Phylogenetic Relationships among Whiteflies in the Bemisia tabaci (Gennadius) Species Complex from Major Cassava Growing Areas in Kenya
Abstract
:1. Introduction
1.1. Whiteflies as Insect Vectors
1.2. Bemisia tabaci Species Complex
1.3. Host Range
1.4. Species Molecular Markers
2. Materials and Methods
2.1. Whitefly Collection
2.2. DNA Extraction
2.3. PCR Amplification of mtCOI DNA
2.4. Gel Electrophoresis and DNA Sequencing
2.5. Phylogenetic Analysis of mtCOI Sequences
3. Results
Distribution of Bemisia tabaci Putative Species on Cassava in Kenya
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A


References
- Cossa, N.S. Epidemiology of Cassava Mosaic Disease in Mozambique. Ph.D. Thesis, University of Witwatersrand, Johannesburg, South Africa, 2011. [Google Scholar]
- Nweke, F.I. A Cash Crop in Africa. COSCA Working Paper No. 14. In Collaborative Study of Cassava in Africa; International Institute of Tropical Agriculture: Ibadan, Nigeria, 1996. [Google Scholar]
- Mwango’mbe, A.W.; Mbugua, S.K.; Olubayo, F.O.; Ngugi, E.K.; Mwinga, R.; Munga, T.; Muiru, W.M. Challenges and Opportunities in Cassava Production among the Rural Households in Kilifi County in the Coastal Region of Kenya. J. Biol. Agric. Healthc. 2013, 3, 30–35. [Google Scholar]
- Were, H.K.; Winter, S.; Maiss, E. Characterisation and distribution of cassava viruses in Kenya. Afr. Crop Sci. Conf. Proc. 2007, 8, 909–912. [Google Scholar]
- Sseruwagi, P. Molecular variability of cassava Bemisia tabaci and its effects on the epidemiology of cassava mosaic Geminiviruses in Uganda. Ph.D. Thesis, University of Witwatersrand, Johannesburg, South Africa, 2005. [Google Scholar]
- Wyatt, S.D.; Brown, J.K. Detection of subgroup III Geminivirus isolated in leaf extracts by degenerate primers and polymerase chain reaction. Phytopathology 1996, 86, 1288–1293. [Google Scholar] [CrossRef]
- International Institute of Tropical Agriculture. Preemptive Management of the Virulent Cassava Mosaic Disease through an Integrated Cassava Development Approach for Enhanced Rural Sector Economy in the South-South and South-East Zones of Nigeria; International Institute of Tropical Agriculture: Ibadan, Nigeria, 2003. [Google Scholar]
- Mugerwa, H.; Marie, E.C.; Alicai, T.; Ateka, E.; Atuncha, H.; Ndunguru, J.; Sseruwagi, P. Genetic Diversity and Geographic Distribution of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) Genotypes Associated with Cassava in East Africa. Ecol. Evol. 2012, 2, 2749–2762. [Google Scholar] [CrossRef] [PubMed]
- Simon, E.B.; Rob, W.B.; William, S.S.; Kahiu, N.; Markham, P.G.; John, S. Genetic diversity and phylogeography of cassava mosaic viruses in Kenya. J. Gen. Virol. 2006, 87, 3053–3065. [Google Scholar]
- Aduwo, J.R.; Guy, A. Assessing the Performance of two Artificial Neural Networks in the Classification of Cassava Mosaic Disease. Available online: http://cit.mak.ac.ug/iccir/downloads/ICCIR_10/Jennifer%20ose%20Aduwo%20and%20Acellam%20Guy.pdf (accessed on 13 September 2016).
