From Prebiotics to Probiotics: The Evolution and Functions of tRNA Modifications
Abstract
:1. Before There Was RNA, There Were Modifications
2. Nucleotide Modifications in the Prebiotic World
3. Barriers to the Incorporation and Maintenance of Modifications
4. Influence of RNA Modifications on the Evolution of the Genetic Code
5. Establishing Translational Accuracy
6. Reading Frame Maintenance
7. Evolution of tRNA Modification Enzymes
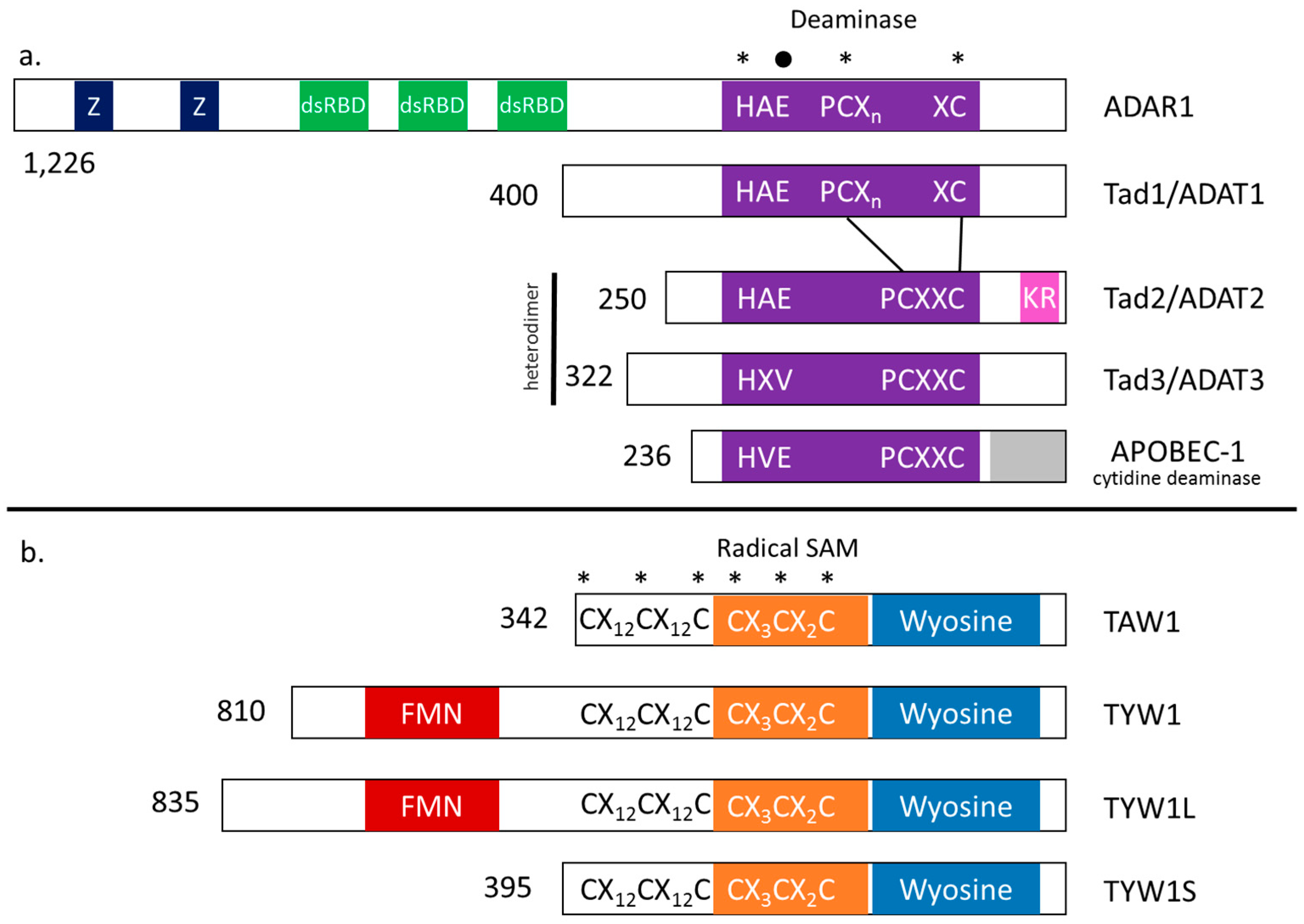
8. Modularity of tRNA Modification Enzymes
9. Concluding Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Crick, F.H.C. The origin of the genetic code. J. Mol. Biol. 1968, 38, 367–379. [Google Scholar] [CrossRef]
- Orgel, L.E. Evolution of the genetic apparatus. J. Mol. Biol. 1968, 38, 381–393. [Google Scholar] [CrossRef]
- Woese, C.R. The Genetic Code: The Molecular Basis for Genetic Expression; Harper and Row: New York, NY, USA, 1967. [Google Scholar]
- Gilbert, W. Origin of life: The RNA world. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Orgel, L.E. Prebiotic chemistry and the origin of the RNA world. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 99–123. [Google Scholar] [PubMed]
- Lazcano, A.; Miller, S.L. The origin and early evolution of life: Prebiotic chemistry, the pre-RNA world, and time. Cell 1996, 85, 793–798. [Google Scholar] [CrossRef]
- Bada, J.L.; Lazcano, A. Origin of life. Some like it hot, but not the first biomolecules. Science 2002, 296, 1982–1983. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.L.; Urey, H.C. Origin of Life. Science 1959, 130, 1622–1624. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.L.; Urey, H.C. Organic compound synthes on the primitive earth. Science 1959, 130, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Oró, J.; Kimball, A.P. Synthesis of purines under possible primitive earth conditions. II. Purine intermediates from hydrogen cyanide. Arch. Biochem. Biophys. 1962, 96, 293–313. [Google Scholar] [CrossRef]
- Oró, J.; Kimball, A.P. Synthesis of purines under possible primitive earth conditions. I. Adenine from hydrogen cyanide. Arch. Biochem. Biophys. 1961, 94, 217–227. [Google Scholar] [CrossRef]
- Levy, M.; Miller, S.L.; Oró, J. Production of guanine from NH4CN polymerizations. J. Mol. Evol. 1999, 49, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.; Miller, S.L. The prebiotic synthesis of modified purines and their potential role in the RNA world. J. Mol. Evol. 1999, 48, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Robertson, M.P.; Levy, M.; Miller, S.L. Prebiotic synthesis of diaminopyrimidine and thiocytosine. J. Mol. Evol. 1996, 43, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Benner, S.A.; Ellington, A.D.; Tauer, A. Modern metabolism as a palimpsest of the RNA world. Proc. Natl. Acad. Sci. USA 1989, 86, 7054–7058. [Google Scholar] [CrossRef] [PubMed]
- Martínez Giménez, J.A.; Sáez, G.T.; Seisdedos, R.T. On the function of modified nucleosides in the RNA world. J. Theor. Biol. 1998, 194, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Levisohn, R.; Spiegelman, S. Further extracellular Darwinian experiments with replicating RNA molecules: Diverse variants isolated under different selective conditions. Proc. Natl. Acad. Sci. USA 1969, 63, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Mills, D.R.; Peterson, R.L.; Spiegelman, S. An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule. Proc. Natl. Acad. Sci. USA 1967, 58, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Pace, N.R.; Spiegelman, S. In vitro synthesis of an infectious mutant RNA with a normal RNA replicase. Science 1966, 153, 64–67. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Chen, X.; Robertson, M.; Syrett, A. Evolutionary origins and directed evolution of RNA. Int. J. Biochem. Cell Biol. 2009, 41, 254–265. [Google Scholar] [CrossRef] [PubMed]
- Uphoff, K.W.; Bell, S.D.; Ellington, A.D. In vitro selection of aptamers: The dearth of pure reason. Curr. Opin. Struct. Biol. 1996, 6, 281–288. [Google Scholar] [CrossRef]
- Unrau, P.J.; Bartel, D.P. RNA-catalysed nucleotide synthesis. Nature 1998, 395, 260–263. [Google Scholar] [PubMed]
