary mechanics of domain organization in proteomes and the rise of modularity in the protein world. Structure 2009, 17, 66–78. [Google Scholar] [CrossRef]
- Makarova, K.S.; Yutin, N.; Bell, S.D.; Koonin, E.V. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat. Rev. Microbiol. 2010, 8, 731–741. [Google Scholar] [CrossRef]
- Wickstead, B.; Gull, K. The evolution of the cytoskeleton. J. Cell Biol. 2011, 194, 513–525. [Google Scholar] [CrossRef]
- Roberts, E.; Sethi, A.; Montoya, J.; Woese, C.R.; Luthey-Schulten, Z. Molecular signatures of ribosomal evolution. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 13953–13958. [Google Scholar]
- Vishwanath, P.; Favaretto, P.; Hartman, H.; Mohr, F.C.; Smith, T.F. Ribosomal protein-sequence block structure suggests complex prokaryotic evolution with implications for the origin of eukaryotes. Mol. Phylogenet. Evol. 2004, 33, 615–625. [Google Scholar] [CrossRef]
- Kirschvink, J.L.,; Gaidos, E.J.; Bertani, L.E.; Beukes, N.J.; Gutzmer, J.; Maepa, L.N.; Steinberger, R.E. Paleoproterozoic snowball Earth: extreme climatic and geochemical global change and its biological consequences. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 1400–1405. [Google Scholar]
- Bengston, S. Origins and early evolution of predation. In The fossil record of predation; Kowalewski, M., Kelley, P.H., Eds.; The Paleontological Society: Boulder, CO, USA, 2002; (Paleontol. Soc. Papers 8), pp. 289-317. [Google Scholar]
- Brocks, J.J.; Logan, G.A.; Buick, R.; Summons, R. Archean molecular fossils and the early rise of eukaryotes. Science 1999, 285, 1033–1036. [Google Scholar] [CrossRef]
- George, S.C.; Volk, H.; Dutkiewicz, A.; Ridley, J.; Buick, R. Preservation of hydrocarbons and biomarkers in oil trapped inside fluid inclusions for <2 billion years. Geochim. Cosmochim. Acta 2008, 72, 844–870. [Google Scholar] [CrossRef]
- Glansdorff, N. About the last common ancestor, the universal lifetree and lateral gene transfer: a reappraisal. Mol. Microbiol. 2000, 38, 177–185. [Google Scholar] [CrossRef]
- Glansdorff, N.; Xu, Y.; Labedan, B. The Last Universal Common Ancestor: emergence, constitution and genetic legacy of an elusive forerunner. Biol. Direct 2008, 3, 29–35. [Google Scholar] [CrossRef]
- Glansdorff, N.; Xu, Y.; Labedan, B. The origin of life and the last universal common ancestor: do we need a change of perspective? Res. Microbiol. 2009, 160, 522–528. [Google Scholar] [CrossRef]
- Glansdorff, N.; Xu, Y.; Labedan, B. The conflict between horizontal gene transfer and the safeguard of identity: origin of meiotic sexuality. J. Mol. Evol. 2009, 9, 470–480. [Google Scholar] [CrossRef]
- Darwin, C. On the origin of species by means of natural selection; Murray: London, UK, 1859. [Google Scholar]
- Lake, J.A.; Henderson, E.; Oakes, M.; Clark, M.W. Eocytes: A new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 1984, 81, 3786–3790. [Google Scholar]
- Cox, C.J.; Foster, P.G.; Hirt, R.P.; Harris, S.R.; Embley, T.M. The archaebacterial origin of eukaryotes. Proc Natl Acad Sci U. S. A. 2008, 105, 20356–20361. [Google Scholar]
