The Distinctive Regulation of Cyanobacterial Glutamine Synthetase
Abstract
:1. Introduction
1.1. GS Catalyzes a Core Reaction of N Assimilation in Bacteria
1.2. GS Regulation Is Diverse among Distinct Organisms
2. Cyanobacteria Evolved Exceptional GS Regulatory Mechanisms
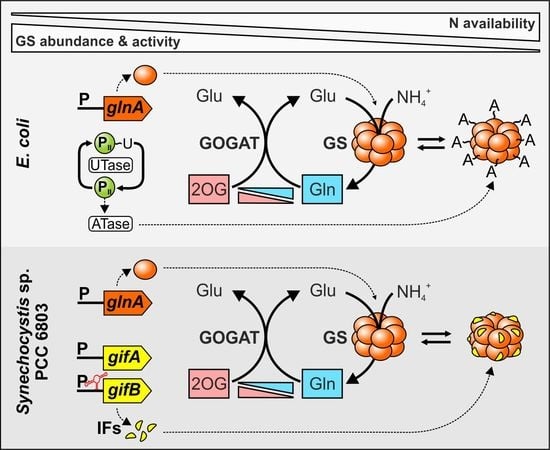
2.1. Sensing of the Cellular N Status and Transcriptional regulation of the glnA Gene in Cyanobacteria
2.2. Cyanobacterial GS Is Inactivated by Interaction with Small Proteins
2.3. The Biochemical Mechanism of GS Inactivation by Protein-Protein Interaction
2.4. GS Inactivating Factors Are Common in Cyanobacteria and Transcriptionally Regulated by NtcA
2.5. A Small Regulatory RNA Interacts with the gifA mRNA and Interferes with IF7 Production
2.6. A Glutamine Riboswitch in the gifB mRNA Regulates IF17 Synthesis
2.7. The Regulation of GS in Marine Picocyanobacteria is Unclear
2.8. GlnN Represents Another GS in Cyanobacteria
3. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Dixon, R.; Kahn, D. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2004, 2, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Forchhammer, K. Glutamine signalling in bacteria. Front. Biosci. J. Virtual Libr. 2007, 12, 358–370. [Google Scholar] [CrossRef]
- Zehr, J.P.; Ward, B.B. Nitrogen cycling in the ocean: New perspectives on processes and paradigms. Appl. Environ. Microbiol. 2002, 68, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Helling, R.B. Why does Escherichia coli have two primary pathways for synthesis of glutamate? J. Bacteriol. 1994, 176, 4664–4668. [Google Scholar] [CrossRef] [PubMed]
- Helling, R.B. Pathway choice in glutamate synthesis in Escherichia coli. J. Bacteriol. 1998, 180, 4571–4575. [Google Scholar] [PubMed]
- Halpern, Y.S.; Umbarger, H.E. Conversion of ammonia to amino groups in Escherichia coli. J. Bacteriol. 1960, 80, 285–288. [Google Scholar] [PubMed]
- Tempest, D.W.; Meers, J.L.; Brown, C.M. Synthesis of glutamate in Aerobacter aerogenes by a hitherto unknown route. Biochem. J. 1970, 117, 405–407. [Google Scholar] [CrossRef] [PubMed]
- Temple, S.J.; Vance, C.P.; Stephen Gantt, J. Glutamate synthase and nitrogen assimilation. Trends Plant Sci. 1998, 3, 51–56. [Google Scholar] [CrossRef]
- Kumada, Y.; Benson, D.R.; Hillemann, D.; Hosted, T.J.; Rochefort, D.A.; Thompson, C.J.; Wohlleben, W.; Tateno, Y. Evolution of the glutamine synthetase gene, one of the oldest existing and functioning genes. Proc. Natl. Acad. Sci. USA 1993, 90, 3009–3013. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.; Cullimore, J. Expression of three plant glutamine synthetase cDNA in Escherichia coli. Eur. J. Biochem. 1990, 193, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.R.; Masuchi, Y.; Robb, F.T.; Doolittlle, W.F. Evolutionary relationships of bacterial and archaeal glutamine synthetase genes. J. Mol. Evol. 1994, 38, 566–576. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.C.; Florencio, F.J. A new type of glutamine synthetase in cyanobacteria: The protein encoded by the glnN gene supports nitrogen assimilation in Synechocystis sp. strain PCC 6803. J. Bacteriol. 1994, 176, 1260–1267. [Google Scholar] [CrossRef] [PubMed]
- Southern, J.; Parker, J.R.; Woods, D.R. Novel structure, properties and inactivation of glutamine synthetase cloned from Bacteroides fragilis. Microbiology 1987, 133, 2437–2446. [Google Scholar] [CrossRef]
- Mathis, R.; Gamas, P.; Meyer, Y.; Cullimore, J.V. The presence of GSI-like genes in higher plants: Support for the paralogous evolution of GSI and GSII genes. J. Mol. Evol. 2000, 50, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.L.; Smith, G.J.; Alberte, R.S. Glutamine synthetase in marine algae: New surprises from an old enzyme. J. Phycol. 2002, 37, 793–795. [Google Scholar] [CrossRef]
- Darrow, R.A.; Knotts, R.R. Two forms of glutamine synthetase in free-living root-nodule bacteria. Biochem. Biophys. Res. Commun. 1977, 78, 554–559. [Google Scholar] [CrossRef]
