Epitranscriptome and FMRP Regulated mRNA Translation
Abstract
:1. Introduction
2. RNA Methylation and Neuronal Function
3. RNA Editing
4. Dendritic Local Translation and Neurological Disorders
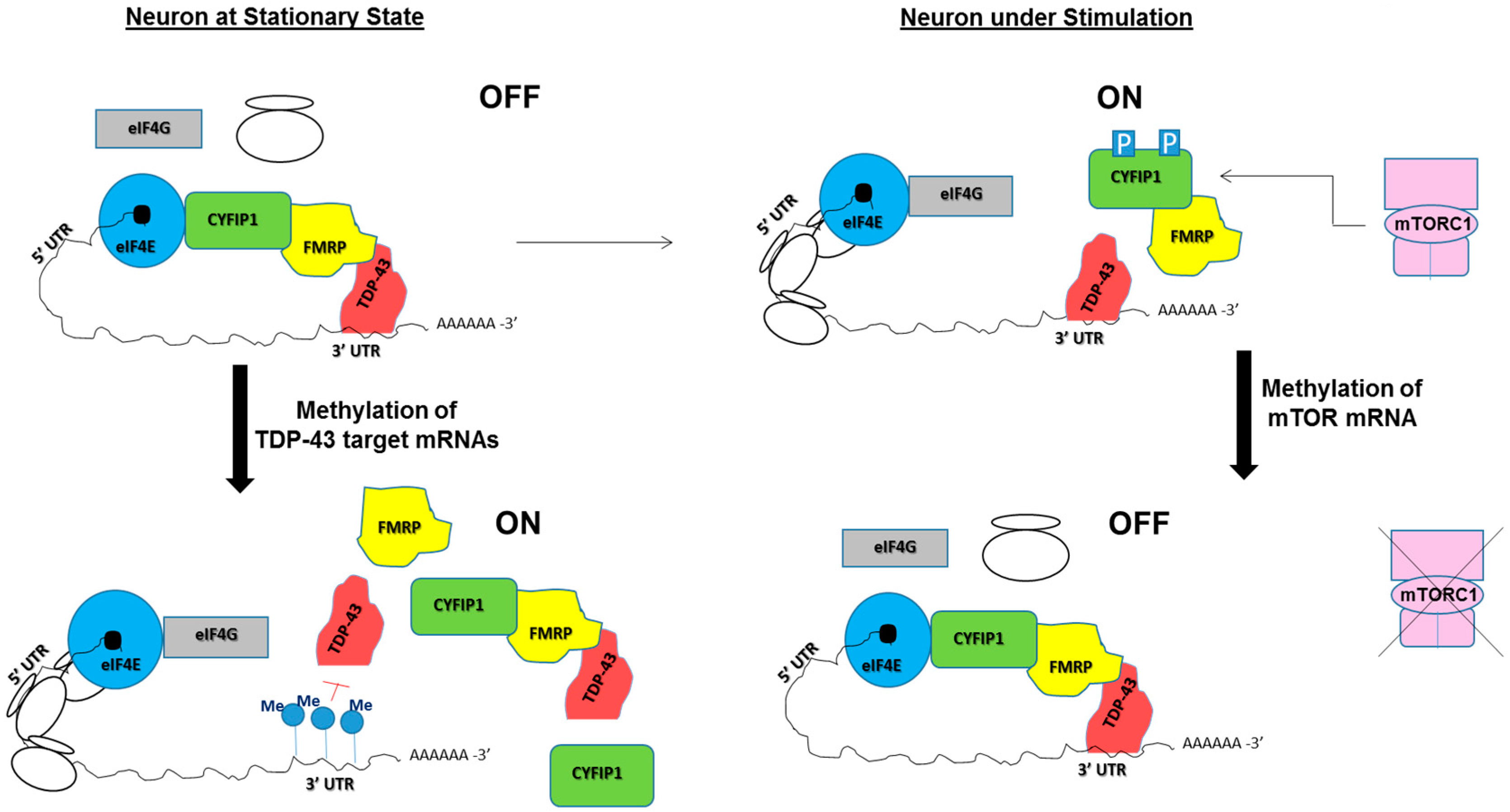
5. Potential Roles of RNA Modifications in TDP-43/FMRP-Associated Neurological Diseases
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Agris, P.C.P.; Rozenski, J.; Fabris, D.; Vendeix, F. The RNA Modification Database. Available online: http://mods.rna.albany.edu (accessed on 27 June 2017).
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Squires, J.E.; Patel, H.R.; Nousch, M.; Sibbritt, T.; Humphreys, D.T.; Parker, B.J.; Suter, C.M.; Preiss, T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012, 40, 5023–5033. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Sajini, A.A.; Blanco, S.; Dietmann, S.; Lombard, P.; Sugimoto, Y.; Paramor, M.; Gleeson, J.G.; Odom, D.T.; Ule, J.; et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013, 4, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Khoddami, V.; Cairns, B.R. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 2013, 31, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Liu, J.; He, C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015, 29, 1343–1355. [Google Scholar] [CrossRef] [PubMed]
- Bujnicki, J.M.; Feder, M.; Ayres, C.L.; Redman, K.L. Sequence-structure-function studies of tRNA:M5c methyltransferase trm4p and its relationship to DNA:M5c and RNA:M5u methyltransferases. Nucleic Acids Res. 2004, 32, 2453–2463. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jia, G. Methylation modifications in eukaryotic messenger RNA. J. Genet. Genom. 2014, 41, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Li, J.B.; Church, G.M. Deciphering the functions and regulation of brain-enriched A-to-I RNA editing. Nat. Neurosci. 2013, 16, 1518–1522. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, J.J.; Seeburg, P.H. A-to-I RNA editing: Effects on proteins key to neural excitability. Neuron 2012, 74, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.; Bernstein, D.A.; Mumbach, M.R.; Jovanovic, M.; Herbst, R.H.; Leon-Ricardo, B.X.; Engreitz, J.M.; Guttman, M.; Satija, R.; Lander, E.S.; et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014, 159, 148–162. [Google Scholar] [CrossRef] [PubMed]
- Carlile, T.M.; Rojas-Duran, M.F.; Zinshteyn, B.; Shin, H.; