Mobility of a Mononucleotide within a Lipid Matrix: A Neutron Scattering Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.1.1. Lipid Extraction
2.1.2. Preparation of Matrix/AMP Complexes
2.2. Experiments
- The cold Neutron Backscattering Spectrometer IN16b is a high energy resolution instrument, so it is particularly devoted to discriminating slow molecular motions; in particular, IN16b provides an energy resolution of ∆E ≈ 0.8 µeV Full Width at Half Maximum (FWHM), corresponding to an observable time scale of ≈1 ns [18]. The scattering vector Q, given by the vectorial sum of the incident and scattered neutron wave vectors and corresponding to the analogous in the reciprocal Fourier space of the distance in the direct space, is measured on IN16b in the range (0.3 < Q < 1.8 Å−1), while the incident wavelength has been fixed to λ = 6.27 Å; elastic scans were performed stepwise in the temperature range (20 < T < 310 K).
- The thermal Neutron Backscattering Spectrometer IN13 is similar to the former, but with different energy resolution and observable Q range, ∆E ≈ 8 µeV FWHM leading to t ≈ 0.1 ns and (0.5 < Q < 4.9 Å−1). By using the CaF2 monochromator, the incident wavelength has been fixed to the value λ = 2.23 Å; elastic scans were performed stepwise in the temperature range (20 < T < 310 K).
- The disk Chopper Time-of-flight Spectrometer IN5 is a direct geometry time-of-flight instrument primarily devoted to inelastic/quasielastic neutron scattering measurements (INS/QENS), thus allowing to measure low-energy transfer processes as a function of the momentum transfer. For this experiment, QENS measurements have been performed for all the samples with two incident wavelengths: (a) λ = 5 Å, with an accessible Q interval of (0.4 < Q < 2.2 Å−1), providing an energy resolution of ∆E ≈ 100 µeV FWHM and an observable time-scale of ≈5 ps; (b) λ = 10 Å, with (0.1 < Q < 1 Å−1), ∆E ≈ 10 