- Ekland, E.H.; Szostak, J.W.; Bartel, D.P. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 1995, 269, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Ekland, E.H.; Bartel, D.P. RNA-catalysed RNA polymerization using nucleoside triphosphates. Nature 1996, 382, 373–376. [Google Scholar] [CrossRef] [PubMed]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- White, H.B. Coenzymes as fossils of an earlier metabolic state. J. Mol. Evol. 1976, 7, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Roth, A.; Winkler, W.C.; Regulski, E.E.; Lee, B.W.K.; Lim, J.; Jona, I.; Barrick, J.E.; Ritwik, A.; Kim, J.N.; Welz, R.; et al. A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat. Struct. Mol. Biol. 2007, 14, 308–317. [Google Scholar] [CrossRef] [PubMed]
- McCrate, N.E.; Varner, M.E.; Kim, K.I.; Nagan, M.C. Molecular dynamics simulations of human tRNA UUU Lys,3: The role of modified bases in mRNA recognition. Nucleic Acids Res. 2006, 34, 5361–5368. [Google Scholar] [CrossRef] [PubMed]
- Bajji, A.C.; Davis, D.R. Synthesis and biophysical characterization of tRNA(Lys,3) anticodon stem-loop RNAs containing the mcm(5)s(2)U nucleoside. Org. Lett. 2000, 2, 3865–3868. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, M.; Durant, P.C.; Davis, D.R. Hypermodified nucleosides in the anticodon of tRNA(Lys) stabilize a canonical U-turn structure. Biochemistry 2000, 39, 12575–12584. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Li, L.; Engelhart, A.E.; Gan, J.; Wang, J.; Szostak, J.W. Structural insights into the effects of 2′-5′ linkages on the RNA duplex. Proc. Natl. Acad. Sci. USA 2014, 111, 3050–3055. [Google Scholar] [CrossRef] [PubMed]
- Engelhart, A.E.; Powner, M.W.; Szostak, J.W. Functional RNAs exhibit tolerance for non-heritable 2′-5′ versus 3′-5′ backbone heterogeneity. Nat. Chem. 2013, 5, 390–394. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R. On the evolution of cells. Proc. Natl. Acad. Sci. USA 2002, 99, 8742–8747. [Google Scholar] [CrossRef] [PubMed]
- Urbonavičius, J.; Qian, Q.; Durand, J.M.B.; Hagervall, T.G.; Björk, G.R. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001, 20, 4863–4873. [Google Scholar] [CrossRef] [PubMed]
- Björk, G.R.; Jacobsson, K.; Nilsson, K.; Johansson, M.J.O.; Byström, A.S.; Persson, O.P. A primordial tRNA modification required for the evolution of life? 2001, 20, 231–239. [Google Scholar]
- Machnicka, M.A.; Milanowska, K.; Oglou, O.O.; Purta, E.; Kurkowska, M.; Olchowik, A.; Januszewski, W.; Kalinowski, S.; Dunin-Horkawicz, S.; Rother, K.M.; et al. MODOMICS: A database of RNA modification pathways—2013 Update. Nucleic Acids Res. 2013, 41, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Jackman, J.E.; Alfonzo, J.D. Transfer RNA modifications: Nature’s combinatorial chemistry playground. Wiley Interdiscip. Rev. RNA 2013, 4, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Swinehart, W.; Jackman, J. Diversity in mechanism and function of tRNA methyltransferases. RNA Biol. 2015, 12, 398–411. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.C.K.; Mao, D.Y.L.; Neculai, D.; Strecker, J.; Chiovitti, D.; Kurinov, I.; Poda, G.; Thevakumaran, N.; Yuan, F.; Szilard, R.K.; et al. Reconstitution and characterization of eukaryotic N6-threonylcarbamoylation of tRNA using a minimal enzyme system. Nucleic Acids Res. 2013, 41, 6332–6346. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, C.; El Yacoubi, B.; de Crécy-Lagard, V.; Iwata-Reuyl, D. Biosynthesis of threonylcarbamoyl adenosine (t6A), a universal tRNA nucleoside. J. Biol. Chem. 2012, 287, 13666–13673. [Google Scholar] [CrossRef] [PubMed]
- Thiaville, P.C.; Iwata-Reuyl, D.; de Crécy-Lagard, V. Diversity of the biosynthesis pathway for Threonylcarbamoyladenosine (t6A), a universal modification of tRNA. RNA Biol. 2014, 11, 1529–1539. [Google Scholar] [CrossRef] [PubMed]
