Integral Phylogenomic Approach over Ilex L. Species from Southern South America
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Chloroplasts DNA Extraction and Processing
2.3. Genomic DNA Extraction and Processing
2.4. Plastome Characterization
2.5. Non-Coding Marker Selection
2.6. Chloroplast Phylogenomic Analyses
2.7. Nuclear Sequences Analyses
2.8. Supernetwork
2.9. Total Evidence and Character Analyses
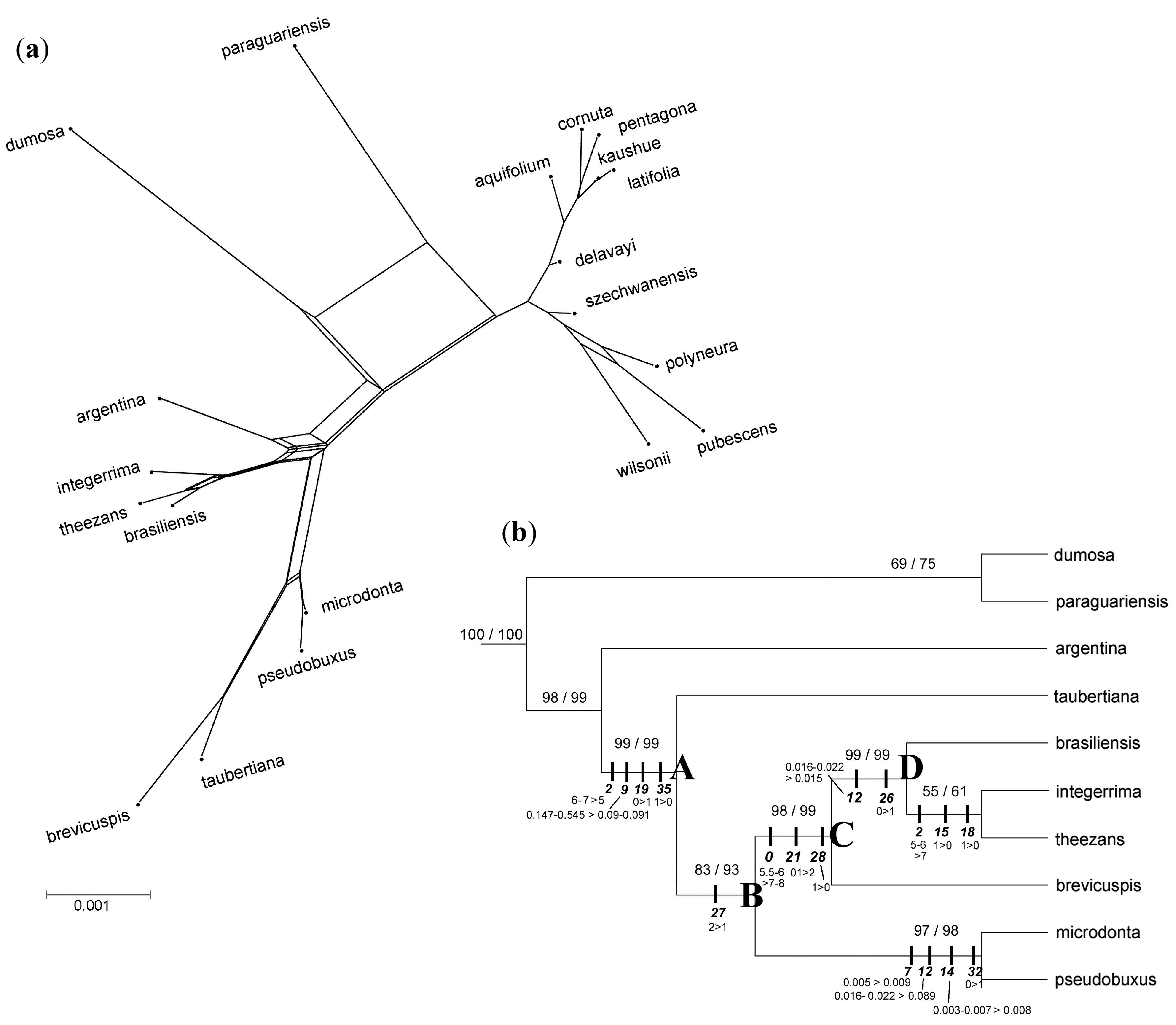
3. Results
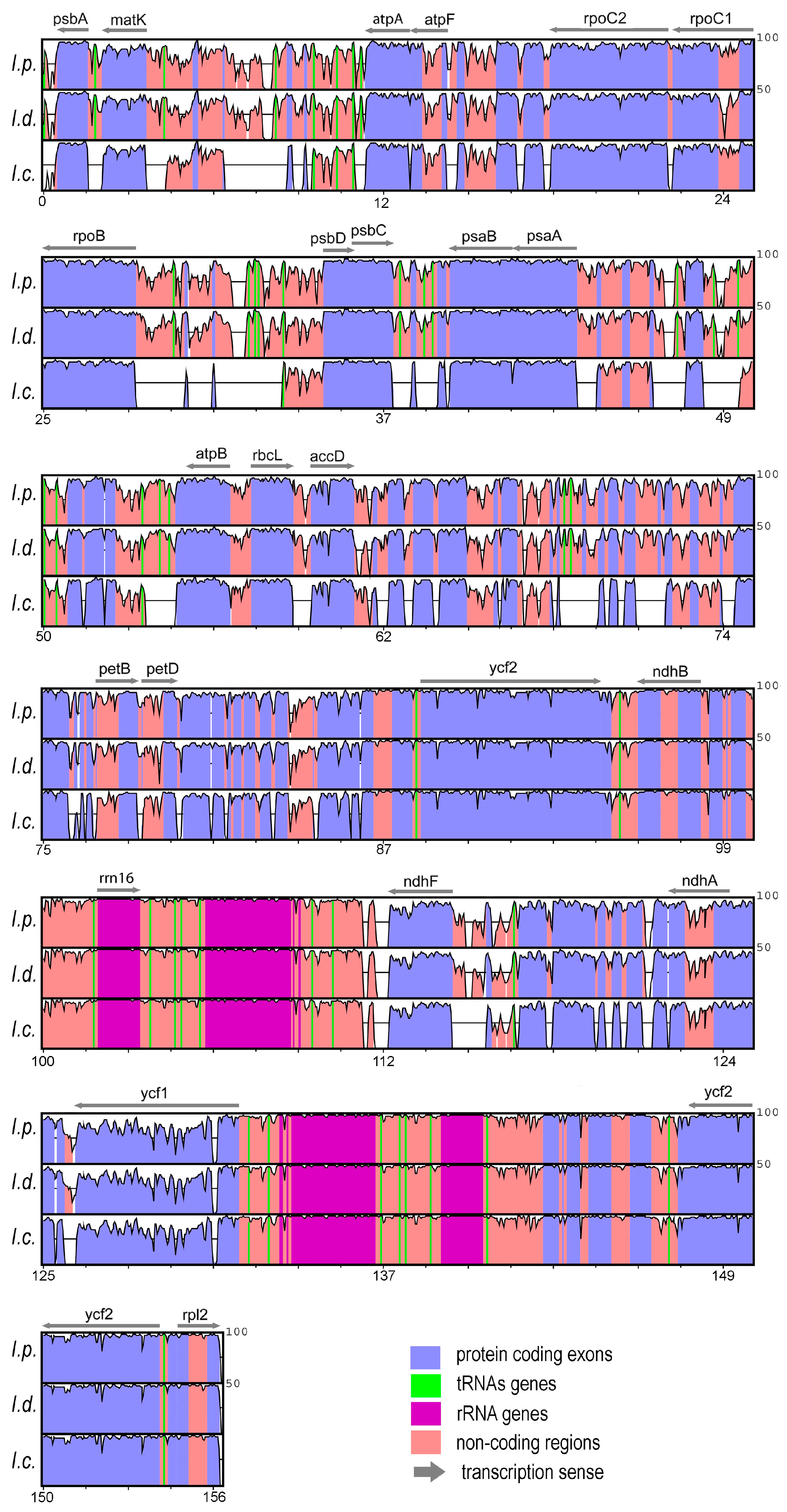
3.1. Analyses and Characterization of Plastomes
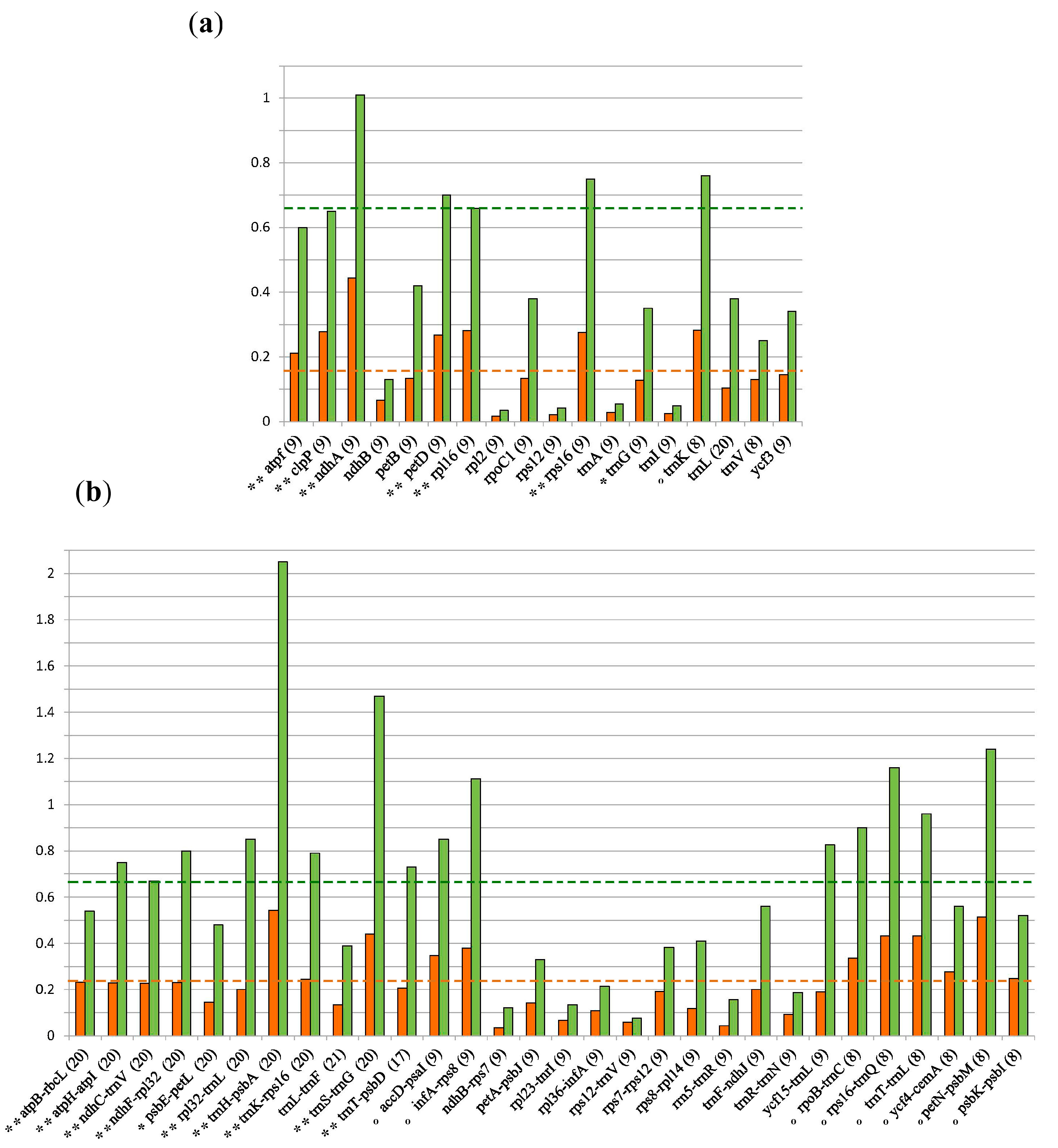
3.2. Marker Selection
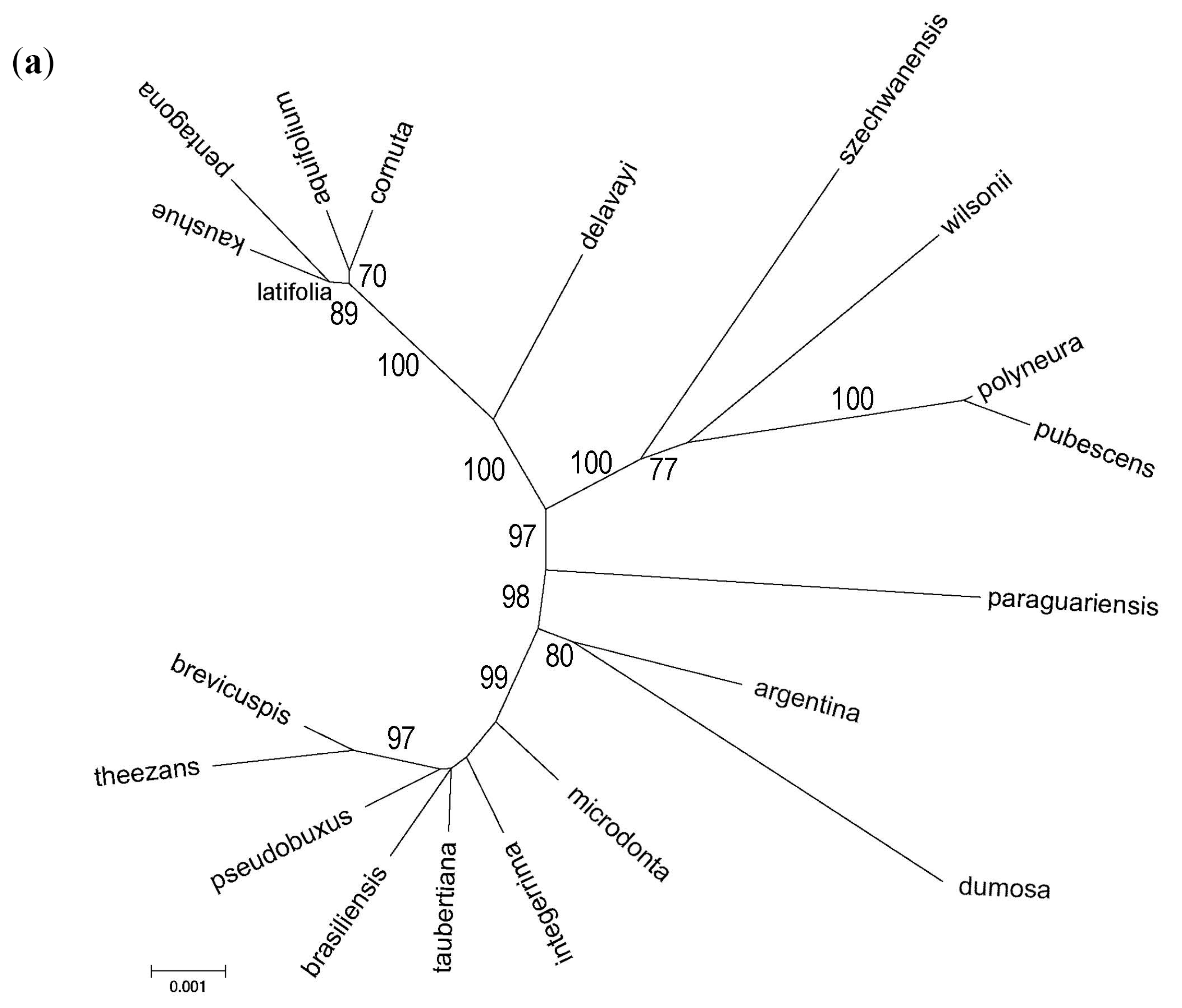
3.3. Phylogenomic Analyses
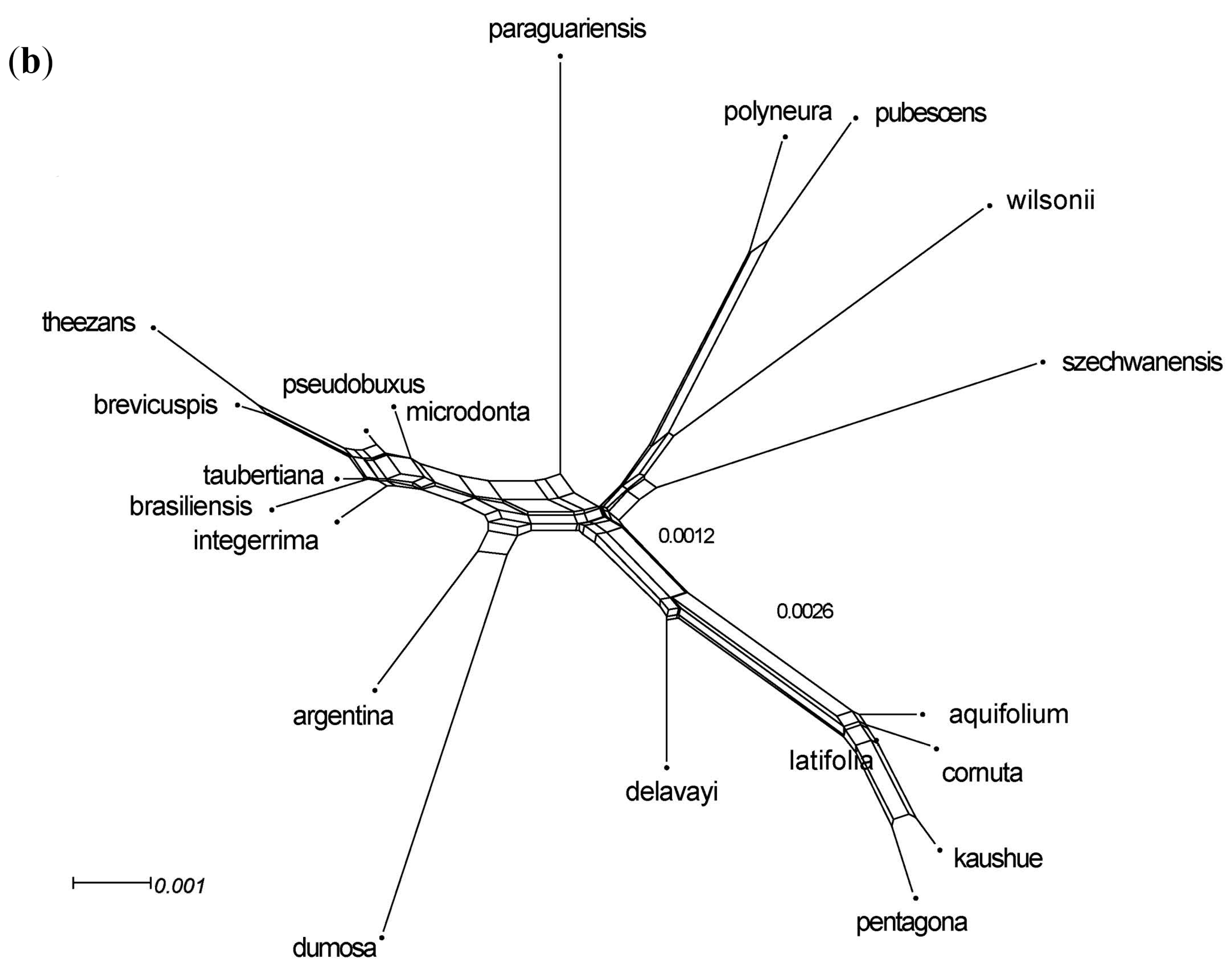
3.4. Nuclear Sequence Analyses
3.5. Integrated Data
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Loizeau, P.A.; Barriera, G.; Manen, J.F.; Broennimann, O. Towards an understanding of the distribution of Ilex L. (Aquifoliaceae) on a World-wide scale. Biol. Skr. 2005, 55, 501–520. [Google Scholar]
- Loizeau, P.A.; Savolainen, V.; Andrews, S.; Spichiger, R. Aquifoliaceae. In Flowering Plants. Eudicots; The Families and Genera of Vascular Plants; Kadereit, J.W., Bittrich, V., Eds.; Springer International Publishing: Basel, Switzerland, 2016; Volume XIV, pp. 31–36. ISBN 978-3-319-28532-0. [Google Scholar]
- Keller, H.A.; Giberti, G.C. Primer registro para la flora Argentina de Ilex affinis (Aquifoliaceae), sustituto de la ‘yerba mate’. Bol. Soc. Argent. Bot. 2011, 46, 187–194. [Google Scholar]
- Giberti, G.C.; Gurni, A.A. Anatomía floral comparada de once especies sudamericanas de Ilex L. (Aquifoliaceae) relacionadas con la yerba mate. Dominguezia 2008, 24, 77–94. [Google Scholar]
- Giberti, G.C. La ‘yerba mate’ (Ilex paraguariensis, Aquifoliaceae) en tempranos escritos rioplatenses de bonpland y su real distribución geográfica en sudamerica austral. Bonplandia 2011, 20, 203–212. [Google Scholar]
- Giberti, G.C. Diferentes Aspectos del Género Ilex (Aquifoliaceae). Corología, Arquitectura Floral, Posición Sistemática. Ph.D. Thesis, Universidad de Buenos Aires, Buenos Aires, Argentina, 2001. [Google Scholar]