- Legg, P.J.; Shirima, R.; Tajebe, S.L.; Guastella, D.; Boniface, S.; Jeremiah, S.; Nsami, E.; Chikoti, P.; Rapisarda, C. Biology and management of Bemisia whitefly vectors of cassava virus pandemics in Africa. Pest Manag. Sci. 2014, 70, 1446–1453. [Google Scholar] [CrossRef] [PubMed]
- Legg, J.P.; Sseruwagi, P.; Boniface, S.; Okao-Okuja, G.; Shirima, R.; Bigirimana, S.; Gashaka, G.; Herrmann, H.W.; Jeremiah, S.; Obiero, H.; et al. Spatio-temporal patterns of genetic change amongst populations of cassava Bemisia tabaci whiteflies driving virus pandemics in East and Central Africa. Virus Res. 2013, 186, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Legg, J.P.; Owor, B.; Sseruwagi, P.; Ndunguru, J. Cassava mosaic virus disease in East and Central Africa: Epidemiology and management of a regional pandemic. Adv. Virus Res. 2006, 67, 355–418. [Google Scholar] [PubMed]
- Ndunguru, J.; Sseruwagi, P.; Tairo, F.; Stomeo, F.; Maina, S.; Djinkeng, A.; Kehoe, M.; Boykin, L.M. Analyses of Twelve New Whole Genome Sequences of Cassava Brown Streak Viruses and Ugandan Cassava Brown Streak Viruses from East Africa: Diversity, Supercomputing and Evidence for Further Speciation. PLoS ONE 2015, 10, e0139321. [Google Scholar]
- Alicai, T.; Ndunguru, J.; Sseruwagi, P.; Tairo, F.; Okao-Okuja, G.; Nanvubya, R.; Kiiza, L.; Kubatko, K.; Kehoe, M.A.; Boykin, L.M. Cassava brown streak virus has a rapidly evolving genome: Implications for virus speciation, variability, diagnosis and host resistance. Sci. Rep. 2016. [Google Scholar] [CrossRef] [PubMed]
- Ntawuruhunga, P.; Legg, J. New Spread of Cassava Brown Streak Virus Disease and its Implications for the Movement of Cassava Germplasm in the East and Central African Region. Available online: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.547.7713&rep=rep1&type=pdf (accessed on 13 September 2016).
- Mohammed, I.U.; Abarshi, M.M.; Muli, B.; Hillocks, R.J.; Maruthi, M.N. The Symptom and Genetic Diversity of Cassava Brown Streak Viruses Infecting Cassava in East Africa. Adv. Virol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Palaniswami, M.S.; Henneberry, T.J. Bemisia tabaci (Genn.): Biotypes and Cassava Mosaic Virus in India. In The Whitefly, Bemisia tabaci (Homoptera: Aleyrodidae) Interaction with Geminivirus-Infected Host Plants; Springer: Houten, The Netherlands, 2011; pp. 121–163. [Google Scholar]
- Martin, J.H. Whiteflies of Belize (Hemiptera: Aleyrodidae). Part 1: Introduction and account of the subfamilify Aleurodidae Quintance and Baker. Zootaxa 2004, 681, 1–119. [Google Scholar]
- Navas-Castillo, J.; Fiallo-Olive, E.; Sanchez-Campos, S. Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 2011, 49, 219–248. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.A.; Seijo, E.T.; Vallad, E.G.; Peres, A.N.; Druffel, L.K. Evaluating Weeds as Hosts of Tomato yellow leaf curl virus. Environ. Entomol. 2015, 44, 1101–1107. [Google Scholar] [CrossRef] [PubMed]
- Thompson, W.M.O. The Whitefly, Bemisia tabaci (Homoptera: Aleyrodidae) Interaction with Geminivirus-Infected Host Plants; Springer: Houten, The Netherlands, 2011. [Google Scholar]
- Data sheets on quarantine organisms No. 178, Bemisia tabaci. Available online: https://www.eppo.int/QUARANTINE/data_sheets/insects/BEMITA_ds.pdf (accessed on 13 September 2016).
