1. Introduction
Hypertension (HTN) and its end-organ manifestations including stroke, coronary heart disease, and chronic renal failure are major contributors to morbidity and mortality in the United States and globally [
1,
2]. On average, life expectancy is reduced by five years among those with HTN, which is responsible for nearly one in every eight deaths worldwide [
3]. Multiple important individual, societal, and environmental variables contribute to an individual’s risk of developing HTN [
4]. Particularly noteworthy is the persistence of racial disparities in HTN prevalence, control, and untoward outcomes between African Americans (AA) and Caucasians (CAU), despite the fact that a higher proportion of AAs are both aware of and receive treatment for HTN [
5].
Typical intervention strategies used to reduce blood pressure (BP) include implementing strategies at various levels of patient influence (patient, family, healthcare provider, community level) [
6] and in some cases implementing strategies to enhance control among specific groups, such as African Americans [
7,
8]. Such interventions aim to reduce BP by improving medication adherence, guiding better lifestyle choices, using home BP monitors, addressing clinical inertia in intensifying anti-hypertensive treatment, using team-based approaches to improve HTN management, and other strategies [
7,
8,
9,
10,
11].
Additional factors of interest in the study of HTN include advancing our understanding of how genes associate with both the presence of HTN and the responsiveness to interventions aiming to reduce BP, and how genes interact with the many other contributing factors, such as advancing age, that influence the prevalence of HTN. A better understanding of these genetic influences may inform the implementation of targeted and personalized therapies that mitigate the untoward consequences of sustained HTN.
Genome-wide association studies (GWAS) identified associations between specific genetic loci, mapped by the presence of single nucleotide polymorphisms (SNPs) that represent genetic variation among populations, and the prevalence of HTN [
12,
13,
14,
15,
16,
17,
18,
19]. Remarkably, Simino and colleagues [
12] developed a unique approach to analyzing cross-sectional GWAS data by stratifying hypertension-SNP association data into age brackets, which provided results suggesting that some SNPs associate with BP, but the magnitude and direction of this association varied by age. However, it is not clear whether these data, often obtained from large studies of well-defined populations near major medical institutions, are applicable to subjects in rural areas that often suffer from health disparities. Moreover, it is not known whether these data are germane given the multifactorial nature and numerous different environmental modifiers that interact with genes to influence BP [
20,
21], a phenomenon seen with other chronic diseases as well [
22,
23,
24]. The multifactorial nature of chronic diseases distinguishes them and their study from Mendelian diseases, such as cystic fibrosis, sickle cell anemia, phenylketonuria, and others, where the presence of specific risk alleles are sufficient to cause a disease phenotype [
25,
26,
27].
Our team developed and implemented a two-year multi-level intervention, called the Heart Healthy Lenoir (HHL) project, to improve clinical management of HTN, with a specific focus on reducing racial disparities in BP levels. The primary outcome of the intervention was the change in systolic blood pressure (SBP) from baseline to 12-month follow-up (hereafter denoted by ΔSBP, calculated as follow-up minus baseline). Five hundred and twenty-five participants with a clinical diagnosis of uncontrolled hypertension participated in the HHL high BP study. We recruited patients whose last recorded SBP was ≥150 mmHg in order to enhance the probability of the subjects having uncontrolled HTN (SBP ≥ 140 mmHg or diastolic blood pressure (DBP) ≥ 90 mmHg) at their study enrollment visit. Along with baseline survey and biometric data, participants were invited to provide blood samples for genetic analyses [
28]. Our goal was to determine if precision medicine approaches in a rural population can provide insight into BP regulation and possible responsiveness to a hypertension intervention.