- Giberti, G.C. Aquifoliaceae. In Catálogo de las Plantas Vasculares del Cono Sur (Argentina, Sur de Brasil, Chile, Paraguay y Uruguay), Vol. 2. Dicotyledoneae: Acanthaceae–Fabaceae (Abarema–Schizolobium); Monographs in Systematic Botany from the Missouri Botanical Garden; Zuloaga, F., Morrone, O., Belgrano, M., Eds.; Missouri Botanical Garden Press: St. Louis, MO, USA, 2008; pp. 1143–1146. [Google Scholar]
- Grela, I.A. Geografía Florística de Las Especies Arbóreas de Uruguay: Propuesta Para la Delimitación de Dendrofloras. Master’s Thesis, Universidad de la República, Montevideo, Uruguay, 2004. [Google Scholar]
- Anesini, C.; Turner, S.; Cogoi, L.; Filip, R. Study of the participation of caffeine and polyphenols on the overall antioxidant activity of mate (Ilex paraguariensis). LWT Food Sci. Technol. 2012, 45, 299–304. [Google Scholar] [CrossRef]
- Heck, C.I.; de Mejía, E.G. Yerba mate tea (Ilex paraguariensis): A comprehensive review on chemistry, health implications, and technological considerations. J. Food Sci. 2007, 72, R138–R151. [Google Scholar] [CrossRef] [PubMed]
- Berté, K.A.S.; Beux, M.R.; Spada, P.K.W.D.S.; Salvador, M.; Hoffmann-Ribani, R. Chemical composition and antioxidant activity of yerba-mate (Ilex paraguariensis A. St.-Hil., Aquifoliaceae) extract as obtained by spray drying. J. Agric. Food Chem. 2011, 59, 5523–5527. [Google Scholar] [CrossRef] [PubMed]
- Giberti, G.C. Los parientes silvestres de la yerba mate y el problema de su adulteración. Dominguezia 1989, 7, 1–22. [Google Scholar]
- Filip, R.; López, P.; Coussio, J.; Ferraro, G. Mate substitutes or adulterants: Study of xanthine content. Phyther. Res. 1998, 12, 129–131. [Google Scholar] [CrossRef]
- Filip, R.; López, P.; Giberti, G.C.; Coussio, J.; Ferraro, G. Phenolic compounds in seven South American Ilex species. Fitoterapia 2001, 72, 774–778. [Google Scholar] [CrossRef]
- Rafiee Seyed, M.A. Determinación de Arbutina en las Especies de Ilex. Master’s Thesis, Universidad de Belgrano, Buenos Aires, Argentina, 2012. [Google Scholar]
- Ricco, R.A.; Wagner, M.L.; Gurni, A.A. Estudio comparativo de flavonoides en seis especies austrosudamericanas del género Ilex. Acta Farm. Bonaer. 1991, 10, 29–35. [Google Scholar]
- Ricco, R.A.; Giberti, G.C.; Wagner, M.L.; Gurni, A.A. Two unusual maté substitutes (Ilex pseudobuxus and I. taubertiana): Morphology, flavonoids and geographical distribution. Rojasiana 2013, 12, 77–90. [Google Scholar]