- Edmands, J.; Noridge, N.A.; Benson, D.R. The actinorhizal root-nodule symbiont Frankia sp. strain CpI1 has two glutamine synthetases. Proc. Natl. Acad. Sci. USA 1987, 84, 6126–6130. [Google Scholar] [CrossRef] [PubMed]
- Kumada, Y.; Takano, E.; Nagaoka, K.; Thompson, C.J. Streptomyces hygroscopicus has two glutamine synthetase genes. J. Bacteriol. 1990, 172, 5343–5351. [Google Scholar] [CrossRef] [PubMed]
- Woolfolk, C.A.; Stadtman, E.R. Cumulative feedback inhibition in the multiple end product regulation of glutamine synthetase activity in Escherichia coli. Biochem. Biophys. Res. Commun. 1964, 17, 313–319. [Google Scholar] [CrossRef]
- Woolfolk, C.A.; Stadtman, E.R. Regulation of glutamine synthetase: III. Cumulative feedback inhibition of glutamine synthetase from Escherichia coli. Arch. Biochem. Biophys. 1967, 118, 736–755. [Google Scholar] [CrossRef]
- Woolfolk, C.A.; Shapiro, B.; Stadtman, E.R. Regulation of glutamine synthetase. I. Purification and properties of glutamine synthetase from Escherichia coli. Arch. Biochem. Biophys. 1966, 116, 177–192. [Google Scholar] [CrossRef]
- Liaw, S.H.; Pan, C.; Eisenberg, D. Feedback inhibition of fully unadenylylated glutamine synthetase from Salmonella typhimurium by glycine, alanine, and serine. Proc. Natl. Acad. Sci. USA 1993, 90, 4996–5000. [Google Scholar] [CrossRef] [PubMed]
- Liaw, S.H.; Jun, G.; Eisenberg, D. Interactions of nucleotides with fully unadenylylated glutamine synthetase from Salmonella typhimurium. Biochemistry 1994, 33, 11184–11188. [Google Scholar] [CrossRef] [PubMed]
- Adler, S.P.; Purich, D.; Stadtman, E.R. Cascade control of Escherichia coli glutamine synthetase. Properties of the PII regulatory protein and the uridylyltransferase-uridylyl-removing enzyme. J. Biol. Chem. 1975, 250, 6264–6272. [Google Scholar] [PubMed]
- Brown, M.S.; Segal, A.; Stadtman, E.R. Modulation of glutamine synthetase adenylylation and deadenylylation is mediated by metabolic transformation of the PII-regulatory protein. Proc. Natl. Acad. Sci. USA 1971, 68, 2949–2953. [Google Scholar] [CrossRef] [PubMed]
- Kingdon, H.S.; Shapiro, B.M.; Stadtman, E.R. Regulation of glutamine synthetase. VIII. ATP: Glutamine synthetase adenylyltransferase, an enzyme that catalyzes alterations in the regulatory properties of glutamine synthetase. Proc. Natl. Acad. Sci. USA 1967, 58, 1703–1710. [Google Scholar] [CrossRef] [PubMed]
- Stadtman, E.R. Oxidation of free amino acids and amino acid residues in proteins by radiolysis and by metal-catalyzed reactions. Annu. Rev. Biochem. 1993, 62, 797–821. [Google Scholar] [CrossRef] [PubMed]
- Garcia, E.; Rhee, S.G. Cascade control of Escherichia coli glutamine synthetase. Purification and properties of PII uridylyltransferase and uridylyl-removing enzyme. J. Biol. Chem. 1983, 258, 2246–2253. [Google Scholar] [PubMed]
- Mangum, J.H.; Magni, G.; Stadtman, E.R. Regulation of glutamine synthetase adenylylation and deadenylylation by the enzymatic uridylylation and deuridylylation of the PII regulatory protein. Arch. Biochem. Biophys. 1973, 158, 514–525. [Google Scholar] [CrossRef]
- Rhee, S.G.; Chock, P.B. Purification and characterization of uridylylated and unuridylylated forms of regulatory protein PII involved in the glutamine synthetase regulation in Escherichia coli. Isozymes 1983, 8, 141–153. [Google Scholar] [PubMed]
- Anderson, W.B.; Stadtman, E.R. Glutamine synthetase deadenylylation: A phosphorolytic reaction yielding ADP as nucleotide product. Biochem. Biophys. Res. Commun. 1970, 41, 704–709. [Google Scholar] [CrossRef]
- Shapiro, B.M. Glutamine synthetase deadenylylating enzyme system from Escherichia coli. Resolution into two components, specific nucleotide stimulation, and cofactor requirements. Biochemistry 1969, 8, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, B.M.; Stadtman, E.R. Glutamine synthetase deadenylylating enzyme. Biochem. Biophys. Res. Commun. 1968, 30, 32–37. [Google Scholar] [CrossRef]