- Boykin, L.M.; Armstrong, K.F.; Kubatko, L.; de Barro, P. Species Delimitation and Global Biosecurity. Evol. Bioinform. 2012, 8, 1–37. [Google Scholar] [CrossRef] [PubMed]
- Dinsdale, A.; Cook, L.; Riginos, C.; Buckley, Y.M.; De Barro, P. Refined Global Analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) Mitochondrial Cytochrome Oxidase 1 to Identify Species Level Genetic Boundaries. Ann. Entomol. Soc. Am. 2010, 103, 196–208. [Google Scholar] [CrossRef]
- Lee, W.; Park, J.; Lee, G.-S.; Lee, S.; Akimoto, S.-I. Taxonomic Status of the Bemisia tabaci Complex (Hemiptera: Aleyrodidae) and Reassessment of the Number of Its Constituent Species. PLoS ONE 2013, 8, e63817. [Google Scholar]
- Boykin, L.M.; Barro, P.J.D. A practical guide to identifying members of the Bemisia tabaci species complex: And other morphologically identical species. Front. Ecol. Evol. 2014, 45, 1–5. [Google Scholar] [CrossRef]
- Legg, J.P.; French, R.; Rogan, D.; Okao-Okuja, G.; Brown, J.K. A distinct, Bemisia tabaci (Gennadius) Hemiptera: Sternorrhyncha: Aleyrodidae) genotype cluster is associated with the epidemic of severe cassava mosaic virus disease in Uganda. Mol. Ecol. 2002, 11, 1219–1229. [Google Scholar] [CrossRef] [PubMed]
- Riis, L.; Njuguna, T.; Raini, R.; Lohr, B.; Bob, M.A. Whiteflies and Whitefly-Borne Viruses in the Tropics; International Center of insect Physiology and ecology (ICIPE): Nairobi, Kenya, 2000. [Google Scholar]
- Brown, J.K.; Frohlich, D.R.; Rosell, R.C. The sweetpotato or silverleaf whiteflies: Biotypes of Bemisia tabaci or a species complex? Ann. Rev. Entomol. 1995, 40, 511–534. [Google Scholar] [CrossRef]
- Sseruwagi, P.; Maruthi, M.N.; Colvin, J.; Rey, M.; Brown, J.K.; Legg, J.P. Colonization of non-cassava plant species by cassava whiteflies (Bemisia tabaci) in Uganda. Entomol. Exp. Appl. 2006, 19, 145–153. [Google Scholar] [CrossRef]
- Caterino, M.S.; Cho, S.; Sperling, F.A.H. The Current State Of Insect Molecular Systematics: A thriving Tower Of Babel. Ann. Rev. Entomol. 2000, 45, 1–54. [Google Scholar] [CrossRef] [PubMed]
- De Barro, P.J. Genetic structure of the whitefly Bemisia tabaci in the Asia-Pacific region revealed using microsatellite markers. Mol. Ecol. 2005, 14, 3695–3718. [Google Scholar] [CrossRef] [PubMed]
- Boykin, L.M.; Shatters, R.G., Jr.; Rosell, R.C.; McKenzie, C.L.; Bagnall, R.A.; De Barro, P. Global relationships of Bemisia tabaci (Hemiptera: Aleyrodidae) revealed using Bayesian analysis of mitochondrial COI DNA sequences. Mol. Phylogenet. Evol. 2007, 44, 1306–1319. [Google Scholar] [CrossRef] [PubMed]
- Frohlich, D.R.; Torres-Jerez, I.; Bedford, I.D.; Markham, P.G.; Brown, J.K. A phylogeographical analysis of the Bemisia tabaci species complex based on mitochondrial DNA markers. Mol. Ecol. 1999, 8, 1683–1691. [Google Scholar] [CrossRef] [PubMed]
- De Barro, P.J.; Driver, F.; Trueman, J.H.; Curran, J. Phylogenetic relationships of world populations of Bemisia tabaci (Gennadius) using ribosomal ITS1. Mol. Phys. Evol. 2000, 16, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Orodho, A.B. Country Pasture/Forage Resource Profiles, Kenya; National Agricultural Research Station: Kitale, Kenya, 2006.