- Maiocchi, M.G.; Del Vitto, L.A.; Petenatti, M.E.; Marchevsky, E.J.; Pellerano, R.G.; Petenatti, E.M. Multielemental composition and nutritional value of and their commercial mixture in different forms of use. Rev. Fac. Ciencias Agrar. 2016, 48, 145–159. [Google Scholar]
- Cuénod, P.; Del Pero Martinez, M.A.; Loizeau, P.A.; Spichiger, R.; Andrews, S.; Manen, J.F. Molecular phylogeny and biogeography of the genus Ilex L. (Aquifoliaceae). Ann. Bot. 2000, 85, 111–122. [Google Scholar] [CrossRef]
- Gottlieb, A.M.; Giberti, G.C.; Poggio, L. Molecular analyses of the genus Ilex (Aquifoliaceae) in southern South America, evidence from AFLP and its sequence data. Am. J. Bot. 2005, 92, 352–369. [Google Scholar] [CrossRef] [PubMed]
- Manen, J.F.; Boulter, M.C.; Naciri-Graven, Y. The complex history of the genus Ilex L. (Aquifoliaceae): Evidence from the comparison of plastid and nuclear DNA sequences and from fossil data. Plant Syst. Evol. 2002, 235, 79–98. [Google Scholar] [CrossRef]
- Manen, J.F.; Barriera, G.; Loizeau, P.A.; Naciri, Y. The history of extant Ilex species (Aquifoliaceae): Evidence of hybridization within a Miocene radiation. Mol. Phylogenet. Evol. 2010, 57, 961–977. [Google Scholar] [CrossRef] [PubMed]
- Manen, J.F. Are both sympatric species Ilex perado and Ilex canariensis secretly hybridizing? Indication from nuclear markers collected in Tenerife. BMC Evol. Biol. 2004, 4, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selbach-Schnadelbach, A.; Smith Cavalli, S.; Manen, J.F.; Coelho, G.C.; De Souza-Chies, T.T. New information for Ilex phylogenetics based on the plastid psbA-trnH intergenic spacer (Aquifoliaceae). Bot. J. Linn. Soc. 2009, 159, 182–193. [Google Scholar] [CrossRef]
- Setoguchi, H.; Watanabe, I. Intersectional gene flow between insular endemics of Ilex (Aquifoliaceae) on the Bonin Islands and the Ryukyu Islands. Am. J. Bot. 2000, 87, 793–810. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Li, N.; Wang, S.; Zhou, Y.; Huang, W.; Ma, Y.Y.Y.; Zhou, R. Molecular evidence for the hybrid origin of Ilex dabieshanensis (Aquifoliaceae). PLoS ONE 2016, 11, e0147825. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.-C.; Gu, X.; Xiao, P.-G.; Liang, Z.; Xu, L.; Peng, Y. Research progress in the phytochemistry and biology of Ilex pharmaceutical resources. Acta Pharm. Sin. B 2013, 3, 8–19. [Google Scholar] [CrossRef]