- Shapiro, B.M.; Kingdon, H.S.; Stadtman, E.R. Regulation of glutamine synthetase. VII. Adenylyl glutamine synthetase: A new form of the enzyme with altered regulatory and kinetic properties. Proc. Natl. Acad. Sci. USA 1967, 58, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.A.; Dodsworth, J.A. Nitrogen regulation in bacteria and archaea. Annu. Rev. Microbiol. 2007, 61, 349–377. [Google Scholar] [CrossRef] [PubMed]
- Anderson, W.B.; Hennig, S.B.; Ginsburg, A.; Stadtman, E.R. Association of ATP: Glutamine synthetase adenylyltransferase activity with the PI component of the glutamine synthetase deadenylylation system. Proc. Natl. Acad. Sci. USA 1970, 67, 1417–1424. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.G.; Park, S.C.; Koo, J.H. The role of adenylyltransferase and uridylyltransferase in the regulation of glutamine synthetase in Escherichia coli. In Current Topics in Cellular Regulation; Shaltiel, S., Chock, P.B., Eds.; Modulation by Covalent Modification; Elsevier: Amsterdam, Netherlands, 1985; Volume 27, pp. 221–232. [Google Scholar]
- Stadtman, E.R. The story of glutamine synthetase regulation. J. Biol. Chem. 2001, 276, 44357–44364. [Google Scholar] [CrossRef] [PubMed]
- Stadtman, E.R. Regulation of glutamine synthetase activity. EcoSal Plus 2004, 1. [Google Scholar] [CrossRef] [PubMed]
- Weiss, V.; Kramer, G.; Dünnebier, T.; Flotho, A. Mechanism of regulation of the bifunctional histidine kinase NtrB in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2002, 4, 229–233. [Google Scholar] [PubMed]
- Merrick, M.J.; Edwards, R.A. Nitrogen control in bacteria. Microbiol. Rev. 1995, 59, 604–622. [Google Scholar] [PubMed]
- Wray, L.V.; Zalieckas, J.M.; Fisher, S.H. Bacillus subtilis glutamine synthetase controls gene expression through a protein-protein interaction with transcription factor TnrA. Cell 2001, 107, 427–435. [Google Scholar] [CrossRef]
- Fedorova, K.; Kayumov, A.; Woyda, K.; Ilinskaja, O.; Forchhammer, K. Transcription factor TnrA inhibits the biosynthetic activity of glutamine synthetase in Bacillus subtilis. FEBS Lett. 2013, 587, 1293–1298. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.J.; Zhulin, I.B. ANTAR: An RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem. Sci. 2002, 27, 3–5. [Google Scholar] [CrossRef]
- Stutz, H.E.; Quixley, K.W.M.; McMaster, L.D.; Reid, S.J. Co-regulation of the nitrogen-assimilatory gene cluster in Clostridium saccharobutylicum. Microbiol. Read. Engl. 2007, 153, 3081–3090. [Google Scholar] [CrossRef] [PubMed]
- Ehlers, C.; Weidenbach, K.; Veit, K.; Forchhammer, K.; Schmitz, R.A. Unique mechanistic features of post-translational regulation of glutamine synthetase activity in Methanosarcina mazei strain Gö1 in response to nitrogen availability. Mol. Microbiol. 2005, 55, 1841–1854. [Google Scholar] [CrossRef] [PubMed]
- Pedro-Roig, L.; Camacho, M.; Bonete, M.-J. Regulation of ammonium assimilation in Haloferax mediterranei: Interaction between glutamine synthetase and two GlnK proteins. Biochim. Biophys. Acta BBA Proteins Proteom. 2013, 1834, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.R.; Castenholz, R.W. Evolution of thermotolerance in hot spring cyanobacteria of the genus Synechococcus. Appl. Environ. Microbiol. 2000, 66, 4222–4229. [Google Scholar] [CrossRef] [PubMed]
- Pandey, K.D.; Shukla, S.P.; Shukla, P.N.; Giri, D.D.; Singh, J.S.; Singh, P.; Kashyap, A.K. Cyanobacteria in Antarctica: Ecology, physiology and cold adaptation. Cell. Mol. Biol. 2004, 50, 575–584. [Google Scholar] [PubMed]
- Reed, R.H.; Chudek, J.A.; Foster, R.; Stewart, W.D.P. Osmotic adjustment in cyanobacteria from hypersaline environments. Arch. Microbiol. 1984, 138, 333–337. [Google Scholar] [CrossRef]
- Skulberg, O. Part 9. Terrestrial and Limnic Algae and Cyanobacteria. Available online: https://eurekamag.com/research/009/158/009158962.php (accessed on 6 July 2018).