- Mware, B.; Olubayo, F.; Narla, R.; Songa, J.; Amata, R.; Kyamanywa, S.; Ateka, E. First Record of Spiraling Whitefly in Coastal Kenya: Emergence, Host Range, Distribution and Association with Cassava Brown Streak Virus Disease. Int. J. Agric. Biol. 2012, 470, 411–415. [Google Scholar]
- Mehta, P.; Wyman, J.A.; Nakhla, M.K.; Maxwell, D.P. Transmission of tomato yellow leaf curl geminivirus by Bemisia tabaci (Homoptera: Aleyrodidae). J. Econ. Entomol. 1994, 87, 1291–1297. [Google Scholar] [CrossRef]
- Muturi, C.N.; Ouma, J.O.; Malele, I.I.; Ngure, R.M.; Rutto, J.J.; Mithofer, M.; Enyaru, J.; Masiga, D.K. Tracking the Feeding Patterns of Tsetse Flies (Glossina Genus) by Analysis of Blood meals Using Mitochondrial cytochromes Genes. PLoS ONE 2011, 6, e17284. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighing, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Peter, J.G. Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika 1995, 82, 711–732. [Google Scholar]
- Ayres, D.L.; Darling, A.; Zwickl, D.J.; Beerli, P.; Holder, M.T.; Lewis, P.O.; Huelsenbeck, J.P.; Ronquist, F.; Swofford, D.L.; Cummings, M.P.; et al. BEAGLE: An application programming interface and high-performance computing library for statistical phylogenetics. Syst. Biol. 2012, 61, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Boykin, L.M. Bemisia tabaci Nomenclature: Lessons learned. Pest Manag. Sci. 2014, 70, 1454–1459. [Google Scholar] [CrossRef] [PubMed]
- Maruthi, M.N.; Hillocks, R.J.; Mtunda, K.; Raya, M.D.; Muhanna, M.; Kiozia, H.; Rekha, A.R.; Colvin, J.; Thresh, J.M. Transmission of Cassava brown streak virus by Bemisia tabaci (Gennadius). J. Phytopathol. 2005, 153, 307–312. [Google Scholar] [CrossRef]
- Esterhuizen, L.L.; Mabasa, K.G.; van Heerden, S.W.; Czosnek, H.; Brown, J.K.; van Heerden, H. Genetic identification of members of the Bemisia tabaci cryptic species complex from South Africa reveals native and introduced haplotypes. J. Appl. Entomol. 2012, 136, 1–14. [Google Scholar]
- Legg, J.P.; Jeremiah, S.C.; Obiero, H.M.; Maruthi, M.N.; Ndyetabula, I.; Okao-Okuja, G.; Bouwmeester, H.; Bigirimana, S.; Tata-Hangy, W.; Gashaka, G.; et al. Comparing the regional epidemiology of the Cassava Mosaic and Cassava Brown Streak pandemics in Africa. Virus Res. 2011, 159, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.K. The Bemisia tabaci complex: Genetic and phenotypic variation and relevance to TYLCV-vector interactions. In Tomato Yellow Leaf Curl Virus Disease: Management, Molecular Biology, Breeding for Resistance; Czosnek, H., Ed.; Springer: Dordrecht, The Netherlands, 2007. [Google Scholar]
- Thompson, W.M.O. A new host plant species for the cassava biotype of Bemisia tabaci (Gennadius) (Hom., Aleyrodidae). J. Appl. Entomol. 2003, 127, 374–376. [Google Scholar] [CrossRef]
- Legg, J.P. Epidemiology of a whitefly-transmitted cassava mosaic geminivirus pandemic in Africa. In Bemisia: Bionomics and Management of a Global Pest; Stansly, P.A., Naranjo, S.E., Eds.; Springer: Houten, The Netherlands, 2010. [Google Scholar]
- De Barro, P.J. The Bemisia species complex: Questions to guide future research. J. Integr. Agric. 2012, 11, 187–196. [Google Scholar] [CrossRef]
- Otim-Nape, G.W.; Bua, A.; Thresh, J.M.; Baguma, Y.; Ogwal, S.; Semakula, G.N.; Acola, G.; Byabakama, B.; Martin, A. Cassava Mosaic Virus Disease in Uganda: The Current Pandemic and Approaches to Control; Natural Resources Institute: Chatham, UK, 1997.