- Choi, Y.H.; Sertic, S.; Kim, H.K.; Wilson, E.G.; Michopoulos, F.; Lefeber, A.W.M.; Erkelens, C.; Kricun, S.D.P.; Verpoorte, R. Classification of Ilex species based on metabolomic fingerprinting using nuclear magnetic resonance and multivariate data analysis. J. Agric. Food Chem. 2005, 53, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.M.; Tarragó, J.R. Anatomical structure and secretion compounds of colleters in nine Ilex species (Aquifoliaceae) from southern South America. Bot. J. Linn. Soc. 2009, 160, 197–210. [Google Scholar] [CrossRef]
- Borsch, T.; Quandt, D. Mutational dynamics and phylogenetic utility of noncoding chloroplast DNA. Plant Syst. Evol. 2009, 282, 169–199. [Google Scholar] [CrossRef]
- Provan, J.; Powell, W.; Hollingsworth, P.M. Chloroplast microsatellites: New tools for studies in plant ecology and evolution. Trends Ecol. Evol. 2001, 16, 142–147. [Google Scholar] [CrossRef]
- Provan, J.; Biss, P.M.; McMeel, D.; Mathews, S. Universal primers for the amplification of chloroplast microsatellites in grasses (Poaceae). Mol. Ecol. Notes 2004, 4, 262–264. [Google Scholar] [CrossRef]
- Rendell, S.; Ennos, R.A. Chloroplast DNA diversity of the dioecious European tree Ilex aquifolium L. (English holly). Mol. Ecol. 2003, 12, 2681–2688. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Liu, J.; Yu, J.; Wang, L.; Zhou, S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE 2012, 7, e35071. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; DePamphilis, C.W.; Leebens-Mack, J.; Muller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-J.; Lee, H.-L. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004, 11, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Prince, L.M. Plastid primers for angiosperm phylogenetics and phylogeography. Appl. Plant Sci. 2015, 3, 1400085. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Lickey, E.B.; Beck, J.T.; Farmer, S.B.; Liu, W.; Miller, J.; Siripun, K.C.; Winder, C.T.; Schilling, E.E.; Small, R.L. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005, 92, 142–166. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Shafer, H.L.; Rayne Leonard, O.; Kovach, M.J.; Schorr, M.; Morris, A.B. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: The tortoise and the hare IV. Am. J. Bot. 2014, 101, 1987–2004. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.J.; Soltis, P.S.; Bell, C.D.; Burleigh, J.G.; Soltis, D.E. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. USA 2010, 107, 4623–4628. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Liu, Y.-Y.; Tan, Y.-H.; Song, Y.; Corlett, R.T. The complete chloroplast genome sequence of Helwingia himalaica (Helwingiaceae, Aquifoliales) and a chloroplast phylogenomic analysis of the Campanulidae. PeerJ 2016, 4, e2734. [Google Scholar] [CrossRef] [PubMed]
- Debat, H.J.; Grabiele, M.; Aguilera, P.M.; Bubillo, R.E.; Otegui, M.B.; Ducasse, D.A.; Zapata, P.D.; Marti, D.A. Exploring the genes of yerba mate (Ilex paraguariensis A. St.-Hil.) by NGS and de novo transcriptome assembly. PLoS ONE 2014, 9, e109835. [Google Scholar] [CrossRef] [PubMed]
- Cascales, J.; Bracco, M.; Poggio, L.; Gottlieb, A.M. Genetic diversity of wild germplasm of ‘yerba mate’ (Ilex paraguariensis St. Hil.) from Uruguay. Genetica 2014, 142, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Diekmann, K.; Hodkinson, T.R.; Fricke, E.; Barth, S. An optimized chloroplast DNA extraction protocol for grasses (Poaceae) proves suitable for whole plastid genome sequencing and SNP detection. PLoS ONE 2008, 3, e2813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, C.; Hu, N.; Huang, H.; Gao, J.; Zhao, Y.J.; Gao, L.Z. An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. PLoS ONE 2012, 7, e31468. [Google Scholar] [CrossRef] [PubMed]
- Myers, E.W.; Sutton, G.G.; Delcher, A.L.; Dew, I.M.; Fasulo, D.P.; Flanigan, M.J.; Kravitz, S.A.; Mobarry, C.M.; Reinert, K.H.; Remington, K.A.; et al. A whole-genome assembly of Drosophila. Science 2000, 287, 2196–2204. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucl. Acids Res. 2012, 40. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-B.; Yang, S.X.; Li, H.T.; Yang, J.; Li, D.Z. Comparative chloroplast genomes of Camellia species. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primer for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shi, L.; Zhu, Y.; Chen, H.; Zhang, J.; Lin, X.; Guan, X. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 2012, 13, 715. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z. Comparative Analyses of Land Plant Plastid Genomes. Ph.D. Thesis, University of Texas, Austin, TX, USA, November 2010. [Google Scholar]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005, 33, W686–W689. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Brudno, M.; Do, C.B.; Cooper, G.M.; Kim, M.F.; Davydov, E.; Green, E.D.; Sidow, A.; Batzoglou, S. LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003, 13, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Tan, Y.-H.; Liu, Y.-Y.; Song, Y.; Yang, J.-B.; Corlett, R.T. Chloroplast genome structure in Ilex (Aquifoliaceae). Sci. Rep. 2016, 6, 28559. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Bryant, D.; Moulton, V. Neighbor-Net: An agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 2004, 21, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Kluge, A.G. A concern for evidence and a phylogenetic hypothesis of relationships among Epicrates (Boidae, serpentes). Syst. Zool. 1989, 38, 7–25. [Google Scholar] [CrossRef]
- Giberti, G.C. Aquifoliaceae. In Flora del Paraguay; Spichiger, R., Ramella, L., Eds.; Editions des Conservatoire et Jardin botaniques de la Ville de Geneva: Missouri Botanical Garden, Switzerland, 1994; p. 24. [Google Scholar]
- Giberti, G.C. Aquifoliaceae. In Flora Fanerogámica de Argentina; Programa PROFLORA (CONICET): Córdoba, Argentina, 1994. [Google Scholar]
- Giberti, G.C. Ilex theezans, especie confirmada para nuestra flora. Clave de las especies argentinas des genero Ilex (Aquifoliaceae). Bol. Soc. Argent Bot. 1990, 26, 159–162. [Google Scholar]
- Giberti, G.C. Hallazgo de Ilex brasiliensis (Aquifoliaceae) en la Argentina. Bol. Soc. Argent Bot. 1998, 33, 137–140. [Google Scholar]
- Filip, R.; Giberti, G.; Coussio, J.; Acevedo, C.; Ferraro, G. Estudio fitoquímico y farmacológico de Ilex theezans C. Martius ex Reisseck. Dominguezia 2000, 16, 47–53. [Google Scholar]
- Goloboff, P.A.; Farris, J.S.; Nixon, K.C. TNT, a free program for phylogenetic analysis. Cladistics 2008, 24, 774–786. [Google Scholar] [CrossRef]
- Escapa, I.H.; Catalano, S.A. Phylogenetic analysis of Araucariaceae: Integrating molecules, morphology, and fossils. Int. J. Plant Sci. 2013, 174, 1153–1170. [Google Scholar] [CrossRef] [Green Version]
- Tavaré, S. Some probabilistic and statistical problems in the analysis of DNA sequences. In Lectures on Mathematics in the Life Sciences; Miura, R.M., Ed.; American Mathematical Society: Providence, RI, USA, 1986; pp. 57–86. [Google Scholar]