- Phizicky, E.M.; Hopper, A.K. tRNA biology charges to the front. Genes Dev. 2010, 24, 1832–1860. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F.; Vendeix, F.A.P.; Graham, W.D. tRNA’s wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007, 366, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, A.; Chernyakov, I.; Gu, W.; Hiley, S.L.; Hughes, T.R.; Grayhack, E.J.; Phizicky, E.M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 2006, 21, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Helm, M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006, 34, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Kadaba, S.; Krueger, A.; Trice, T.; Krecic, A.M.; Hinnebusch, A.G.; Anderson, J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004, 18, 1227–1240. [Google Scholar] [CrossRef] [PubMed]
- Hagervall, T.G.; Ericson, J.U.; Esberg, K.B.; Ji-nong, L.; Björk, G.R. Role of tRNA modification in translational fidelity. Biochim. Biophys. Acta 1990, 1050, 263–266. [Google Scholar] [CrossRef]
- Helm, M.; Alfonzo, J.D. Posttranscriptional RNA modifications: Playing metabolic games in a cell’s chemical legoland. Chem. Biol. 2014, 21, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Guy, M.P.; Phizicky, E.M. Two-subunit enzymes involved in eukaryotic post-transcriptional tRNA modification. RNA Biol. 2014, 6286, 1608–1618. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F. Decoding the genome: A modified view. Nucleic Acids Res. 2004, 32, 223–238. [Google Scholar] [CrossRef] [PubMed]
- El Yacoubi, B.; Bailly, M.; de Crécy-Lagard, V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012, 46, 69–95. [Google Scholar] [CrossRef] [PubMed]
- Gustilo, E.M.; Vendeix, F.A.; Agris, P.F. tRNA’s modifications bring order to gene expression. Curr. Opin. Microbiol. 2008, 11, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, H.; de Crécy-Lagard, V.; Marck, C. Deciphering synonymous codons in the three domains of life: Co-evolution with specific tRNA modification enzymes. FEBS Lett. 2010, 584, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F. Wobble position modified nucleosides evolved to select transfer RNA codon recognition: A modified-wobble hypothesis. Biochimie 1991, 73, 1345–1349. [Google Scholar] [CrossRef]
- Fabret, C.; Dervyn, E.; Dalmais, B.; Guillot, A.; Marck, C.; Grosjean, H.; Noirot, P. Life without the essential bacterial tRNA Ile2-lysidine synthetase TilS: A case of tRNA gene recruitment in Bacillus subtilis. Mol. Microbiol. 2011, 80, 1062–1074. [Google Scholar] [CrossRef] [PubMed]
- Yarian, C.; Townsend, H.; Czestkowski, W.; Sochacka, E.; Malkiewicz, A.J.; Guenther, R.; Miskiewicz, A.; Agris, P.F. Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem. 2002, 277, 16391–16395. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.A.; Ellis, S.R.; True, H.L. The Sua5 protein is essential for normal translational regulation in yeast. Mol. Cell. Biol. 2010, 30, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Larsen, A.; Heuberger, B.D.; Blain, J.C.; Szostak, J.W. Crystal structure studies of RNA duplexes containing s2U:A and s2U:U base pairs. J. Am. Chem. Soc. 2014, 136, 13916–13924. [Google Scholar] [CrossRef] [PubMed]
- Larsen, A.T.; Fahrenbach, A.C.; Sheng, J.; Pian, J.; Szostak, J.W. Thermodynamic insights into 2-thiouridine-enhanced RNA hybridization. Nucleic Acids Res. 2015, 43, 7675–7687. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F.; Sierzputowska-Gracz, H.; Smith, W.; Malkiewicz, A.; Sochacka, E.; Nawrot, B.; Malkiewicqt, A.; Nawrott, B. Thiolation of uridine carbon-2 restricts the motional dynamics of the transfer RNA wobble position nucleoside. J. Am. Chem. Soc. 1992, 114, 2652–2656. [Google Scholar] [CrossRef]
- Shigi, N. Biosynthesis and functions of sulfur modifications in tRNA. Front. Genet. 2014, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.J.O.; Esberg, A.; Huang, B.; Björk, G.R.; Byström, A.S. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol. Cell. Biol. 2008, 28, 3301–3312. [Google Scholar] [CrossRef] [PubMed]
- Nedialkova, D.D.; Leidel, S.A. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 2015, 161, 1608–1618. [Google Scholar] [CrossRef] [PubMed]
