Developing a Prototype System for Integrating Pharmacogenomics Findings into Clinical Practice
Abstract
:1. Introduction
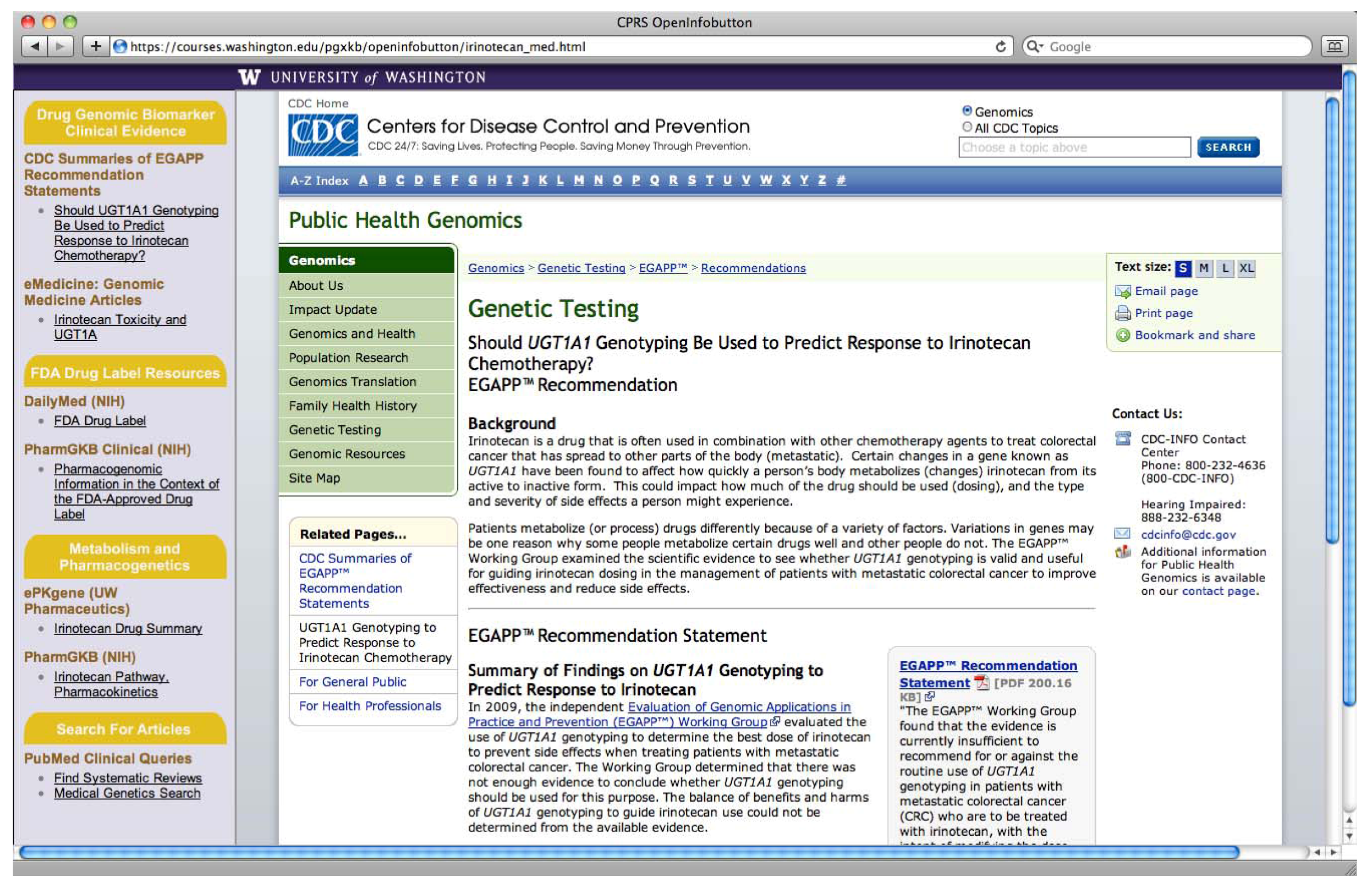
2. Methods
Developing a Prototype Model for Pharmacogenomics Clinical Decision Support
| CDS delivery method | Description |
|---|---|
| (CDS implementation type) | |
| Method 1: PGx link to e-resources | The clinician selects the medication they wish to prescribe and a context-specific link to PGx e-resources appears. |
| (semi-active CDS) | A context-specific link to PGx e-resources appears next to the genetic test results of interest. |