µeV FWHM and an observable time-scale of ≈50 ps. Elastic scans were determined stepwise in the temperature range (20 < T < 310 K); quasi-elastic spectra were taken at T = 310 K. Furthermore, IN5 also allows us to access diffraction patterns for qualitative investigations. Thus, diffraction scans were acquired at variable relative humidity and temperature. Moreover, the empty cell contribution has been removed.
2.3. Elastic and Quasi-Elastic Incoherent Neutron Scattering Data Analysis
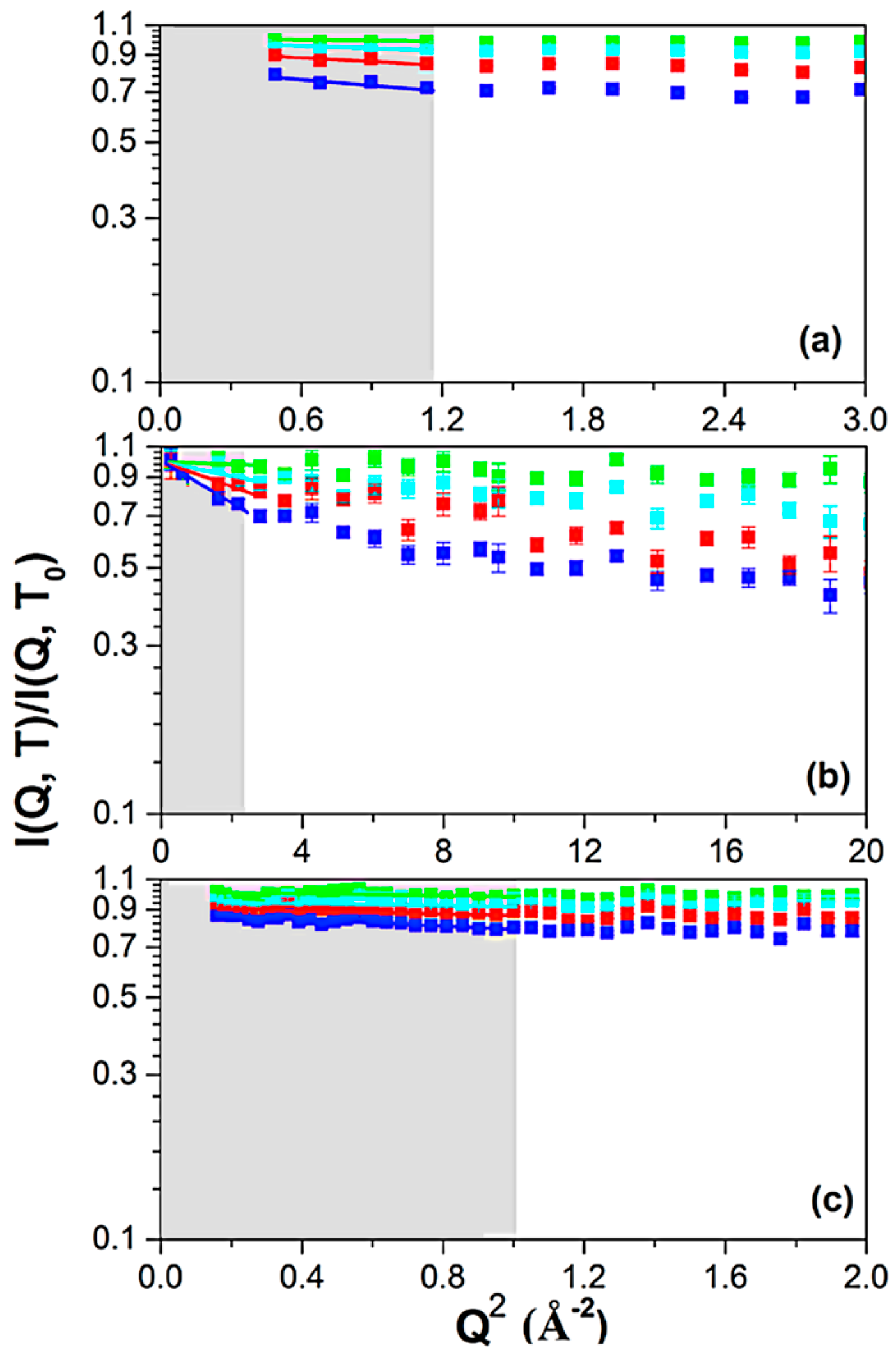
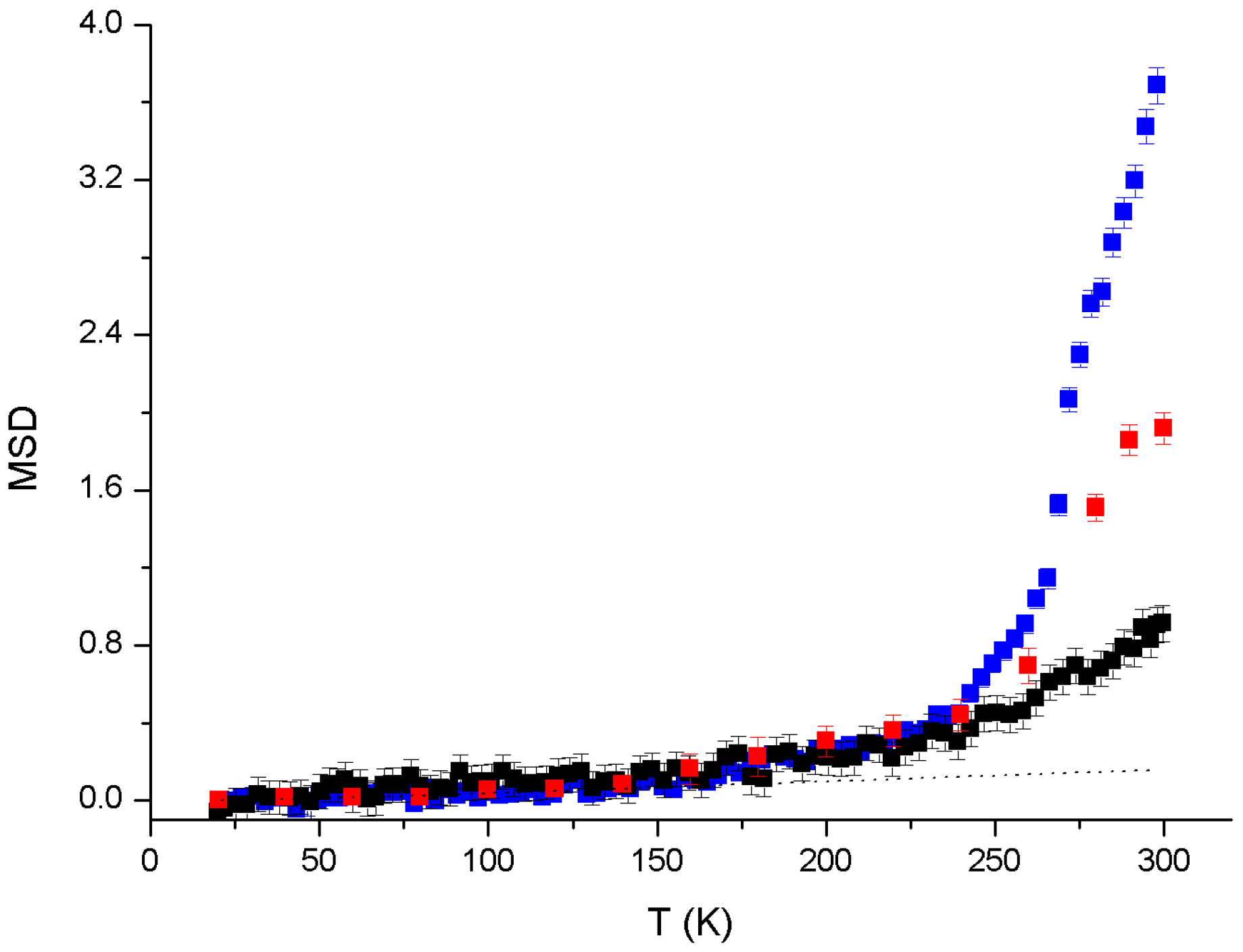
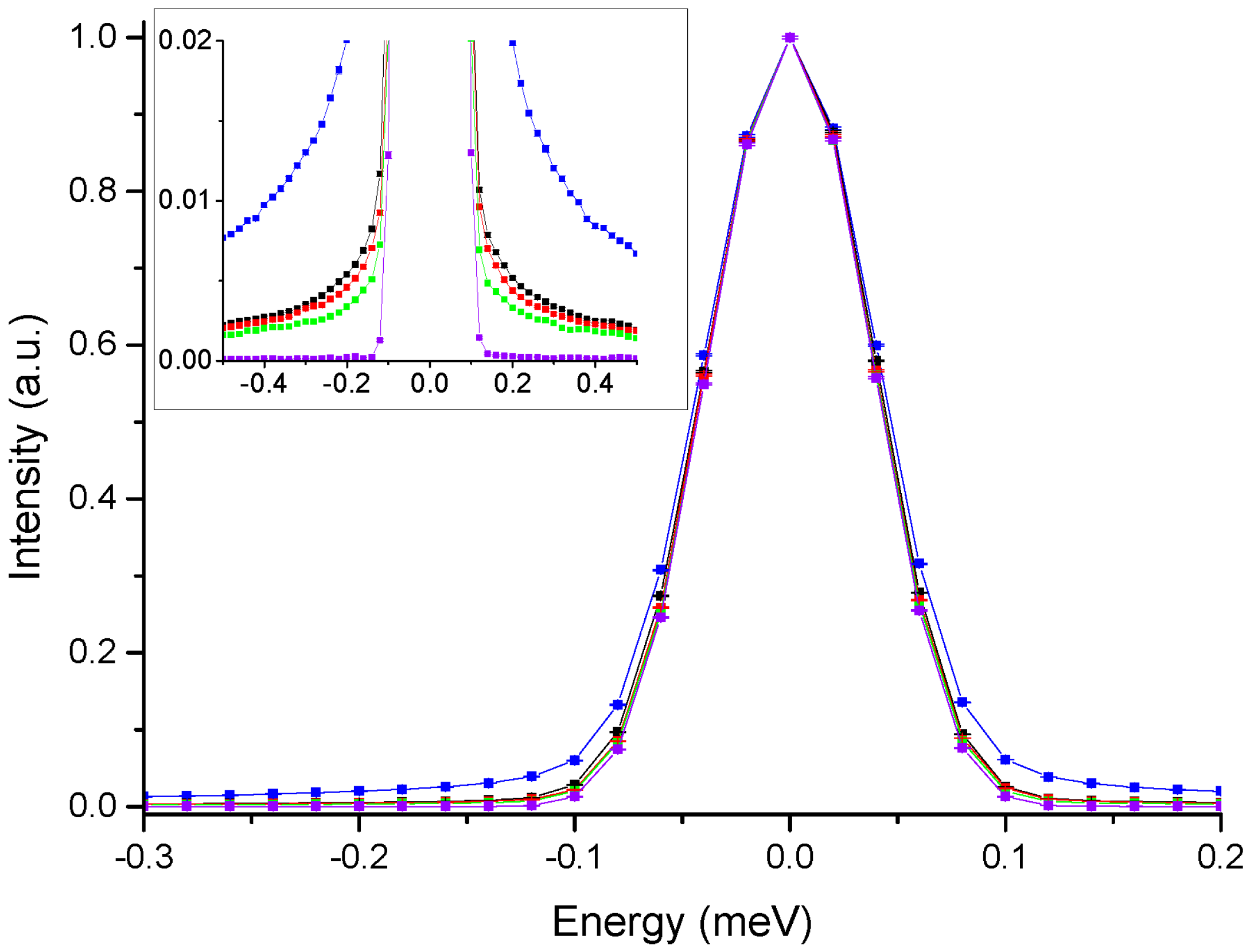
3. Results
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AMP | 5′-adenosine monophosphate |
| dMPL | deuterated multilamellar phospholipid liposomes |
| DNA | Deoxyribonucleic acid |
| EINS | Elastic incoherent neutron scattering |
| FWHM | Full Width at Half Maximum |
| HWHM | Half Width at Half Maximum |
| MDPI | Multidisciplinary Digital Publishing Institute |
| MSD | Mean Square Displacement |
| Q, Q2 | Moment transfer and its square |
| QENS | Quasi-elastic neutron scattering |
| RH | Relative humidity |
| RNA | Ribonucleic acid |
References
- Paleos, C.M. A decisive step toward the origin of life. Trends Biochem. Sci. 2015, 40, 487–488. [Google Scholar] [CrossRef] [PubMed]
- Woese, C. The Genetic Code, the Molecular Basis for Genetic Expression; Harper and Row: New York, NY, USA, 1967. [Google Scholar]
- Crick, F.H.C. The Origin of the genetic code. J. Mol. Biol. 1968, 38, 367–379. [Google Scholar] [CrossRef]
- Orgel, L.E. Evolution of the genetic apparatus. J. Mol. Biol. 1968, 38, 381–393. [Google Scholar] [CrossRef]
- Gilbert, W. Origin of life: The RNA world. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Rajamani, S.; Vlassov, A.; Benner, S.; Coombs, A.; Olasagasti, F.; Deamer, D. Lipid-assisted synthesis of RNA-like polymers from mononucleotides. Orig. Life Evol. Biosph. 2008, 38, 57–74. [Google Scholar] [CrossRef] [PubMed]