- Murphy, F.V.; Ramakrishnan, V.; Malkiewicz, A.; Agris, P.F. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat. Struct. Mol. Biol. 2004, 11, 1186–1191. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, Y.; Kitahara, K.; Suzuki, T. The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon. EMBO J. 2008, 27, 2194–2203. [Google Scholar] [CrossRef] [PubMed]
- Sonawane, K.D.; Sambhare, S.B. Influence of hypermodified nucleosides lysidine and t6A to recognize AUA codon instead of AUG: A molecular dynamics simulation study. Integr. Biol. 2015, 7, 1387–1395. [Google Scholar] [CrossRef] [PubMed]
- Machnicka, M.A.; Olchowik, A.; Grosjean, H.; Bujnicki, J.M. Distribution and frequencies of post-transcriptional modifications in transfer RNAs. RNA Biol. 2014, 11, 1619–1629. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.G.; Piñeyro, D.; Filonava, L.; Stracker, T.H.; Batlle, E.; Ribas de Pouplana, L. A-to-I editing on tRNAs: Biochemical, biological and evolutionary implications. FEBS Lett. 2014, 588, 4279–4286. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Numata, T. Convergent evolution of AUA decoding in bacteria and archaea. RNA Biol. 2014, 11, 1586–1596. [Google Scholar] [CrossRef] [PubMed]
- Köhrer, C.; Mandal, D.; Gaston, K.W.; Grosjean, H.; Limbach, P.A.; Rajbhandary, U.L. Life without tRNAIle-lysidine synthetase: Translation of the isoleucine codon AUA in Bacillus subtilis lacking the canonical tRNA2Ile. Nucleic Acids Res. 2014, 42, 1904–1915. [Google Scholar] [CrossRef] [PubMed]
- Curran, J.F. Decoding with the A:I wobble pair is inefficient. Nucleic Acids Res. 1995, 23, 683–688. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, T.; Miyauchi, K.; Nakane, D.; Miyata, M.; Muto, A.; Nishimura, S.; Suzuki, T. Decoding system for the AUA codon by tRNAIle with the UAU anticodon in Mycoplasma mobile. Nucleic Acids Res. 2013, 41, 2621–2631. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, T.L.; de Crécy-Lagard, V.; Schimmel, P. Incorporation of nonnatural amino acids into proteins. Annu. Rev. Biochem. 2004, 73, 147–176. [Google Scholar] [CrossRef] [PubMed]
- Lajoie, M.J.; Rovner, A.J.; Goodman, D.B.; Aerni, H.R.R.; Haimovich, A.D.; Kuznetsov, G.; Mercer, J.A.; Wang, H.H.; Carr, P.A.; Mosberg, J.A.; et al. Genomically recoded organisms expand biological functions. Science 2013, 342, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Park, H.S.; Hohn, M.J.; Umehara, T.; Guo, L.T.; Osborne, E.M.; Benner, J.; Noren, C.J.; Rinehart, J.; Söll, D. Expanding the genetic code of Escherichia coli with phosphoserine. Science 2011, 333, 1151–1154. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Schultz, P.G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010, 79, 413–444. [Google Scholar] [PubMed]
- Ibba, M.; Stathopoulos, C.; Söll, D. Protein synthesis: Twenty three amino acids and counting. Curr. Biol. 2001, 11, R563–R565. [Google Scholar] [CrossRef]
- Bohlke, N.; Budisa, N. Sense codon emancipation for proteome-wide incorporation of noncanonical amino acids: Rare isoleucine codon AUA as a target for genetic code expansion. FEMS Microbiol. Lett. 2014, 351, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Kwon, I.; Kirshenbaum, K.; Tirrell, D.A. Breaking the degeneracy of the genetic code. J. Am. Chem. Soc. 2003, 125, 7512–7513. [Google Scholar] [CrossRef] [PubMed]
- Link, A.J.; Tirrell, D.A. Reassignment of sense codons in vivo. Methods 2005, 36, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Yamaguchi, A.; Ohtake, K.; Takahashi, M.; Hayashi, A.; Iraha, F.; Kira, S.; Yanagisawa, T.; Yokoyama, S.; Hoshi, H.; et al. Reassignment of a rare sense codon to a non-canonical amino acid in Escherichia coli. Nucleic Acids Res. 2015, 43, 8111–8122. [Google Scholar] [CrossRef] [PubMed]