- Jarvis, A.; Ramirez-Villegas, J.; Campo, B.V.H.; Navarro-Racines, C. Is cassava the answer to African climate change adaptation? Trop. Plant Biol. 2012, 5, 9–29. [Google Scholar] [CrossRef]

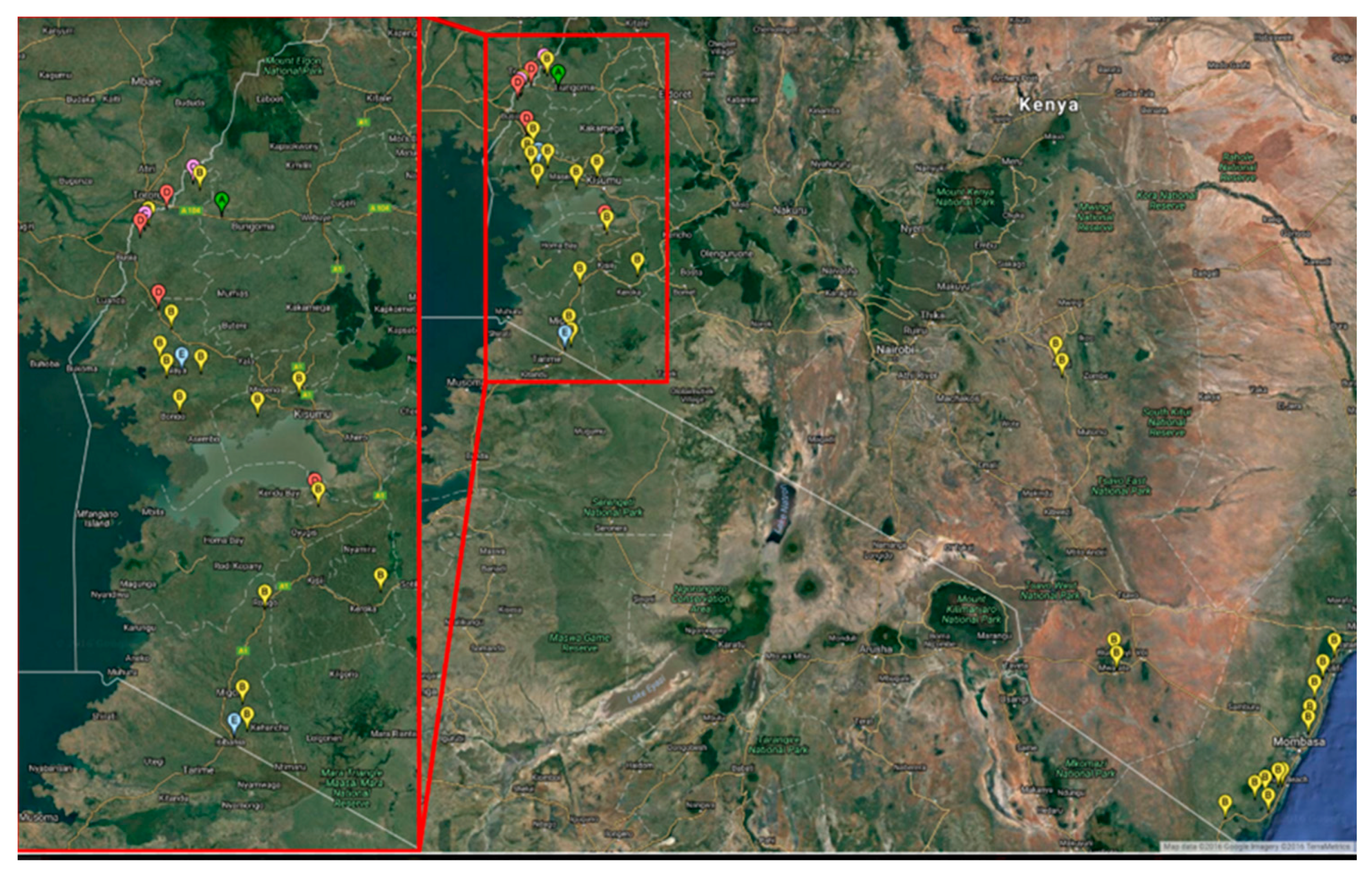
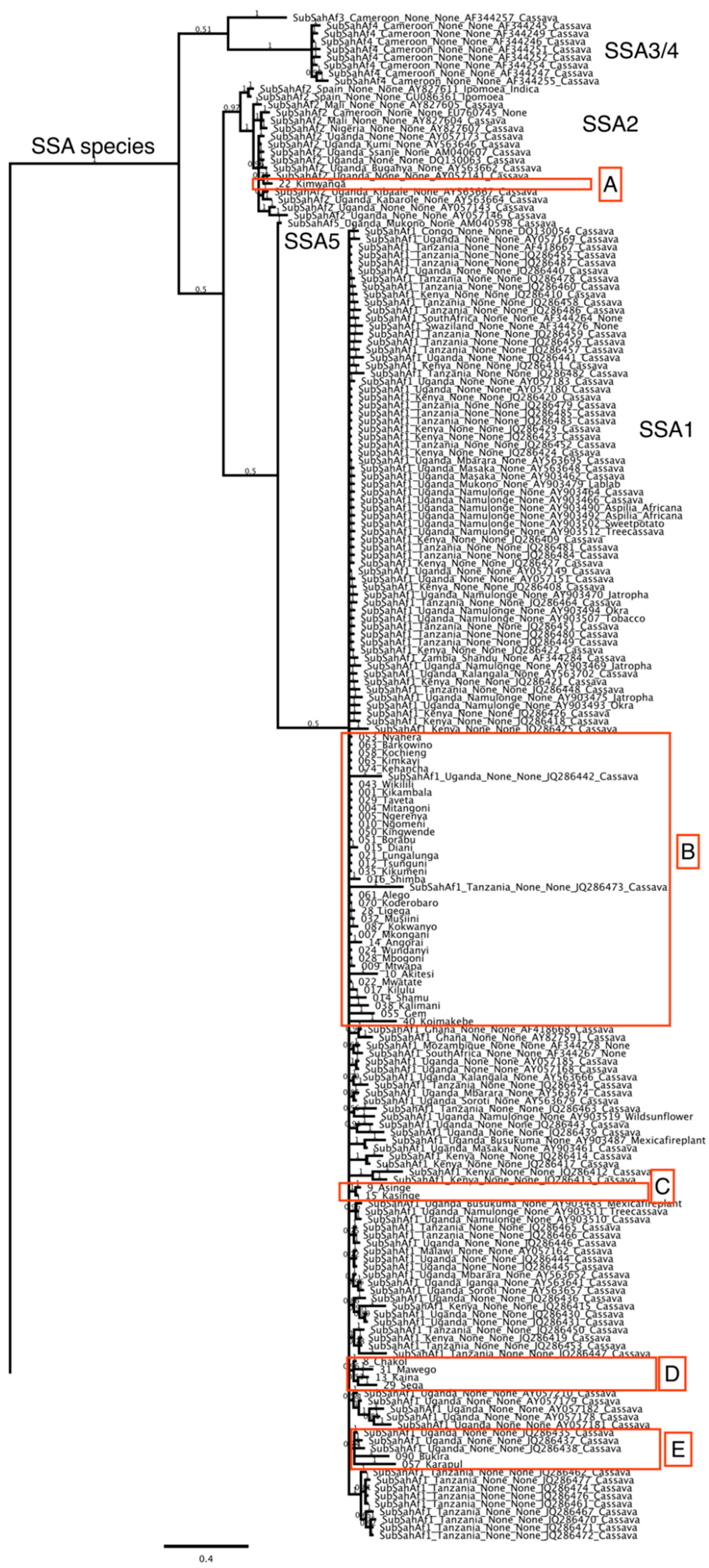
| Sample Number | County | Area | Field | Geographic Location | Altitude | Genetic Group | Accession No. |
|---|---|---|---|---|---|---|---|
| 1 | Siaya | Ligega | NF3 | N00.20321, E034.27061 | 1330 | SSA-B | KY523872 |
| 2 | Siaya | Sega | NF4 | N00.27314, E034.22346 | 1243 | SSA-D | KY523885 |
| 3 | Siaya | Gem | NF5 | N00.03174, E034.25409 | 1957 | SSA-B | KY523894 |
| 4 | Siaya | Karapul | NF8 | N00.05285, E034.30654 | 3132 | SSA-E | KY523892 |
| 5 | Kisumu | Nyahera | NF2 | S00.03051, E034.71960 | 1459 | SSA-B | KY523852 |
| 6 | Siaya | Kochieng | NF10 | N00.09266, E034.23024 | 1256 | SSA-B | KY523854 |
| 7 | Siaya | Alego | NF14 | N00.04531, E034.37395 | 1409 | SSA-B | KY523870 |
| 8 | Kisumu | Kit mikayi | NF22 | S00.10358, E034.57415 | 1176 | SSA-B | KY523855 |
| 9 | Siaya | Barwino | NF16 | S00.09508, E034.29937 | 1509 | SSA-B | KY523853 |
| 10 | Homabay | Koderobaro | NF33 | S00.78241, E034.59921 | 1416 | SSA-B | KY523871 |
| 11 | Migori | Bukira West | NF34 | S01.23.442, E034.49201 | 2363 | SSA-E | KY523887 |
| 12 | Homabay | Kokwanyo | NF27 | S00.42202, E034.78882 | 1275 | SSA-B | KY523875 |
| 13 | Busia | Chakol | WF12 | N00.52575, E034.16062 | 1152 | SSA-D | KY523877 |
| 14 | Busia | Asing’e | WF13 | N00.54886, E034.17672 | 1152 | SSA-C | KY523880 |
| 15 | Busia | Aktesi | WF14 | N00.56531, E034.18919 | 1153 | SSA-B | KY523886 |
| 16 | Busia | Kaina | WF17 | N00.62388, E034.25398 | 1177 | SSA-D | KY523876 |
| 17 | Busia | Ang’orai | WF18 | N00.69299, E034.37015 | 1230 | SSA-B | KY523879 |
| 18 | Busia | Kasing’e | WF19 | N00.71344, E03434693 | 1393 | SSA-C | KY523881 |
| 19 | Bungoma | Kimwanga | WF26 | N00.59644, E034.44701 | 1233 | SSA-A | KY523874 |
| 20 | Taita Taveta | Wundanyi | CF30 | S03.39443, E038.36485 | 1339 | SSA-B | KY523882 |
| 21 | Homabay | Mawego | NF31 | S0038946, E034.77587 | 1424 | SSA-D | KY523888 |
| 22 | Migori | Kiomakebe | NF40 | S01.21158, E034.53714 | 1562 | SSA-B | KY523895 |
| 23 | Migori | Kehancha | NF41 | S01.11733, E034.52141 | 1418 | SSA-B | KY523856 |
| 24 | Nyamira | Borabu | NF44 | S00.72273, E035.00735 | 1926 | SSA-B | KY523864 |
| 25 | Kilifi | Kikambala | CF1 | S03.86207, E039.74467 | 86 | SSA-B | KY523858 |
| 26 | Kilifi | Mitangoni | CF5 | S03.69020, E039.77895 | 61 | SSA-B | KY523860 |