- DeGuzman, V.; Vercoutere, W.; Shenasa, H.; Deamer, D. Generation of oligonucleotides under hydrothermal conditions by non-enzymatic polymerization. J. Mol. Evol. 2014, 78, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, L.; Maurel, M.-C.; Deamer, D. Salt-promoted synthesis of RNA-like molecules in simulated hydrothermal conditions. J. Mol. Evol. 2015, 80, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Blumstein, A.; Kitagawa, N.; Blumstein, R. Polymerization of p-methacry-loyloxy benzoic acid within liquid crystalline media. Mol. Cryst. Liq. Cryst. 1971, 12, 215–227. [Google Scholar] [CrossRef]
- Barrall, E.M.; Johnson, J.F. A review of the status of polymerization in thermotropic liquid crystal media and liquid crystalline monomers. J. Macromol. Sci. Part C 1979, 17, 137–170. [Google Scholar] [CrossRef]
- Deamer, D. Liquid-crystalline nanostructures: Organizing matrices for non-enzymatic nucleic acid polymerization. Chem. Soc. Rev. 2012, 41, 5375–5379. [Google Scholar] [CrossRef] [PubMed]
- Toppozini, L.; Dies, H.; Deamer, D.W.; Rheinstädter, M.C. Adenosine Monophosphate Forms Ordered Arrays in Multilamellar Lipid Matrices: Insights into Assembly of Nucleic Acid for Primitive Life. PLoS ONE 2013, 8, e62810. [Google Scholar] [CrossRef] [PubMed]
- De Ghellinck, A.; Schaller, H.; Laux, V.; Haertlein, M.; Sferrazza, M.; Maréchal, E.; Wacklin, H.; Jouhet, J.; Fragneto, G. Production and analysis of perdeuterated lipids from Pichia pastoris cells. PLoS ONE 2014, 9, e92999. [Google Scholar] [CrossRef] [PubMed]
- Jacrot, B. The study of biological structures by neutron scattering from solution. Rep. Prog. Phys. 1976, 39, 911–953. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Sloane-Stanley, G. A simple method for the isolation and purification of total lipids from animals’ tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [PubMed]