- Krishnakumar, R.; Ling, J. Experimental challenges of sense codon reassignment: An innovative approach to genetic code expansion. FEBS Lett. 2014, 588, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.M.; Reynolds, N.M.; Rivera, K.; Connolly, M.; Guo, L.T.; Ling, J.; Pappin, D.J.; Church, G.M.; Soll, D.D. Efficient reassignment of a frequent serine codon in wild-type Escherichia coli. ACS Synth. Biol. 2016, 5, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Wang, W.; Liu, W.R. Towards reassigning the rare AGG codon in Escherichia coli. ChemBioChem 2014, 15, 1750–1754. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, A.R.; Guimarães, A.R.; Santos, M.A.S. Non-standard genetic codes define new concepts for protein engineering. Life 2015, 5, 1610–1628. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Wang, Y.; Nakamura, A.; Eiler, D.; Kavran, J.M.; Wong, M.; Kiessling, L.L.; Steitz, T.A.; O’Donoghue, P.; Söll, D. Polyspecific pyrrolysyl-tRNA synthetases from directed evolution. Proc. Natl. Acad. Sci. USA 2014, 111, 16724–16729. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, K.; Kimura, S.; Suzuki, T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat. Chem. Biol. 2013, 9, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Sylvers, L.A.; Rogers, K.C.; Shimizu, M.; Ohtsuka, E.; Söll, D. A 2-thiouridine derivative in tRNAGlu is a positive determinant for aminoacylation by Escherichia coli glutamyl-tRNA synthetase. Biochemistry 1993, 32, 3836–3841. [Google Scholar] [CrossRef] [PubMed]
- Madore, E.; Florentz, C.; Giegé, R.; Sekine, S.; Yokoyama, S.; Lapointe, J. Effect of modified nucleotides on Escherichia coli tRNAGlu structure and on its aminoacylation by glutamyl-tRNA synthetase. Predominant and distinct roles of the mnm5 and s2 modifications of U34. Eur. J. Biochem. 1999, 266, 1128–1135. [Google Scholar] [CrossRef] [PubMed]
- Beuning, P.J.; Musier-Forsyth, K. Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers 1999, 52, 1–28. [Google Scholar] [CrossRef]
- Abbott, J.A.; Francklyn, C.S.; Robey-Bond, S.M. Transfer RNA and human disease. Front. Genet. 2014, 5, 158. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Nagao, A.; Suzuki, T. Human Mitochondrial tRNAs: Biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 2011, 45, 299–329. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, S.; Ignatova, Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015, 16, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Beebe, K.; Nangle, L.A.; Jang, J.; Longo-Guess, C.M.; Cook, S.A.; Davisson, M.T.; Sundberg, J.P.; Schimmel, P.; Ackerman, S.L. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 2006, 443, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Bacher, J.M.; de Crecy-Lagard, V.; Schimmel, P.R. Inhibited cell growth and protein functional changes from an editing-defective tRNA synthetase. Proc. Natl. Acad. Sci. USA 2005, 102, 1697–1701. [Google Scholar] [CrossRef] [PubMed]
- Björk, G.R.; Durand, J.M.B.; Hagervall, T.G.; Leipuviene, R.; Lundgren, H.K.; Nilsson, K.; Chen, P.; Qian, Q.; Urbonavičius, J. Transfer RNA modification: Influence on translational frameshifting and metabolism. FEBS Lett. 1999, 452, 47–51. [Google Scholar] [CrossRef]
- Girstmair, H.; Saffert, P.; Rode, S.; Czech, A.; Holland, G.; Bannert, N.; Ignatova, Z. Depletion of cognate charged transfer RNA causes translational frameshifting within the expanded CAG stretch in huntingtin. Cell Rep. 2013, 3, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Schimmel, P. Development of tRNA synthetases and connection to genetic code and disease. Protein Sci. 2008, 17, 1643–1652. [Google Scholar] [CrossRef] [PubMed]
- Tükenmez, H.; Xu, H.; Esberg, A.; Byström, A.S. The role of wobble uridine modifications in +1 translational frameshifting in eukaryotes. Nucleic Acids Res. 2015, 43, 9489–9499. [Google Scholar] [CrossRef] [PubMed]
- Björk, G.R.; Wikström, P.M.; Byström, A.S. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science 1989, 244, 986–989. [Google Scholar] [CrossRef] [PubMed]
- Hagervall, T.G.; Tuohy, T.M.; Atkins, J.F.; Björk, G.R. Deficiency of 1-methylguanosine in tRNA from Salmonella typhimurium induces frameshifting by quadruplet translocation. J. Mol. Biol. 1993, 232, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Paris, Z.; Horáková, E.; Rubio, M.A.T.; Sample, P.; Fleming, I.M.C.; Armocida, S.; Lukes, J.; Alfonzo, J.D. The T. brucei TRM5 methyltransferase plays an essential role in mitochondrial protein synthesis and function. RNA 2013, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Esberg, B.; Li, J.; Björk, G.R.; Curran, J.F. Three modified nucleosides present in the anticodon stem and loop influence the in vivo aa-tRNA selection in a tRNA-dependent manner. J. Mol. Biol. 1997, 271, 209–221. [Google Scholar]
- Maehigashi, T.; Dunkle, J.A.; Miles, S.J.; Dunham, C.M. Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops. Proc. Natl. Acad. Sci. USA 2014, 111, 12740–12745. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Kramer, G.; Graham, D.E.; Appling, D.R. Yeast mitochondrial initiator tRNA is methylated at guanosine 37 by the Trm5-encoded tRNA (guanine-N1-)-methyltransferase. J. Biol. Chem. 2007, 282, 27744–27753. [Google Scholar] [CrossRef] [PubMed]
- El Yacoubi, B.; Hatin, I.; Deutsch, C.; Kahveci, T.; Rousset, J.P.; Iwata-Reuyl, D.; Murzin, A.G.; de Crécy-Lagard, V. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J. 2011, 30, 882–893. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Hernandez, A.; Spears, J.L.; Gaston, K.W.; Limbach, P.A.; Gamper, H.; Hou, Y.M.; Kaiser, R.; Agris, P.F.; Perona, J.J. Structural and mechanistic basis for enhanced translational efficiency by 2-thiouridine at the tRNA anticodon wobble position. J. Mol. Biol. 2013, 425, 3888–3906. [Google Scholar] [CrossRef] [PubMed]
- Lecointe, F.; Namy, O.; Hatin, I.; Simos, G.; Rousset, J.P.; Grosjean, H. Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. J. Biol. Chem. 2002, 277, 30445–30453. [Google Scholar] [CrossRef] [PubMed]
- Bekaert, M.; Rousset, J.P. An extended signal involved in eukaryotic −1 frameshifting operates through modification of the E site tRNA. Mol. Cell 2005, 17, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Bishop, A.C. Identification of the tRNA-dihydrouridine synthase family. J. Biol. Chem. 2002, 277, 25090–25095. [Google Scholar] [CrossRef] [PubMed]
- Kasprzak, J.M.; Czerwoniec, A.; Bujnicki, J.M. Molecular evolution of dihydrouridine synthases. BMC Bioinform. 2012, 13, 153. [Google Scholar] [CrossRef] [PubMed]
- Byrne, R.T.; Jenkins, H.T.; Peters, D.T.; Whelan, F.; Stowell, J.; Aziz, N.; Kasatsky, P.; Rodnina, M.V.; Koonin, E.V.; Konevega, A.L.; et al. Major reorientation of tRNA substrates defines specificity of dihydrouridine synthases. Proc. Natl. Acad. Sci. USA 2015, 112, 6033–6037. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Tanaka, Y.; Yamashita, K.; Suzuki, T.; Nakamura, A.; Hirano, N.; Suzuki, T.; Yao, M.; Tanaka, I. Molecular basis of dihydrouridine formation on tRNA. Proc. Natl. Acad. Sci. USA 2011, 108, 19593–19598. [Google Scholar] [CrossRef] [PubMed]
- Rider, L.W.; Ottosen, M.B.; Gattis, S.G.; Palfey, B.A. Mechanism of dihydrouridine synthase 2 from yeast and the importance of modifications for efficient tRNA reduction. J. Biol. Chem. 2009, 284, 10324–10333. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.; Hiley, S.L.; Hughes, T.R.; Phizicky, E.M. The specificities of four yeast dihydrouridine synthases for cytoplasmic tRNAs. J. Biol. Chem. 2004, 279, 17850–17860. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.; Martzen, M.R.; Phizicky, E.M. A conserved family of Saccharomyces cerevisiae synthases effects dihydrouridine modification of tRNA. RNA 2002, 8, 370–381. [Google Scholar] [CrossRef] [PubMed]
- Savage, D.F.; de Crécy-Lagard, V.; Bishop, A.C. Molecular determinants of dihydrouridine synthase activity. FEBS Lett. 2006, 580, 5198–5202. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Yu, J.; Tanaka, Y.; Tanaka, M.; Tanaka, I.; Yao, M. Structure of dihydrouridine synthase C (DusC) from Escherichia coli. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Christian, T.; Hou, Y.M. Distinct determinants of tRNA recognition by the TrmD and Trm5 methyl transferases. J. Mol. Biol. 2007, 373, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Schubert, H.L.; Blumenthal, R.M.; Cheng, X. Many paths to methyltransfer: A chronicle of convergence. Trends Biochem. Sci. 2003, 28, 329–335. [Google Scholar] [CrossRef]
- Zegers, I.; Gigot, D.; van Vliet, F.; Tricot, C.; Aymerich, S.; Bujnicki, J.M.; Kosinski, J.; Droogmans, L. Crystal structure of Bacillus subtilis TrmB, the tRNA (m7G46) methyltransferase. Nucleic Acids Res. 2006, 34, 1925–1934. [Google Scholar] [CrossRef] [PubMed]