- Stanier, R.Y.; Kunisawa, R.; Mandel, M.; Cohen-Bazire, G. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 1971, 35, 171–205. [Google Scholar] [PubMed]
- Puente-Sánchez, F.; Arce-Rodríguez, A.; Oggerin, M.; García-Villadangos, M.; Moreno-Paz, M.; Blanco, Y.; Rodríguez, N.; Bird, L.; Lincoln, S.A.; Tornos, F.; et al. Viable cyanobacteria in the deep continental subsurface. Proc. Natl. Acad. Sci. USA 2018, 201808176. [Google Scholar] [CrossRef] [PubMed]
- Flombaum, P.; Gallegos, J.L.; Gordillo, R.A.; Rincón, J.; Zabala, L.L.; Jiao, N.; Karl, D.M.; Li, W.K.W.; Lomas, M.W.; Veneziano, D.; et al. Present and future global distributions of the marine cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. USA 2013, 110, 9824–9829. [Google Scholar] [CrossRef] [PubMed]
- Dismukes, G.C.; Klimov, V.V.; Baranov, S.V.; Kozlov, Y.N.; DasGupta, J.; Tyryshkin, A. The origin of atmospheric oxygen on Earth: The innovation of oxygenic photosynthesis. Proc. Natl. Acad. Sci. USA 2001, 98, 2170–2175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores, E.; Herrero, A. Assimilatory nitrogen metabolism and its regulation. In The Molecular Biology of Cyanobacteria; Advances in Photosynthesis; Springer: Dordrecht, The Netherlands, 1994; pp. 487–517. ISBN 978-0-7923-3273-2. [Google Scholar]
- Herrero, A.; Muro-Pastor, A.M.; Flores, E. Nitrogen control in cyanobacteria. J. Bacteriol. 2001, 183, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Sauer, J.; Dirmeier, U.; Forchhammer, K. The Synechococcus strain PCC 7942 glnN product (Glutamine Synthetase III) helps recovery from prolonged nitrogen chlorosis. J. Bacteriol. 2000, 182, 5615–5619. [Google Scholar] [CrossRef] [PubMed]
- Dharmawardene, M.W.; Haystead, A.; Stewart, W.D. Glutamine synthetase of the nitrogen-fixing alga Anabaena cylindrica. Arch. Mikrobiol. 1973, 90, 281–295. [Google Scholar] [CrossRef] [PubMed]
- Lea, P.J.; Miflin, B.J. Glutamate synthase in blue-green algae. Biochem. Soc. Trans. 1975, 3, 381–384. [Google Scholar] [CrossRef] [PubMed]
- Neilson, A.H.; Doudoroff, M. Ammonia assimilation in blue-green algae. Arch. Mikrobiol. 1973, 89, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Meeks, J.C.; Wolk, C.P.; Thomas, J.; Lockau, W.; Shaffer, P.W.; Austin, S.M.; Chien, W.S.; Galonsky, A. The pathways of assimilation of 13NH4+ by the cyanobacterium, Anabaena cylindrica. J. Biol. Chem. 1977, 252, 7894–7900. [Google Scholar] [PubMed]
- Meeks, J.C.; Wolk, C.P.; Lockau, W.; Schilling, N.; Shaffer, P.W.; Chien, W.S. Pathways of assimilation of [13N]N2 and 13NH4+ by cyanobacteria with and without heterocysts. J. Bacteriol. 1978, 134, 125–130. [Google Scholar] [PubMed]
- Wolk, C.P.; Thomas, J.; Shaffer, P.W.; Austin, S.M.; Galonsky, A. Pathway of nitrogen metabolism after fixation of 13N-labeled nitrogen gas by the cyanobacterium Anabaena cylindrica. J. Biol. Chem. 1976, 251, 5027–5034. [Google Scholar] [PubMed]
- Chávez, S.; Lucena, J.M.; Reyes, J.C.; Florencio, F.J.; Candau, P. The presence of glutamate dehydrogenase is a selective advantage for the cyanobacterium Synechocystis sp. strain PCC 6803 under nonexponential growth conditions. J. Bacteriol. 1999, 181, 808–813. [Google Scholar] [PubMed]
- Fisher, R.; Tuli, R.; Haselkorn, R. A cloned cyanobacterial gene for glutamine synthetase functions in Escherichia coli, but the enzyme is not adenylylated. Proc. Natl. Acad. Sci. USA 1981, 78, 3393–3397. [Google Scholar] [CrossRef] [PubMed]
- Mérida, A.; Leurentop, L.; Candau, P.; Florencio, F.J. Purification and properties of glutamine synthetases from the cyanobacteria Synechocystis sp. strain PCC 6803 and Calothrix sp. strain PCC 7601. J. Bacteriol. 1990, 172, 4732–4735. [Google Scholar] [CrossRef] [PubMed]
- Orr, J.; Haselkorn, R. Kinetic and inhibition studies of glutamine synthetase from the cyanobacterium Anabaena 7120. J. Biol. Chem. 1981, 256, 13099–13104. [Google Scholar] [PubMed]
- Stacey, G.; Tabita, F.R.; Baalen, C.V. Nitrogen and ammonia assimilation in the cyanobacteria: Purification of glutamine synthetase from Anabaena sp. Strain CA. J. Bacteriol. 1977, 132, 596–603. [Google Scholar] [PubMed]
- Bottomley, P.J.; Stewart, W.D.P. ATP pools and transients in the blue-green alga, Anabaena cylindrica. Arch. Microbiol. 1976, 108, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Kupiec, R.; Gurevitz, M.; Zilberstein, A. Expression of glnA in the cyanobacterium Synechococcus sp. strain PCC 7942 is initiated from a single nif-like promoter under various nitrogen conditions. J. Bacteriol. 1993, 175, 7727–7731. [Google Scholar] [CrossRef] [PubMed]
- Elmorjani, K.; Liotenberg, S.; Houmard, J.; de Marsac, N.T. Molecular characterization of the gene encoding glutamine synthetase in the cyanobacterium Calothrix sp. PCC 7601. Biochem. Biophys. Res. Commun. 1992, 189, 1296–1302. [Google Scholar] [CrossRef]