- Maurel, M.C.; Da Silva, L.; Deamer, D.; Golub, M.; Misuraca, L.; Natali, F.; Ollivier, J.; Peters, J.; Seydel, T.; Zaccai, G. Self-Organisation of RNA In Lipid Multilayers; Institut Laue-Langevin (ILL): Grenoble, France, 2015. [Google Scholar] [CrossRef]
- Maurel, M.C.; Da Silva, L.; Deamer, D.; Deme, B.; Misuraca, L.; Natali, F.; Ollivier, J.; Peters, J.; Zaccai, G. Self-Organisation of RNA in Salty Environment; Institut Laue-Langevin (ILL): Grenoble, France, 2015. [Google Scholar] [CrossRef]
- Frick, B.; Mamontov, E.; van Eijck, L.; Seydel, T. Recent backscattering instrument developments at the ILL and SNS. Z. Phys. Chem. 2010, 224, 33. [Google Scholar] [CrossRef]
- Schiró, G.; Caronna, C.; Natali, F.; Cupane, A. Direct evidence of the amino acid side chain and backbone contributions to protein anharmonicity. J. Am. Chem. Soc. 2010, 132, 1371–1376. [Google Scholar] [CrossRef] [PubMed]
- Gabel, F.; Bicout, D.; Lehnert, U.; Tehei, M.; Weik, M.; Zaccai, G. Protein dynamics studied by neutron scattering. Q. Rev. Biophys. 2002, 35, 327–367. [Google Scholar] [CrossRef] [PubMed]
- Zaccai, G. How soft is a protein? A protein dynamics force constant measured by neutron scattering. Science 2000, 288, 1604–1607. [Google Scholar] [CrossRef] [PubMed]
- Doster, W.; Cusack, S.; Petry, W. Dynamical transition of myoglobin revealed by inelastic neutron scattering. Nature 1989, 337, 754. [Google Scholar] [CrossRef] [PubMed]
- Roh, J.H.; Novikov, V.N.; Gregory, R.B.; Curtis, J.E.; Chowdhuri, Z.; ASokolov, A.P. Onsets of anharmonicity in protein dynamics. Phys. Rev. Lett. 2005, 95, 38101. [Google Scholar] [CrossRef] [PubMed]