- Ouzounis, C.A.; Kunin, V.; Darzentas, N.; Goldovsky, L. A minimal estimate for the gene content of the last universal common ancestor-exobiology from a terrestrial perspective. Res. Microbiol. 2006, 157, 57–68. [Google Scholar] [CrossRef]
- Jékely, G. Did the last common ancestor have a biological membrane? Biol. Direct 2006, 1, 35–1. [Google Scholar]
- Mulkidjanian, A.Y.; Galperin, M.Y.; Koonin, E.V. Co-evolution of primordial membranes and membrane proteins. Trends Biochem. Sci. 2009, 34, 206–215. [Google Scholar] [CrossRef]
- Peretó, J.; López-Garcia, P.; Moreira, D. Ancestral lipid biosynthesis and early membrane evolution. Trends Biochem. Sci. 2004, 29, 469–477. [Google Scholar] [CrossRef]
- Matsumi, R.; Atomi, H.; Driessen, A.J.M.; van der Oost, J. Isoprenoid biosynthesis in Archaea-Biochemical and evolutionary implications. Res. Microbiol. 2011, 162, 39–52. [Google Scholar] [CrossRef]
- Boucher, Y.; Kamekura, M.; Doolittle, W.F. Origins and evolution of isoprenoid lipid biosynthesis in archaea. Mol. Microbiol. 2004, 52, 515–527. [Google Scholar] [CrossRef]
- Koga, Y.; Morii, H. Biosynthesis of ether-type polar lipids in Archaea and evolutionary considerations. Microbiol. Mol. Biol. Rev. 2007, 71, 97–120. [Google Scholar] [CrossRef]
- Lee, Y.; Chan, S.I. Effect of lysolecithin on the structure and permeability of lecithin bilayer vescicles. Biochemistry 1977, 16, 1303–1309. [Google Scholar] [CrossRef]
- Stahlberg, H.; Fotiadis, D.; Scheuring, S.; Rémigny, H.; Braun, T.; Mitsuoka, Y.; Fujiyoshi, Y.; Engel, A. Two-dimensional crystals: a powerful approach to assess structure, function and dynamics of membrane proteins. FEBS Lett. 2001, 504, 166–172. [Google Scholar] [CrossRef]
- Bulik, D.; van Ophem, P.; Manning, J.; Shen, Z.; Newburg, D.; Jarroll, E. UDP-N-acetylglucosamine pyrophosphorylase: a key enzyme in encysting Giardia is allosterically regulated. J. Biol. Chem. 2000, 275, 14722–14728. [Google Scholar]
- Berbee, M.L.; Taylor, J.W. Systematics and evolution. In The Mycota; McLaughlin, D.J., McLaughlin, E.G., Lemke, P.A., Eds.; Springer-Verlag: Berlin, Germany, 2001. [Google Scholar]
- Dujon, B. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 2006, 22, 375–387. [Google Scholar] [CrossRef]
- Carlile, M.J. 1980 From prokaryote to eukaryote: gains and losses. In The Eukaryotic Microbial Cell; Gooday, G.W., Lloyd, D., Trinci, A.P.J., Eds.; Cambridge University Press: Cambridge, UK, 1-40. [Google Scholar]
- Pianka, E.R. On r- and K-selection. Am. Nat. 1970, 104, 592–597. [Google Scholar]
- Lynch, M. Streamlining and simplification of microbial genome architecture. Annu. Rev. Microbiol. 2006, 60, 327–349. [Google Scholar] [CrossRef]
- Perkins, T.J.; Swain, P.S. Strategies for cellular decision-making. Mol. Syst. Biol. 2009, 5, 326:1–326:15. [Google Scholar]
- Forterre, P. The universal tree of life and the last universal cellular ancestor: revolution and counterrevolutions. In Evolutionary Genomics and Systems Biology; Caetano-Anollés, G., Ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2010; pp. 43–62. [Google Scholar]
- Wolf, Y.I.; Rogozin, I.B.; Kondrashov, A.S.; Koonin, E.V. Genome alignment, evolution of prokaryotic genome organization and prediction of gene function using genomic context. Genome Res. 2001, 11, 356–372. [Google Scholar] [CrossRef]