- Orr, J.; Haselkorn, R. Regulation of glutamine synthetase activity and synthesis in free-living and symbiotic Anabaena spp. J. Bacteriol. 1982, 152, 626–635. [Google Scholar] [PubMed]
- Tumer, N.E.; Robinson, S.J.; Haselkorn, R. Different promoters for the Anabaena glutamine synthetase gene during growth using molecular or fixed nitrogen. Nature 1983, 306, 337–342. [Google Scholar] [CrossRef]
- Wagner, S.J.; Thomas, S.P.; Kaufman, R.I.; Nixon, B.T.; Stevens, S.E. The glnA gene of the cyanobacterium Agmenellum quadruplicatum PR-6 is nonessential for ammonium assimilation. J. Bacteriol. 1993, 175, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.C.; Muro-Pastor, M.I.; Florencio, F.J. Transcription of glutamine synthetase genes (glnA and glnN) from the cyanobacterium Synechocystis sp. strain PCC 6803 is differently regulated in response to nitrogen availability. J. Bacteriol. 1997, 179, 2678–2689. [Google Scholar] [CrossRef] [PubMed]
- Vega-Palas, M.A.; Flores, E.; Herrero, A. NtcA, a global nitrogen regulator from the cyanobacterium Synechococcus that belongs to the Crp family of bacterial regulators. Mol. Microbiol. 1992, 6, 1853–1859. [Google Scholar] [CrossRef] [PubMed]
- Herrero, A.; Paz-Yepes, J.; Vázquez-Bermúdez, M.F.; Flores, E. The NtcA-activated amt1 gene encodes a permease required for uptake of low concentrations of ammonium in the cyanobacterium Synechococcus sp. PCC 7942. Microbiology 2002, 148, 861–869. [Google Scholar] [CrossRef]
- Ohashi, Y.; Shi, W.; Takatani, N.; Aichi, M.; Maeda, S.; Watanabe, S.; Yoshikawa, H.; Omata, T. Regulation of nitrate assimilation in cyanobacteria. J. Exp. Bot. 2011, 62, 1411–1424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osanai, T.; Tanaka, K. Keeping in touch with PII: PII-interacting proteins in unicellular cyanobacteria. Plant Cell Physiol. 2007, 48, 908–914. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, J.; Forchhammer, K.; Burillo, S.; Contreras, A. Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol. Microbiol. 2006, 61, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, J.; Rodríguez-Mateos, F.; Salinas, P.; Lanza, V.F.; Dixon, R.; de la Cruz, F.; Contreras, A. PipX, the coactivator of NtcA, is a global regulator in cyanobacteria. Proc. Natl. Acad. Sci. USA 2014, 111, E2423–E2430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forcada-Nadal, A.; Forchhammer, K.; Rubio, V. SPR analysis of promoter binding of Synechocystis PCC6803 transcription factors NtcA and CRP suggests cross-talk and sheds light on regulation by effector molecules. FEBS Lett. 2014, 588, 2270–2276. [Google Scholar] [CrossRef] [PubMed]
- Muro-Pastor, M.I.; Reyes, J.C.; Florencio, F.J. Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J. Biol. Chem. 2001, 276, 38320–38328. [Google Scholar] [CrossRef] [PubMed]
- Tanigawa, R.; Shirokane, M.; Maeda Si, S.; Omata, T.; Tanaka, K.; Takahashi, H. Transcriptional activation of NtcA-dependent promoters of Synechococcus sp. PCC 7942 by 2-oxoglutarate in vitro. Proc. Natl. Acad. Sci. USA 2002, 99, 4251–4255. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.-X.; Jiang, Y.-L.; He, Y.-X.; Chen, Y.-F.; Teng, Y.-B.; Chen, Y.; Zhang, C.-C.; Zhou, C.-Z. Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate. Proc. Natl. Acad. Sci. USA 2010, 107, 12487–12492. [Google Scholar] [CrossRef] [PubMed]
- Giner-Lamia, J.; Robles-Rengel, R.; Hernández-Prieto, M.A.; Muro-Pastor, M.I.; Florencio, F.J.; Futschik, M.E. Identification of the direct regulon of NtcA during early acclimation to nitrogen starvation in the cyanobacterium Synechocystis sp. PCC 6803. Nucleic Acids Res. 2017, 45, 11800–11820. [Google Scholar] [CrossRef] [PubMed]
- Luque, I.; Flores, E.; Herrero, A. Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 1994, 13, 2862–2869. [Google Scholar] [CrossRef] [PubMed]
- Mitschke, J.; Vioque, A.; Haas, F.; Hess, W.R.; Muro-Pastor, A.M. Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc. Natl. Acad. Sci. USA 2011, 108, 20130–20135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vázquez-Bermúdez, M.F.; Herrero, A.; Flores, E. 2-Oxoglutarate increases the binding affinity of the NtcA (nitrogen control) transcription factor for the Synechococcus glnA promoter. FEBS Lett. 2002, 512, 71–74. [Google Scholar] [CrossRef]
- Zeth, K.; Fokina, O.; Forchhammer, K. Structural basis and target-specific modulation of ADP sensing by the Synechococcus elongatus PII signaling protein. J. Biol. Chem. 2014, 289, 8960–8972. [Google Scholar] [CrossRef] [PubMed]
- Mérida, A.; Candau, P.; Florencio, F.J. Regulation of glutamine synthetase activity in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803 by the nitrogen source: Effect of ammonium. J. Bacteriol. 1991, 173, 4095–4100. [Google Scholar] [CrossRef] [PubMed]