| Method 2: Alert message | The clinician enters prescribing information consistent with empirical therapy, clicks the “prescribe” button, and an alert message pops up providing a message relevant to the patients’ genetic test results and the medication being ordered. |
| (active CDS) | |
| Method 3: PGx link to e-resources within an alert message | A context-specific link to PGx e-resources appears within an alert message relevant to the patients’ genetic test results and the medication being ordered. |
| (semi-active CDS that follows active CDS) |
3. Results
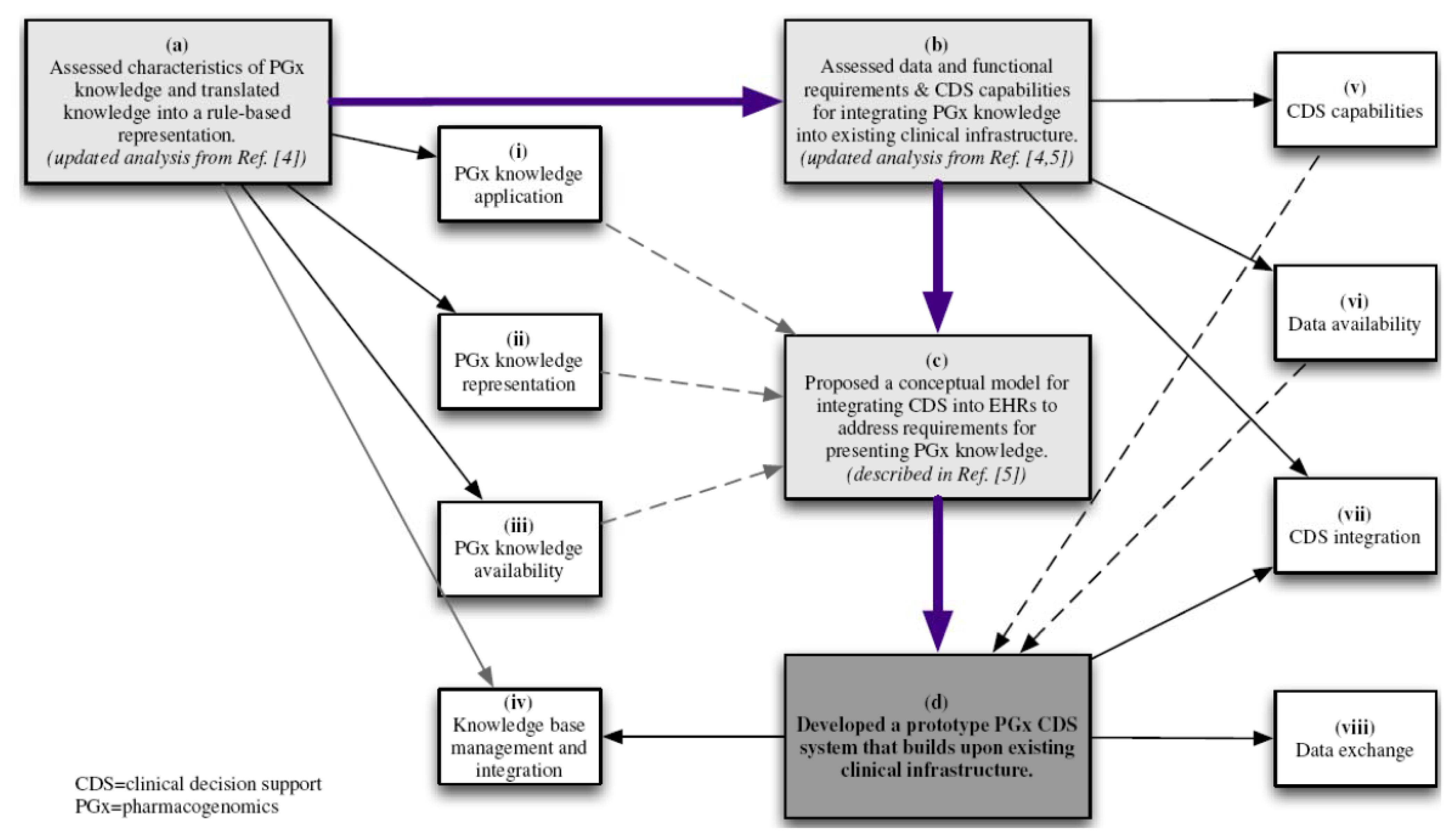
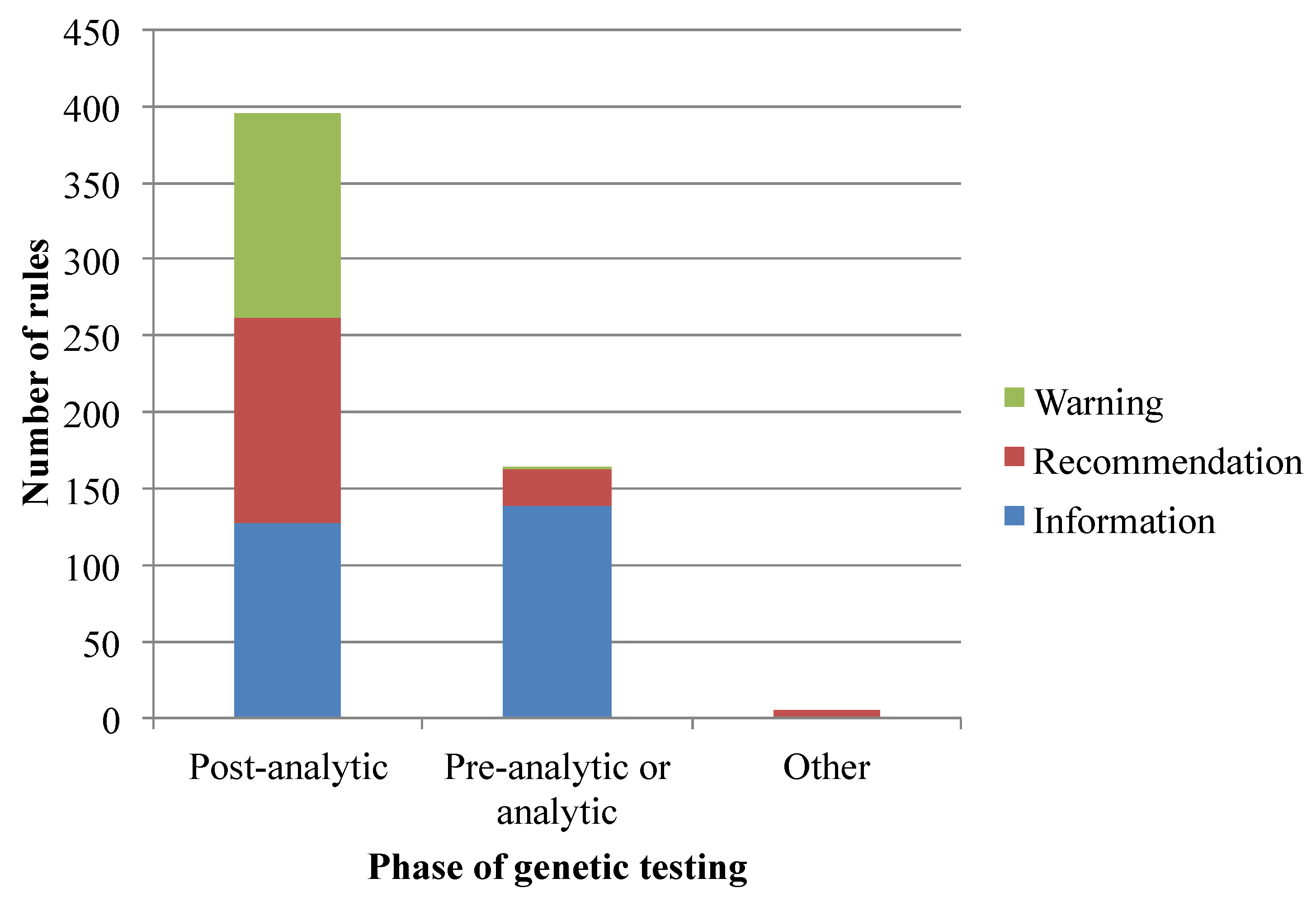
3.1. Translation of Pharmacogenomics Knowledge into a Rule-Based Representation