4. Discussion
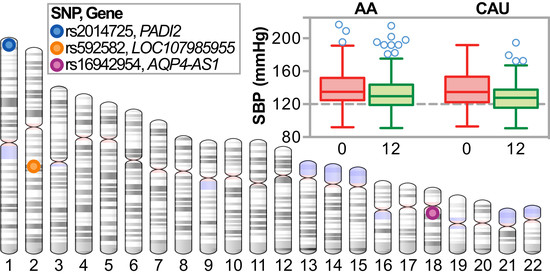
We explored associations of SNPs and BP change in a cohort of African American and Caucasian participants (
Figure 1) during a one-year multi-level intervention aimed to reduce BP in patients with established HTN in Eastern, NC (USA) (
Figure 2). Remarkably, within our small, rural population of study participants in a region of the country that suffers disproportionally from higher cardiovascular disease risk, we associated several known genetic variants with baseline SBP levels (
Table 4) by controlling for age, gender, and smoking. Moreover, the SNP rs592582 also associated with the responsiveness to the intervention. The minor allele of rs592582 associated with higher baseline SBP and lower SBP after one year, suggesting that this SNP not only associates with the presence of hypertension, but also associates with responsiveness to interventions like those employed in the HHL study. The remaining candidate SNPs that associated with baseline SBP did not associate with the responsiveness to the intervention, which prompted us to perform an unbiased genome-wide association analysis of SNPs with ∆SBP in combination with exclusionary data filtering (
Figure 5) to identify other genetic factors that may contribute to the intervention response. We identified four and 15 loci in either our AA or CAU groups that were identified by multiple SNPs with either SNP main effects and/or SNP–age interactions that associated with a change in SBP (
Figure 4,
Table 5 and
Table 6). Additionally, we explored the interaction of participant age and SNPs to evaluate the potential impact of age on the responsiveness to the intervention within a SNP group. Several of these loci identified genotypes in which the BP increases or decreases after one year of the intervention depended on the age group of the participants (
Figure 6,
Table 5 and
Table 6). Our observation of gene–age interactions with BP is consistent with other recent studies that also observed an influence of age on the association of SNPs with cardiovascular risk factors such as SBP or BMI [
12,
51]. Hence, the impact of age on SNP associations with the responsiveness to our HTN intervention may provide further insight into the underlying biology of a why certain individuals respond differently to hypertension treatments and the possible influence of age on the effectiveness of the intervention.
Our approach led us to two genes of interest,
AQP4-AS1 and
PADI2. These two genes and the specific variants we identified also associated with other important cardiovascular disease risk factors such as BMI and blood lipids in other independent cohorts [
48,
49], suggesting that these loci play a role in cardiovascular health risk. Additionally, these SNPs associated strongly with the expression of the gene where they are located (
Figure 7), identifying these as eSNPs and perhaps indicating a functional role for these variants in our interventional response.
AQP-AS1 is a long non-coding RNA comprised of 10 overlapping non-coding RNA (ncRNA) transcripts of unknown function conserved in both mice and zebrafish [
52,
53].
PADI2, however, is known to encode a peptidyl arginine deiminase that catalyzes the post-translational deimination of proteins by converting arginine residues into citrullines, including myelin basic protein, vimentin, actin, and histones [
54]. The eSNPs we identified associated with a robust change in
PADI2 expression in multiple tissues including blood cells, heart, aorta, and both visceral and subcutaneous adipose (
Table S9). Altered PADI2 activity is implicated in neurodegenerative and inflammatory diseases [
54] and most recently, vascular angiogenesis [
55].
PADI2 expression and anti-citrullinated protein antibodies were higher in smokers, suggesting that increased
PADI2 expression, particularly in genetically susceptible subjects (
Figure 7b), promotes more robust pathophysiological responses to environmental stressors [
56,
57,
58]. Our data suggest that these
PADI2 eSNPs and the expression of
PADI2 may be have differential effects on blood pressure regulation depending on age (
Figure 6b) and may be useful in understanding an individual’s response to blood pressure interventions, particularly in smokers.