- Mérida, A.; Candau, P.; Florencio, F.J. In vitro reactivation of in vivo ammonium-inactivated glutamine synthetase from Synechocystis sp. PCC 6803. Biochem. Biophys. Res. Commun. 1991, 181, 780–786. [Google Scholar] [CrossRef]
- Reyes, J.C.; Florencio, F.J. A novel mechanism of glutamine synthetase inactivation by ammonium in the cyanobacterium Synechocystis sp. PCC 6803. Involvement of an inactivating protein. FEBS Lett. 1995, 367, 45–48. [Google Scholar] [CrossRef]
- García-Domínguez, M.; Reyes, J.C.; Florencio, F.J. Glutamine synthetase inactivation by protein–protein interaction. Proc. Natl. Acad. Sci. USA 1999, 96, 7161–7166. [Google Scholar] [CrossRef] [PubMed]
- Saelices, L.; Galmozzi, C.V.; Florencio, F.J.; Muro-Pastor, M.I. Mutational analysis of the inactivating factors, IF7 and IF17 from Synechocystis sp. PCC 6803: Critical role of arginine amino acid residues for glutamine synthetase inactivation. Mol. Microbiol. 2011, 82, 964–975. [Google Scholar] [CrossRef] [PubMed]
- Galmozzi, C.V.; Fernández-Avila, M.J.; Reyes, J.C.; Florencio, F.J.; Muro-Pastor, M.I. The ammonium-inactivated cyanobacterial glutamine synthetase I is reactivated in vivo by a mechanism involving proteolytic removal of its inactivating factors. Mol. Microbiol. 2007, 65, 166–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saelices, L.; Robles-Rengel, R.; Florencio, F.J.; Muro-Pastor, M.I. A core of three amino acids at the carboxyl-terminal region of glutamine synthetase defines its regulation in cyanobacteria. Mol. Microbiol. 2015, 96, 483–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Domínguez, M.; Reyes, J.C.; Florencio, F.J. NtcA represses transcription of gifA and gifB, genes that encode inhibitors of glutamine synthetase type I from Synechocystis sp. PCC 6803. Mol. Microbiol. 2000, 35, 1192–1201. [Google Scholar] [CrossRef] [PubMed]
- Luque, I.; Forchhammer, K. Nitrogen assimilation and C/N balance sensing. In The Cyanobacteria: Molecular Biology, Genetics and Evolution; Herrero, A., Flores, E., Eds.; Caister Academic Press: Norwich, UK, 2008; pp. 335–382. [Google Scholar]
- Galmozzi, C.V.; Saelices, L.; Florencio, F.J.; Muro-Pastor, M.I. Posttranscriptional regulation of glutamine synthetase in the filamentous cyanobacterium Anabaena sp. PCC 7120: Differential expression between vegetative cells and heterocysts. J. Bacteriol. 2010, 192, 4701–4711. [Google Scholar] [CrossRef] [PubMed]
- Waters, L.S.; Storz, G. Regulatory RNAs in bacteria. Cell 2009, 136, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by small RNAs in bacteria: Expanding frontiers. Mol. Cell 2011, 43, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.G.H.; Romby, P. Small RNAs in bacteria and archaea: Who they are, what they do, and how they do it. Adv. Genet. 2015, 90, 133–208. [Google Scholar] [CrossRef] [PubMed]
- Kopf, M.; Hess, W.R. Regulatory RNAs in photosynthetic cyanobacteria. FEMS Microbiol. Rev. 2015, 39, 301–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopf, M.; Klähn, S.; Scholz, I.; Matthiessen, J.K.F.; Hess, W.R.; Voß, B. Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2014, 21, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Mitschke, J.; Georg, J.; Scholz, I.; Sharma, C.M.; Dienst, D.; Bantscheff, J.; Voß, B.; Steglich, C.; Wilde, A.; Vogel, J.; et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. USA 2011, 108, 2124–2129. [Google Scholar] [CrossRef] [PubMed]
- Voss, B.; Georg, J.; Schön, V.; Ude, S.; Hess, W.R. Biocomputational prediction of non-coding RNAs in model cyanobacteria. BMC Genom. 2009, 10, 123. [Google Scholar] [CrossRef] [PubMed]
- Ionescu, D.; Voss, B.; Oren, A.; Hess, W.R.; Muro-Pastor, A.M. Heterocyst-specific transcription of NsiR1, a non-coding RNA encoded in a tandem array of direct repeats in cyanobacteria. J. Mol. Biol. 2010, 398, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Muro-Pastor, A.M. The heterocyst-specific NsiR1 small RNA is an early marker of cell differentiation in cyanobacterial filaments. mBio 2014, 5, e01079-14. [Google Scholar] [CrossRef] [PubMed]
- Klähn, S.; Schaal, C.; Georg, J.; Baumgartner, D.; Knippen, G.; Hagemann, M.; Muro-Pastor, A.M.; Hess, W.R. The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc. Natl. Acad. Sci. USA 2015, 112, E6243–E6252. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, M.; Bryant, D.A. Acclimation of the global transcriptome of the cyanobacterium Synechococcus sp. Strain PCC 7002 to nutrient limitations and different nitrogen sources. Front. Microbiol. 2012, 3, 145. [Google Scholar] [CrossRef] [PubMed]
- Kopf, M.; Klähn, S.; Scholz, I.; Hess, W.R.; Voß, B. Variations in the non-coding transcriptome as a driver of inter-strain divergence and physiological adaptation in bacteria. Sci. Rep. 2015, 5, 9560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogel, J.; Wagner, E.G.H. Target identification of small noncoding RNAs in bacteria. Curr. Opin. Microbiol. 2007, 10, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Escribano, I.; Vioque, A.; Muro-Pastor, A.M. NsrR1, a nitrogen stress-repressed sRNA, contributes to the regulation of nblA in Nostoc sp. PCC 7120. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Prasse, D.; Schmitz, R.A. Small RNAs involved in regulation of nitrogen metabolism. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Mironov, A.S.; Gusarov, I.; Rafikov, R.; Lopez, L.E.; Shatalin, K.; Kreneva, R.A.; Perumov, D.A.; Nudler, E. Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell 2002, 111, 747–756. [Google Scholar] [CrossRef]
- Nahvi, A.; Sudarsan, N.; Ebert, M.S.; Zou, X.; Brown, K.L.; Breaker, R.R. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002, 9, 1043–1049. [Google Scholar] [CrossRef]
- Winkler, W.; Nahvi, A.; Breaker, R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 2002, 419, 952–956. [Google Scholar] [CrossRef] [PubMed]
- Barrick, J.E.; Breaker, R.R. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007, 8, R239. [Google Scholar] [CrossRef] [PubMed]
- Breaker, R.R. Riboswitches: From ancient gene-control systems to modern drug targets. Future Microbiol. 2009, 4, 771–773. [Google Scholar] [CrossRef] [PubMed]
- Dambach, M.D.; Winkler, W.C. Expanding roles for metabolite-sensing regulatory RNAs. Curr. Opin. Microbiol. 2009, 12, 161–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garst, A.D.; Edwards, A.L.; Batey, R.T. Riboswitches: Structures and mechanisms. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, Z.; Wang, J.X.; Bogue, J.; Yang, J.; Corbino, K.; Moy, R.H.; Breaker, R.R. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 2010, 11, R31. [Google Scholar] [CrossRef] [PubMed]
- Kalvari, I.; Argasinska, J.; Quinones-Olvera, N.; Nawrocki, E.P.; Rivas, E.; Eddy, S.R.; Bateman, A.; Finn, R.D.; Petrov, A.I. Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2018, 46, D335–D342. [Google Scholar] [CrossRef] [PubMed]
- Ames, T.D.; Breaker, R.R. Bacterial aptamers that selectively bind glutamine. RNA Biol. 2011, 8, 82–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klähn, S.; Bolay, P.; Wright, P.R.; Atilho, R.M.; Brewer, K.I.; Hagemann, M.; Breaker, R.R.; Hess, W.R. A glutamine riboswitch is a key element for the regulation of glutamine synthetase in cyanobacteria. Nucleic Acids Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, S.W.; Olson, R.J.; Zettler, E.R.; Goericke, R.; Waterbury, J.B.; Welschmeyer, N.A. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature 1988, 334, 340–343. [Google Scholar] [CrossRef]
- Moore, L.R.; Chisholm, S.W. Photophysiology of the marine cyanobacterium Prochlorococcus: Ecotypic differences among cultured isolates. Limnol. Oceanogr. 1999, 44, 628–638. [Google Scholar] [CrossRef]
- Chisholm, S.W.; Frankel, S.L.; Goericke, R.; Olson, R.J.; Palenik, B.; Waterbury, J.B.; West-Johnsrud, L.; Zettler, E.R. Prochlorococcus marinus nov. gen. nov. sp.: An oxyphototrophic marine prokaryote containing divinyl chlorophyll a and b. Arch. Microbiol. 1992, 157, 297–300. [Google Scholar] [CrossRef]
- Rocap, G.; Larimer, F.W.; Lamerdin, J.; Malfatti, S.; Chain, P.; Ahlgren, N.A.; Arellano, A.; Coleman, M.; Hauser, L.; Hess, W.R.; et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 2003, 424, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Dufresne, A.; Garczarek, L.; Partensky, F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005, 6, R14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giovannoni, S.J.; Thrash, J.C.; Temperton, B. Implications of streamlining theory for microbial ecology. ISME J. 2014, 8, 1553–1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steglich, C.; Futschik, M.E.; Lindell, D.; Voss, B.; Chisholm, S.W.; Hess, W.R. The challenge of regulation in a minimal photoautotroph: Non-coding RNAs in Prochlorococcus. PLoS Genet. 2008, 4, e1000173. [Google Scholar] [CrossRef]
- Moore, C.M.; Mills, M.M.; Arrigo, K.R.; Berman-Frank, I.; Bopp, L.; Boyd, P.W.; Galbraith, E.D.; Geider, R.J.; Guieu, C.; Jaccard, S.L.; et al. Processes and patterns of oceanic nutrient limitation. Nat. Geosci. 2013, 6, 701–710. [Google Scholar] [CrossRef] [Green Version]
- Grzymski, J.J.; Dussaq, A.M. The significance of nitrogen cost minimization in proteomes of marine microorganisms. ISME J. 2012, 6, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Read, R.W.; Berube, P.M.; Biller, S.J.; Neveux, I.; Cubillos-Ruiz, A.; Chisholm, S.W.; Grzymski, J.J. Nitrogen cost minimization is promoted by structural changes in the transcriptome of N-deprived Prochlorococcus cells. ISME J. 2017, 11, 2267–2278. [Google Scholar] [CrossRef] [PubMed]