3.2. Data and Functional Requirements of Current Clinical Decision Support Systems
3.3. Developing a Prototype Model for Pharmacogenomics Clinical Decision Support
3.3.1. Limitations to Implementing the Pharmacogenomics Clinical Decision Support Prototype
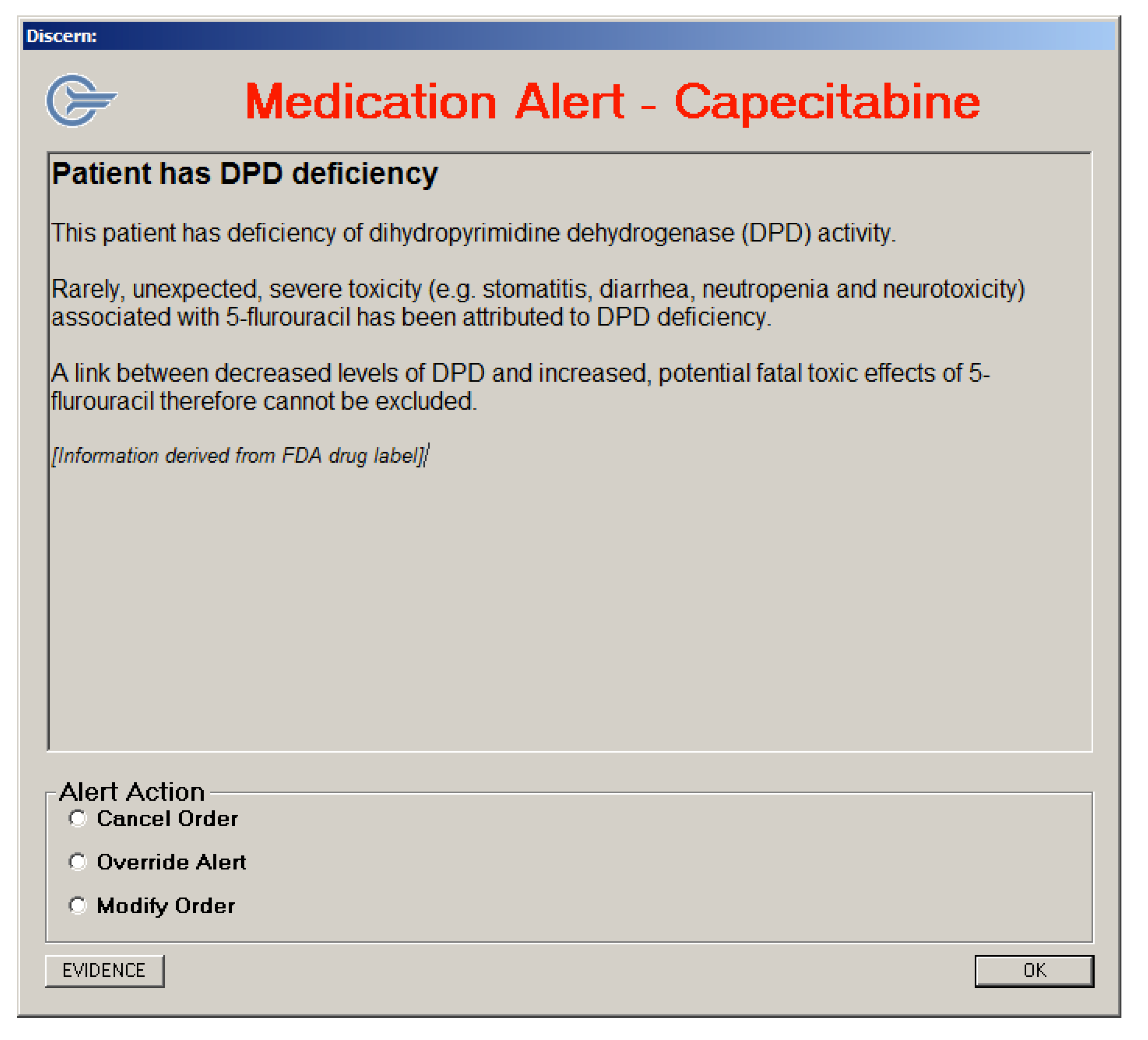
3.3.3. Alert Messages Generated using the Discern Expert Rules Engine
4. Discussion
4.1. Implementing Clinical Decision Support for Drug Therapy Individualization
4.2. Providing Pharmacogenomics Knowledge within Clinical Information Systems
5. Conclusions
Supplementary Files
Acknowledgments
Conflict of Interest
References and Notes
- Baars, M.J.; Henneman, L.; Ten Kate, L.P. Deficiency of knowledge of genetics and genetic tests among general practitioners, gynecologists, and pediatricians: A global problem. Genet. Med. 2005, 7, 605–610. [Google Scholar]
- Menasha, J.; Schechter, C.; Willner, J. Genetic testing: A physician’s perspective. Mt. Sinai J. Med. 2000, 67, 144–151. [Google Scholar]
- US Department of Health and Human Services; Office of the National Coordinator for Health Information Technology. Personalized Healthcare Detailed Use Case. Available online: http://healthit.hhs.gov/portal/server.pt/gateway/PTARGS_0_10731_848111_0_0_18/PHCDetailed.pdf (accessed on 11 October 2012).
- Overby, C.; Tarczy-Hornoch, P.; Hoath, J.; Kalet, I.; Veenstra, D. Feasibility of incorporating genomic knowledge into electronic medical records for pharmacogenomic clinical decision support. BMC Bioinform. 2010, 11 Suppl. 9, S10. [Google Scholar]
- Overby, C.; Tarczy-Hornoch, P.; Hoath, J.; Smith, J.; Fenstermacher, D.; Devine, E. An evaluation of functional and user interface requirements for pharmacogenomic clinical decision support. In Proceedings of 2011 First IEEE International Conference on Health Informatics, Imaging, and Systems Biology (HISB), San Jose, CA, USA, 26–29 July, 2011.
- US Food and Drug Administration. Table of Pharmacogenomic Biomarkers in Drug Labels. (updated May 2011). 2011. Available online: http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm (accessed on 11 October 2012).
- Wright, A.; Goldberg, H.; Hongsermeier, T.; Middleton, B. A description and functional taxonomy of rule-based decision support content at a large integrated delivery network. J. Am. Med. Inform. Assoc. 2007, 14, 489–496. [Google Scholar] [CrossRef]
- Del Fiol, G.; Kawamoto, K.; Cimino, J.J. Open-source, standards-based software to enable decision support. Proc. AMIA Annu. Fall Symp. 2011, 2011, 2127. [Google Scholar]
- Openinfobutton Project Webpage. Available online: http:www.openinfobutton.org (accessed on 11 October 2012).
- Discern Expert®. Cerner Coorporation, Kansas City, MO, USA, 2011.
- US Centers for Disease Control and Prevention. CDC Summaries of EGAPP Recommendation Statements (last updated 13 January 2011). Available online: http://www.cdc.gov/genomics/gtesting/EGAPP/recommend/ (accessed on 11 October 2012).
- Gwinn, M.; Dotson, W.D.; Khoury, M.J. Plos currents: Evidence on genomic tests—At the crossroads of translation. PLoS Curr. 2010, 2. [Google Scholar]
- Relling, M.V.; Klein, T.E. CPIC: Clinical pharmacogenetics implementation consortium of the pharmacogenomics research network. Clin. Pharmacol. Ther. 2011, 89, 464–467. [Google Scholar] [CrossRef]
- Hughes, S. Vanderbilt now also routinely gene testing for clopidogrel metabolizer status. HeartWire 2010. [Google Scholar]
- US National Library of Medicine. DailyMed Website. Available online: http://dailymed.nlm.nih.gov/dailymed/about.cfm (accessed on 11 October 2012).
- The Pharmacogenomics Knowledge Base [PharmGKB] Website. Available online: http://pharmgkb.org (accessed on 18 October 2012).