| 27 | Kilifi | Ngerenya | CF7 | S03.54610, E039.83553 | 53 | SSA-B | KY523861 |
| 28 | Kilifi | Mkongani | CF9 | S03.39686, E039.91546 | 20 | SSA-B | KY523878 |
| 29 | Kilifi | Mtwapa(KARLO) | CF12 | S03.93400, E039.73534 | 38 | SSA-B | KY523884 |
| 30 | Kwale | Ngombeni | CF13 | S04.12535, E039.63130 | 25 | SSA-B | KY523862 |
| 31 | Kwale | Tsunguni | CF15 | S04.18433, E039.55974 | 115 | SSA-B | KY523867 |
| 32 | Kwale | Shamu | CF17 | S04.30154, E039.54972 | 42 | SSA-B | KY523891 |
| 33 | Kwale | Diani | CF18 | S04.30731, E039.51998 | 90 | SSA-B | KY523865 |
| 34 | Kwale | Shimba hills | CF19 | S04.35600, E039.42932 | 104 | SSA-B | KY523869 |
| 35 | Kwale | Kilulu | CF21 | S04.39463, E039.35639 | 85 | SSA-B | KY523890 |
| 36 | Kwale | Kikoneni | CF22 | S04.42855, E039.31791 | 70 | SSA-B | KY523868 |
| 37 | Kwale | Lungalunga | CF25 | S04.53264, E039.14732 | 64 | SSA-B | KY523866 |
| 38 | Kwale | Kingwende | CF27 | S04.48550, E039.45035 | 22 | SSA-B | KY523863 |
| 39 | Taita Taveta | Mwatate | CF28 | S03.48400, E038.38088 | 830 | SSA-B | KY523889 |
| 40 | Taita Taveta | Mbogoni | CF36 | S03.41414, E037.70263 | 729 | SSA-B | KY523883 |
| 41 | Taita Taveta | Taveta | CF37 | S03.46017, E037.69030 | 720 | SSA-B | KY523859 |
| 42 | Kitui | Kalimani | EF18 | S01.31211, E037.95479 | 1232 | SSA-B | KY523893 |