- Wood, K.; Tobias, D.J.; Kessler, B.; Gabel, F.; Oesterhelt, D.; Mulder, F.A.A.; Zaccai, G.; Weik, M. The low-temperature inflection observed in neutron scattering measurements of proteins is due to methyl rotation: direct evidence using isotope labeling and molecular dynamics simulations. J. Am. Chem. Soc. 2010, 132, 4990. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, J.; Bellissent-Funel, M.C. Experimental determination of the nature of diffusive motions of water molecules at low temperatures. Phys. Rev. A 1985, 31, 1913–1917. [Google Scholar] [CrossRef]
- Himbert, S.; Chapman, M.; Deamer, D.W.; Rheinstädter, M.C. Organization of Nucleotides in Different Environments and the formation of Pre-Polymers. Sci. Rep. 2016, 6, 31285. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.S.; Deamer, D. Dry/wet cycling and the thermodynamics and kinetics of prebiotic polymer synthesis. Life 2016, 6, 28. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Misuraca, L.; Natali, F.; Da Silva, L.; Peters, J.; Demé, B.; Ollivier, J.; Seydel, T.; Laux-Lesourd, V.; Haertlein, M.; Zaccai, G.; et al. Mobility of a Mononucleotide within a Lipid Matrix: A Neutron Scattering Study. Life 2017, 7, 2. https://doi.org/10.3390/life7010002
Misuraca L, Natali F, Da Silva L, Peters J, Demé B, Ollivier J, Seydel T, Laux-Lesourd V, Haertlein M, Zaccai G, et al. Mobility of a Mononucleotide within a Lipid Matrix: A Neutron Scattering Study. Life. 2017; 7(1):2. https://doi.org/10.3390/life7010002
Chicago/Turabian StyleMisuraca, Loreto, Francesca Natali, Laura Da Silva, Judith Peters, Bruno Demé, Jacques Ollivier, Tilo Seydel, Valerie Laux-Lesourd, Michael Haertlein, Giuseppe Zaccai, and et al. 2017. "Mobility of a Mononucleotide within a Lipid Matrix: A Neutron Scattering Study" Life 7, no. 1: 2. https://doi.org/10.3390/life7010002
APA StyleMisuraca, L., Natali, F., Da Silva, L., Peters, J., Demé, B., Ollivier, J., Seydel, T., Laux-Lesourd, V., Haertlein, M., Zaccai, G., Deamer, D., & Maurel, M. C. (2017). Mobility of a Mononucleotide within a Lipid Matrix: A Neutron Scattering Study. Life, 7(1), 2. https://doi.org/10.3390/life7010002