- Raubeson, L.A.; Jansen, R.K. Chloroplast genomes of plants. In Plant Diversity and Evolution: Genotypic and Phenotypic Variation in Higher Plants; Henry, R.J., Ed.; CABI: Oxfordshire, UK, 2005; pp. 45–68. ISBN 0851999042. [Google Scholar]
- Li, R.; Ma, P.F.; Wen, J.; Yi, T.S. Complete sequencing of five Araliaceae chloroplast genomes and the phylogenetic implications. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Ruhlman, T.A.; Jansen, R.K. The plastid genomes of flowering plants. In Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2014; Volume 1132, pp. 3–38. ISBN 978-1-62703-994-9. [Google Scholar]
- Timme, R.E.; Kuehl, J.V.; Boore, J.L.; Jansen, R.K. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: Identification of divergent regions and categorization of shared repeats. Am. J. Bot. 2007, 94, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Middleton, C.P.; Senerchia, N.; Stein, N.; Akhunov, E.D.; Keller, B.; Wicker, T.; Kilian, B. Sequencing of chloroplast genomes from wheat, barley, rye and their relatives provides a detailed insight into the evolution of the triticeae tribe. PLoS ONE 2014, 9, e85761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tangphatsornruang, S.; Sangsrakru, D.; Chanprasert, J.; Uthaipaisanwong, P.; Yoocha, T.; Jomchai, N.; Tragoonrung, S. The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: Structural organization and phylogenetic relationships. DNA Res. 2010, 17, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, A.M.; Poggio, L. Quantitative and qualitative genomic characterization of cultivated Ilex L. species. Plant Genet. Resour. Charact. Util. 2014, 1–11. [Google Scholar] [CrossRef]
- Noirot, M.; Barre, P.; Duperray, C.; Louarn, J.; Hamon, S. Effects of caffeine and chlorogenic acid on propidium iodide accessibility to DNA: Consequences on genome size evaluation in coffee tree. Ann. Bot. 2003, 92, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Du, F.K.; Lang, T.; Lu, S.; Wang, Y.; Li, J.; Yin, K. An improved method for chloroplast genome sequencing in non-model forest tree species. Tree Genet. Genomes 2015, 11, 114. [Google Scholar] [CrossRef]
- Dangi, R.; Tamhankar, S.; Choudhary, R.K.; Rao, S. Molecular phylogenetics and systematics of Trigonella L. (Fabaceae) based on nuclear ribosomal ITS and chloroplast trnL intron sequences. Genet. Resour. Crop Evol. 2016, 63, 79–96. [Google Scholar] [CrossRef]