- Martin, W.; Hoffmeister, M.; Rotte, C.; Henze, K. An overview of endosymbiotic models for the origins of eukaryotes, their ATP-producing organelles (mitochondria and hydrogenosomes), and their heterotrophic lifestyle. Biol. Chem. 2001, 382, 1521–1539. [Google Scholar] [CrossRef]
- Koonin, E.V. Temporal order of evolution of DNA replication systems inferred by comparison of cellular and viral DNA polymerases. Biol. Direct 2006, 1, 39:1–39:18. [Google Scholar]
- Koonin, E.V.; Senkevich, T.G.; Dolja, V.V. The ancient Virus World and evolution of cells. Biol. Direct 2006, 1, 29–1. [Google Scholar]
- Claverie, J.M.; Ogata, H.; Audic, S.; Abergel, C.; Suhre, K.; Fournier, P.E. Mimivirus and the emerging concept of “giant” virus. Virus Res. 2006, 117, 133–144. [Google Scholar] [CrossRef]
- Raoult, D.; Forterre, P. Redefining viruses: Lessons from Mimivirus. Nat. Rev. Microbiol. 2008, 6, 315–319. [Google Scholar] [CrossRef]
- Boyer, M.; Madoui, M.A.; Gimenez, G.; La Scola, B.; Raoult, D. Phylogenetic and phyletic studies of informational genes in genomes highlight existence of a 4th Domain of Life including Giant Viruses. PLoS One 2010, 5, e15530:1–e15530:8. [Google Scholar]
- This epigraph is paraphrased from Voltaire, "Le doute n'est pas une condition agréable, mais la certitude est absurde", letter to Frederick II of Prussia, 6 April 1767. Available online: http://en.wikiquote.org/wiki/Voltaire (accessed on 13 January 2012).
- Forterre, P.; Philippe, H. Where is the root of the universal tree of life? BioEssays 1999, 21, 871–879. [Google Scholar] [CrossRef]
- Reanney, D.C. On the origin of prokaryotes. J. Theor. Biol. 1974, 48, 243–251. [Google Scholar] [CrossRef]
- Lane, N.; Martin, W. The energetics of genome complexity. Nature 2010, 467, 929–934. [Google Scholar] [CrossRef]
- Danchin, A. Archives or palimpsests? Bacterial genomes unveil a scenario for the origin of life. Biol. Theory 2007, 2, 52–61. [Google Scholar] [CrossRef]
- Danchin, A.; Fang, G.; Noria, S. The extant core bacterial proteome is an archive of the origin of life. Proteomics 2007, 7, 875–889. [Google Scholar] [CrossRef]
- Origins of Life: The Primal Self-Organization; Egel, R.; Lankenau, D.-H.; Mulkidjanian, A.Y. (Eds.) Springer-Verlag: Heidelberg, Germany, 2011.
- Martin, W. Heinrich-Heine-Universität, Düsseldorf. Personal communication, 17 October 2011. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Egel, R. Primal Eukaryogenesis: On the Communal Nature of Precellular States, Ancestral to Modern Life. Life 2012, 2, 170-212. https://doi.org/10.3390/life2010170
Egel R. Primal Eukaryogenesis: On the Communal Nature of Precellular States, Ancestral to Modern Life. Life. 2012; 2(1):170-212. https://doi.org/10.3390/life2010170
Chicago/Turabian StyleEgel, Richard. 2012. "Primal Eukaryogenesis: On the Communal Nature of Precellular States, Ancestral to Modern Life" Life 2, no. 1: 170-212. https://doi.org/10.3390/life2010170
APA StyleEgel, R. (2012). Primal Eukaryogenesis: On the Communal Nature of Precellular States, Ancestral to Modern Life. Life, 2(1), 170-212. https://doi.org/10.3390/life2010170