- Scanlan, D.J.; Ostrowski, M.; Mazard, S.; Dufresne, A.; Garczarek, L.; Hess, W.R.; Post, A.F.; Hagemann, M.; Paulsen, I.; Partensky, F. Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. MMBR 2009, 73, 249–299. [Google Scholar] [CrossRef] [PubMed]
- El Alaoui, S.; Diez, J.; Humanes, L.; Toribio, F.; Partensky, F.; García-Fernández, J.M. In vivo regulation of glutamine synthetase activity in the marine chlorophyll b-containing cyanobacterium Prochlorococcus sp. strain PCC 9511 (oxyphotobacteria). Appl. Environ. Microbiol. 2001, 67, 2202–2207. [Google Scholar] [CrossRef] [PubMed]
- El Alaoui, S.; Diez, J.; Toribio, F.; Gómez-Baena, G.; Dufresne, A.; García-Fernández, J.M. Glutamine synthetase from the marine cyanobacteria Prochlorococcus spp: Characterization, phylogeny and response to nutrient limitation. Environ. Microbiol. 2003, 5, 412–423. [Google Scholar] [CrossRef] [PubMed]
- Florencio, F.J.; Reyes, J.C. Regulation of ammonium assimilation in cyanobacteria. In Photosynthetic Nitrogen Assimilation and Associated Carbon and Respiratory Metabolism; Advances in Photosynthesis and Respiration; Springer: Dordrecht, The Netherlands, 2002; pp. 93–113. ISBN 978-0-7923-6336-1. [Google Scholar]
- García-Fernández, J.M.; de Marsac, N.T.; Diez, J. Streamlined regulation and gene loss as adaptive mechanisms in Prochlorococcus for optimized nitrogen utilization in oligotrophic environments. Microbiol. Mol. Biol. Rev. 2004, 68, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Tolonen, A.C.; Aach, J.; Lindell, D.; Johnson, Z.I.; Rector, T.; Steen, R.; Church, G.M.; Chisholm, S.W. Global gene expression of Prochlorococcus ecotypes in response to changes in nitrogen availability. Mol. Syst. Biol. 2006, 2, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez-Baena, G.; Diez, J.; García-Fernández, J.M.; El Alaoui, S.; Humanes, L. Regulation of glutamine synthetase by metal-catalyzed oxidative modification in the marine oxyphotobacterium Prochlorococcus. Biochim. Biophys. Acta 2001, 1568, 237–244. [Google Scholar] [CrossRef]
- Gómez-Baena, G.; Domínguez-Martín, M.A.; Donaldson, R.P.; García-Fernández, J.M.; Diez, J. Glutamine synthetase sensitivity to oxidative modification during nutrient starvation in Prochlorococcus marinus PCC 9511. PLoS ONE 2015, 10, e0135322. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.; Sugano, S. Inactivation of Bacillus subtilis glutamine synthetase by metal-catalyzed oxidation. J. Biochem. 1992, 112, 828–833. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.L.; Oliver, C.N.; Fulks, R.M.; Stadtman, E.R. Turnover of bacterial glutamine synthetase: Oxidative inactivation precedes proteolysis. Proc. Natl. Acad. Sci. USA 1981, 78, 2120–2124. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.T.; Parker, J.R.; Goodman, H.J.; Jones, D.T.; Woods, D.R. Molecular analysis of a novel glutamine synthetase of the anaerobe Bacteroides fragilis. J. Gen. Microbiol. 1989, 135, 3271–3279. [Google Scholar] [CrossRef] [PubMed]
- Pesole, G.; Bozzetti, M.P.; Lanave, C.; Preparata, G.; Saccone, C. Glutamine synthetase gene evolution: A good molecular clock. Proc. Natl. Acad. Sci. USA 1991, 88, 522–526. [Google Scholar] [CrossRef] [PubMed]
- García-Domínguez, M.; Reyes, J.C.; Florencio, F.J. Purification and characterization of a new type of glutamine synthetase from cyanobacteria. Eur. J. Biochem. 1997, 244, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Martín, M.A.; Díez, J.; García-Fernández, J.M. Physiological studies of glutamine synthetases I and III from Synechococcus sp. WH7803 reveal differential regulation. Front. Microbiol. 2016, 7, 969. [Google Scholar] [CrossRef] [PubMed]
- Crespo, J.L.; García-Domínguez, M.; Florencio, F.J. Nitrogen control of the glnN gene that codes for GS type III, the only glutamine synthetase in the cyanobacterium Pseudanabaena sp. PCC 6903. Mol. Microbiol. 1998, 30, 1101–1112. [Google Scholar] [CrossRef] [PubMed]



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bolay, P.; Muro-Pastor, M.I.; Florencio, F.J.; Klähn, S. The Distinctive Regulation of Cyanobacterial Glutamine Synthetase. Life 2018, 8, 52. https://doi.org/10.3390/life8040052
Bolay P, Muro-Pastor MI, Florencio FJ, Klähn S. The Distinctive Regulation of Cyanobacterial Glutamine Synthetase. Life. 2018; 8(4):52. https://doi.org/10.3390/life8040052
Chicago/Turabian StyleBolay, Paul, M. Isabel Muro-Pastor, Francisco J. Florencio, and Stephan Klähn. 2018. "The Distinctive Regulation of Cyanobacterial Glutamine Synthetase" Life 8, no. 4: 52. https://doi.org/10.3390/life8040052