- Kim, H.K.; Saifullah; Khan, S.; Wilson, E.G.; Kricun, S.D.P.; Meissner, A.; Goraler, S.; Deelder, A.M.; Choi, Y.H.; Verpoorte, R. Metabolic classification of South American Ilex species by NMR-based metabolomics. Phytochemistry 2010, 71, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Barral, G.; Poggio, L.; Giberti, G.C. Chromosome numbers and DNA content from Ilex argentina (Aquifoliaceae). Bol. Soc. Argent. Bot. 1995, 30, 243–248. [Google Scholar]
- Nahar, L.; Russell, W.R.; Middleton, M.; Shoeb, M.; Sarker, S.D. Antioxidant phenylacetic acid derivatives from the seeds of Ilex aquifolium. Acta. Pharm. 2005, 55, 187–193. [Google Scholar] [PubMed]
- Hao, D.-C.; Xiao, P.-G.; Peng, Y.; Dong, J.; Liu, W. Evaluation of the chloroplast barcoding markers by mean and smallest interspecific distances. Pak. J. Bot. 2012, 44, 1271–1274. [Google Scholar]
- Li, L.; Xu, L.J.; Ma, G.Z.; Dong, Y.M.; Peng, Y.; Xiao, P.-G. The large-leaved Kudingcha (Ilex latifolia Thunb and Ilex kudingcha C.J. Tseng): A traditional Chinese tea with plentiful secondary metabolites and potential biological activities. J. Nat. Med. 2013, 67, 425–437. [Google Scholar] [CrossRef] [PubMed]
- Koop, H.U.; Herz, S.; Golds, T.J.; Nickelsen, J. The genetic transformation of plastids. In Topics in Current Genetics; Bock, R., Ed.; Springer-Verlag: Berlin/Heidelberg, Germany, 2007; Volume 19, pp. 457–510. ISBN 3540753753. [Google Scholar]
- Verma, D.; Daniell, H. Chloroplast vector systems for biotechnology applications. Plant Physiol. 2007, 145, 1129–1143. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cascales, J.; Bracco, M.; Garberoglio, M.J.; Poggio, L.; Gottlieb, A.M. Integral Phylogenomic Approach over Ilex L. Species from Southern South America. Life 2017, 7, 47. https://doi.org/10.3390/life7040047
Cascales J, Bracco M, Garberoglio MJ, Poggio L, Gottlieb AM. Integral Phylogenomic Approach over Ilex L. Species from Southern South America. Life. 2017; 7(4):47. https://doi.org/10.3390/life7040047
Chicago/Turabian StyleCascales, Jimena, Mariana Bracco, Mariana J. Garberoglio, Lidia Poggio, and Alexandra M. Gottlieb. 2017. "Integral Phylogenomic Approach over Ilex L. Species from Southern South America" Life 7, no. 4: 47. https://doi.org/10.3390/life7040047