- Kozbial, P.Z.; Mushegian, A.R. Natural history of S-adenosylmethionine-binding proteins. BMC Struct. Biol. 2005, 5, 19. [Google Scholar] [CrossRef] [PubMed]
- Perche-Letuvée, P.; Molle, T.; Forouhar, F.; Mulliez, E.; Atta, M. Wybutosine biosynthesis: Structural and mechanistic overview. RNA Biol. 2014, 11, 1508–1518. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Masuda, I.; Yoshida, K.; Goto-Ito, S.; Sekine, S.; Suh, S.W.; Hou, Y.M.; Yokoyama, S. Structural basis for methyl-donor–dependent and sequence-specific binding to tRNA substrates by knotted methyltransferase TrmD. Proc. Natl. Acad. Sci. USA 2015, 112, E4197–E4205. [Google Scholar] [CrossRef] [PubMed]
- Phizicky, E.M.; Alfonzo, J.D. Do all modifications benefit all tRNAs? FEBS Lett. 2010, 584, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Gerber, A.; Grosjean, H.; Melcher, T.; Keller, W. Tad1p, a yeast tRNA-specific adenosine deaminase, is related to the mammalian pre-mRNA editing enzymes ADAR1 and ADAR2. EMBO J. 1998, 17, 4780–4789. [Google Scholar] [CrossRef] [PubMed]
- Gerber, A.P.; Keller, W. RNA editing by base deamination: More enzymes, more targets, new mysteries. Trends Biochem. Sci. 2001, 26, 376–384. [Google Scholar] [CrossRef]
- Rubio, M.A.T.; Pastar, I.; Gaston, K.W.; Ragone, F.L.; Janzen, C.J.; Cross, G.A.; Papavasiliou, F.N.; Alfonzo, J.D. An adenosine-to-inosine tRNA-editing enzyme that can perform C-to-U deamination of DNA. Proc. Natl. Acad. Sci. USA 2007, 104, 7821–7826. [Google Scholar] [CrossRef] [PubMed]
- Kuratani, M.; Ishii, R.; Bessho, Y.; Fukunaga, R.; Sengoku, T.; Shirouzu, M.; Sekine, S.; Yokoyama, S. Crystal structure of tRNA adenosine deaminase (TadA) from Aquifex aeolicus. J. Biol. Chem. 2005, 280, 16002–16008. [Google Scholar] [CrossRef] [PubMed]
- Ragone, F.L.; Spears, J.L.; Wohlgamuth-Benedum, J.M.; Kreel, N.; Papavasiliou, F.N.; Alfonzo, J.D. The C-terminal end of the Trypanosoma brucei editing deaminase plays a critical role in tRNA binding. RNA 2011, 1296–1306. [Google Scholar] [CrossRef] [PubMed]
- Gerber, A.P. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 1999, 286, 1146–1149. [Google Scholar] [CrossRef] [PubMed]
- Spears, J.L.; Rubio, M.A.T.; Gaston, K.W.; Wywial, E.; Strikoudis, A.; Bujnicki, J.M.; Papavasiliou, F.N.; Alfonzo, J.D. A single zinc ion is sufficient for an active Trypanosoma brucei tRNA editing deaminase. J. Biol. Chem. 2011, 286, 20366–20374. [Google Scholar] [CrossRef] [PubMed]
- Elias, Y.; Huang, R.H. Biochemical and structural studies of A-to-I editing by tRNA:A34 deaminases at the wobble position of transfer RNA. Biochemistry 2005, 44, 12057–12065. [Google Scholar] [CrossRef] [PubMed]
- Umitsu, M.; Nishimasu, H.; Noma, A.; Suzuki, T.; Ishitani, R.; Nureki, O. Structural basis of AdoMet-dependent aminocarboxypropyl transfer reaction catalyzed by tRNA-wybutosine synthesizing enzyme, TYW2. Proc. Natl. Acad. Sci. USA 2009, 106, 15616–15621. [Google Scholar] [CrossRef] [PubMed]
- Waas, W.F.; de Crécy-Lagard, V.; Schimmel, P. Discovery of a gene family critical to wyosine base formation in a subset of phenylalanine-specific transfer RNAs. J. Biol. Chem. 2005, 280, 37616–37622. [Google Scholar] [CrossRef] [PubMed]
- Noma, A.; Kirino, Y.; Ikeuchi, Y.; Suzuki, T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 2006, 25, 2142–2154. [Google Scholar] [CrossRef] [PubMed]
- Kalhor, H.R.; Penjwini, M.; Clarke, S. A novel methyltransferase required for the formation of the hypermodified nucleoside wybutosine in eucaryotic tRNA. Biochem. Biophys. Res. Commun. 2005, 334, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Noma, A.; Suzuki, T.; Ishitani, R.; Nureki, O. Structural basis of tRNA modification with CO2 fixation and methylation by wybutosine synthesizing enzyme TYW4. Nucleic Acids Res. 2009, 37, 2910–2925. [Google Scholar] [CrossRef] [PubMed]
- Young, A.P.; Bandarian, V. Mechanistic studies of the radical S-Adenosyl-I-methionine enzyme 4-demethylwyosine synthase reveal the site of hydrogen atom abstraction. Biochemistry 2015, 54, 3569–3572. [Google Scholar] [CrossRef] [PubMed]