- Hernandez-Boussard, T.; Whirl-Carrillo, M.; Hebert, J.M.; Gong, L.; Owen, R.; Gong, M.; Gor, W.; Liu, F.; Truong, C.; Whaley, R.; et al. The pharmacogenetics and pharmacogenomics knowledge base: Accentuating the knowledge. Nucleic Acids Res. 2008, 36, D913–D918. [Google Scholar]
- Hachad, H.; Overby, C.L.; Argon, S.; Yeung, C.K.; Ragueneau-Majlessi, I.; Levy, R.H. E-pkgene: A knowledge-based research tool for analysing the impact of genetics on drug exposure. Hum. Genomics 2011, 5, 506–515. [Google Scholar] [CrossRef]
- Del Fiol, G.; Williams, M.; Maram, N.; Rocha, R.; Wood, G.; Mitchell, J. Integrating genetic information resources with an ehr. Proc. AMIA Annu. Symp. 2006, 2006, 904. [Google Scholar]
- Kaihoi, B.; Petersen, C.; Bolander, M. Providing "just-in-time" medical genomics information for patient care. Proc. AMIA Annu. Symp. 2005, 2005, 1003. [Google Scholar]
- Maviglia, S.; Yoon, C.; Bates, D.; Kuperman, G. Knowledgelink: Impact of context-sensitive information retrieval on clinicians’ information needs. J. Am. Med. Inform. Assoc. 2006, 13, 67–73. [Google Scholar]
- US Department of Health and Human Services. Personalized Health Care: Pioneers, Partnerships, Progress. Available online: http:www.hhs.gov/myhealthcare/news/phc_2008_report.pdf (accessed on 11 October 2012).
- Relling, M. Delivery of information: Implementation of pharmacogenomics in clinical practice. In Presented at Integrating Large-Scale Genomic Information into Clinical Practice: A Workshop, Washington, DC, USA, 19 July 2011.
- Vnencak-Jones, C. Pharmacogenetics: From DNA to dosage—Just a click away. In Presented at Operationalizing Innovation: Where the Cutting Edge Meets Current Practice (CAP Foundation Confernce Series IV), Rosemont, IL, USA, 15–17 April 2011.
- Frueh, F.; Amur, S.; Mummaneni, P.; Epstein, R.; Aubert, R.; DeLuca, T.; Verbrugge, R.; Burckart, G.; Lesko, L. Pharmacogenomic biomarker information in drug labels approved by the united states food and drug administration: Prevalence of related drug use. Pharmacotherapy 2008, 28, 992–998. [Google Scholar] [CrossRef]
- Deverka, P.; Doksum, T.; Carlson, R. Integrating molecular medicine into the us health-care system: Opportunities, barriers, and policy challenges. Clin. Pharmacol. Ther. 2007, 82, 427–434. [Google Scholar] [CrossRef]
- Context-aware knowledge retrieval (infobutton) product brief. HL7 International Wiki Site. Available online: http://wiki.hl7.org/index.php?title=Product_Infobutton (accessed on 11 October 2012).
- Health Information Technology: Standards, Implementation Specifications, and Certification Criteria for Electronic Health Record Technology, 2014 edition; revisions to the permanent certification program for health information technology. In 45 CFR Part 170; Office of the National Coordinator for Health Information Technology (ONC), Department of Health and Human Services: Washington, D.C., USA, 2012.
- Del Fiol, G.; Huser, V.; Strasberg, H.R.; Maviglia, S.M.; Curtis, C.; Cimino, J.J. Implementations of the HL7 context-aware knowledge retrieval ("infobutton") standard: Challenges, strengths, limitations, and uptake. J. Biomed. Inform. 2012, 45, 726–735. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Overby, C.L.; Tarczy-Hornoch, P.; Kalet, I.J.; Thummel, K.E.; Smith, J.W.; Fiol, G.D.; Fenstermacher, D.; Devine, E.B. Developing a Prototype System for Integrating Pharmacogenomics Findings into Clinical Practice. J. Pers. Med. 2012, 2, 241-256. https://doi.org/10.3390/jpm2040241
Overby CL, Tarczy-Hornoch P, Kalet IJ, Thummel KE, Smith JW, Fiol GD, Fenstermacher D, Devine EB. Developing a Prototype System for Integrating Pharmacogenomics Findings into Clinical Practice. Journal of Personalized Medicine. 2012; 2(4):241-256. https://doi.org/10.3390/jpm2040241
Chicago/Turabian StyleOverby, Casey Lynnette, Peter Tarczy-Hornoch, Ira J. Kalet, Kenneth E. Thummel, Joe W. Smith, Guilherme Del Fiol, David Fenstermacher, and Emily Beth Devine. 2012. "Developing a Prototype System for Integrating Pharmacogenomics Findings into Clinical Practice" Journal of Personalized Medicine 2, no. 4: 241-256. https://doi.org/10.3390/jpm2040241