| 43 | Kitui | Wakulili | EF25 | S01.42728, E037.99684 | 1135 | SSA-B | KY523857 |
| 44 | Machakos | Musiini | EF4 | S01.42061, E037.36724 | 1413 | SSA-B | KY523873 |
| Location in Kenya | Number of Samples | SSA-A | SSA-B | SSA-C | SSA-D | SSA-E |
|---|---|---|---|---|---|---|
| Western/Lake | 7 | 1 | 2 | 2 | 2 | - |
| Nyanza | 16 | - | 12 | - | 2 | 2 |
| Eastern | 3 | - | 3 | - | - | - |
| Coast | 18 | - | 18 | - | - | - |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manani, D.M.; Ateka, E.M.; Nyanjom, S.R.G.; Boykin, L.M. Phylogenetic Relationships among Whiteflies in the Bemisia tabaci (Gennadius) Species Complex from Major Cassava Growing Areas in Kenya. Insects 2017, 8, 25. https://doi.org/10.3390/insects8010025
Manani DM, Ateka EM, Nyanjom SRG, Boykin LM. Phylogenetic Relationships among Whiteflies in the Bemisia tabaci (Gennadius) Species Complex from Major Cassava Growing Areas in Kenya. Insects. 2017; 8(1):25. https://doi.org/10.3390/insects8010025
Chicago/Turabian StyleManani, Duke M., Elijah M. Ateka, Steven R. G. Nyanjom, and Laura M. Boykin. 2017. "Phylogenetic Relationships among Whiteflies in the Bemisia tabaci (Gennadius) Species Complex from Major Cassava Growing Areas in Kenya" Insects 8, no. 1: 25. https://doi.org/10.3390/insects8010025