- Young, A.P.; Bandarian, V. Pyruvate is the source of the two carbons that are required for formation of the imidazoline ring of 4-demethylwyosine. Biochemistry 2011, 50, 10573–10575. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Noma, A.; Suzuki, T.; Senda, M.; Senda, T.; Ishitani, R.; Nureki, O. Crystal structure of the radical SAM enzyme catalyzing tricyclic modified base formation in tRNA. J. Mol. Biol. 2007, 372, 1204–1214. [Google Scholar] [CrossRef] [PubMed]
- Young, A.P.; Bandarian, V. Radical mediated ring formation in the biosynthesis of the hypermodified tRNA base wybutosine. Curr. Opin. Chem. Biol. 2013, 17, 613–618. [Google Scholar] [CrossRef] [PubMed]
- De Crécy-Lagard, V.; Brochier-Armanet, C.; Urbonavicius, J.; Fernandez, B.; Phillips, G.; Lyons, B.; Noma, A.; Alvarez, S.; Droogmans, L.; Armengaud, J.; et al. Biosynthesis of wyosine derivatives in tRNA: An ancient and highly diverse pathway in archaea. Mol. Biol. Evol. 2010, 27, 2062–2077. [Google Scholar] [CrossRef] [PubMed]
- Sample, P.J.; Kořený, L.; Paris, Z.; Gaston, K.W.; Rubio, M.A.T.; Fleming, I.M.C.; Hinger, S.; Horáková, E.; Limbach, P.A.; Lukeš, J.; et al. A common tRNA modification at an unusual location: The discovery of wyosine biosynthesis in mitochondria. Nucleic Acids Res. 2015, 43, 4262–4273. [Google Scholar] [CrossRef] [PubMed]
- Cermakian, N.; Cedergren, R. Modified nucleosides always were: An evolutionary model. In Modification and Editing of RNA; Grosjean, H., Benne, R., Eds.; American Society of Microbiology Press: Washington, DC, USA, 1998; pp. 535–541. [Google Scholar]
- Zaborske, J.M.; Bauer DuMont, V.L.; Wallace, E.W.J.; Pan, T.; Aquadro, C.F.; Drummond, D.A. A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein coding across an animal genus. PLoS Biol. 2014, 12, e1002015. [Google Scholar] [CrossRef] [PubMed]
- Zallot, R.; Brochier-Armanet, C.; Gaston, K.W.; Forouhar, F.; Limbach, P.A.; Hunt, J.F.; de Crécy-Lagard, V. Plant, animal, and fungal micronutrient queuosine is salvaged by members of the DUF2419 protein family. ACS Chem. Biol. 2014, 9, 1812–1825. [Google Scholar] [CrossRef] [PubMed]
- Pathak, C.; Jaiswal, Y.K.; Vinayak, M. Hypomodification of transfer RNA in cancer with respect to queuosine. RNA Biol. 2005, 2, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Gündüz, U.; Elliott, M.S.; Seubert, P.H.; Houghton, J.A.; Houghton, P.J.; Trewyn, R.W.; Katze, J.R. Absence of tRNA-guanine transglycosylase in a human colon adenocarcinoma cell line. Biochim. Biophys. Acta 1992, 1139, 229–238. [Google Scholar] [CrossRef]
- Landin, R.M.; Pétrissant, G. Queuosine deficient tRNAHis and tRNAAsp from the spleens of young mice, erythroleukemic tumoral spleens and cultured Friend cells. Biochem. Biophys. Res. Commun. 1982, 109, 1140–1147. [Google Scholar] [CrossRef]
- Fergus, C.; Barnes, D.; Alqasem, M.; Kelly, V. The queuine micronutrient: Charting a course from microbe to man. Nutrients 2015, 7, 2897–2929. [Google Scholar] [CrossRef] [PubMed]
- Rakovich, T.; Boland, C.; Bernstein, I.; Chikwana, V.M.; Iwata-Reuyl, D.; Kelly, V.P. Queuosine deficiency in eukaryotes compromises tyrosine production through increased tetrahydrobiopterin oxidation. J. Biol. Chem. 2011, 286, 19354–19363. [Google Scholar] [CrossRef] [PubMed]



© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McKenney, K.M.; Alfonzo, J.D. From Prebiotics to Probiotics: The Evolution and Functions of tRNA Modifications. Life 2016, 6, 13. https://doi.org/10.3390/life6010013
McKenney KM, Alfonzo JD. From Prebiotics to Probiotics: The Evolution and Functions of tRNA Modifications. Life. 2016; 6(1):13. https://doi.org/10.3390/life6010013
Chicago/Turabian StyleMcKenney, Katherine M., and Juan D. Alfonzo. 2016. "From Prebiotics to Probiotics: The Evolution and Functions of tRNA Modifications" Life 6, no. 1: 13. https://doi.org/10.3390/life6010013
APA StyleMcKenney, K. M., & Alfonzo, J. D. (2016). From Prebiotics to Probiotics: The Evolution and Functions of tRNA Modifications. Life, 6(1), 13. https://doi.org/10.3390/life6010013