Other dietary supplement studies and medication studies evaluated SNP associations with BP change in Han Chinese cohorts. Gu and Kelly tested the effectiveness of sodium and potassium supplementation, respectively, on a subsample of hypertensive patients that were part of the Genetic Epidemiology Network of Salt Sensitivity Study [
37,
59]. Gu et al. [
37] examined associations between 11 renin-angiotensin-aldosterone-system candidate genes with SBP, DBP, and mean arterial BP change among 1860 Han Chinese subjects who either had HTN or were the sibling, offspring, or spouse of the individuals with HTN. This cohort consumed a low sodium diet for seven days followed by a high sodium diet for an additional seven days. Five SNPs were independently associated with BP responses to a low sodium diet, while just one was associated with BP response to a high sodium diet. They also shared findings of two additional SNPs that were significantly associated with BP reduction in only males who were exposed to the low sodium diet. The investigators suggested that these genes may play a critical role in the salt sensitivity of BP and could help identify patients that may benefit most from a low sodium diet. Furthermore, using participants from this same GenSalt study, Kelley et al. [
59] performed a separate analysis on 1906 study subjects that were exposed to a high sodium diet for 14 days, but were additionally provided 60 mmols of potassium daily for the last seven days. Their results identified regions on chromosomes 3 and 11 that may harbor susceptibility loci for dietary potassium sensitivity and a novel variant in the angiotensin II receptor suggested to be a strong predictor of BP response to potassium sensitivity. As in Gu’s work, they suggest that such findings may provide insights into the pathophysiology of hypertension and the genetic mechanisms that underlie potassium sensitivity. They too concluded that the ultimate value of these types of discoveries might be in guiding people with specific genotypes to dietary interventions that may provide the greatest impact on BP control.
Multiple anti-hypertensive medication trials have been performed to attempt to identify SNPs associated with responsiveness to individual classes of anti-hypertensives [
60,
61]. Additional studies identified SNPs associated with opposite effects on a subject’s BP with different classes of anti-hypertensive medications [
62]. For example, some SNPs are associated with a BP reduction in response to one class of medication (e.g., angiotensin receptor blockers) and a BP increase in response to other classes (e.g., diuretics). The results from Turner et al. [
62] and our study presented here further the call to develop personalized medicine approaches in treating patients with hypertension, allowing personal (genetic- and age-based) recommendations for specific combinations of drugs. As in our study, few findings in the these aforementioned medication studies reached the traditional level of statistical significance deemed sufficient in GWAS [
63]; however, many of the SNPs identified in these studies and ours are potentially important to both disease etiology and tailoring treatments for HTN.
Limitations: As a cohort study reporting pre-post measures, we cannot rule out the possibility that the observed changes in BP may be due to secular trends or other factors that were not captured in our data collection and were not instigated by the multi-level intervention per se. The pragmatic nature of implementing this kind of an intervention with research-naïve clinical partners was both a limitation and a strength [
29]. We designed and implemented the intervention with broad stakeholder input to maximize feasibility and sustainability, but in a multi-level intervention in “real world” practices, there is no way to disentangle the effect of any particular aspect of the intervention as being more or less important in BP control. Likewise, we did not have a measure of medication adherence beyond self-report, an important component that needs to be addressed for a broad range of diseases if we are to move forward with precision medicine approaches to care. Our research teams and practicing clinicians are keenly interested in including anti-hypertensive medication metabolite data in both trials and routine care as a measure of patient compliance and adherence. This approach could reliably identify which anti-hypertensive medications are being taken by individual patients. We did not find any associations between different measures of medication exposure and the effectiveness of the intervention. However, designing studies that focus both on health disparities and specific anti-hypertensives may identify gene-drug interactions that could ultimately aid in using genetic data as a component of blood pressure care.
Additionally, we tested for genetic variation associations using a platform ensuring broad coverage across the genome that captures variation in both of our ancestral groups, but certainly there could be additional, important genetic variation that contributed to the responsiveness to the intervention that was not represented on our arrays or imputation panels. Due to the small sample size, we used an exploratory
p value to establish significance in this study. Additionally, for our modeling work, not all variables retained in our multiple regression model were independently associated with the outcome within each race. We included variables noted in the literature to be associated with BP outcomes in other papers, but in some cases from very different populations [
20,
37,
64]. Without the data of prior studies on populations, such as that we had in the HHL study, we decided to include the variables listed. Lastly, our study population was from a small region in Eastern NC. Thus, our results should not be generalized to larger populations. However, the genetic ancestry of our study population is reflective of study populations from large US-based GWAS studies; hence, we believe that our population is reasonably representative of the larger African American and Caucasian population in the US.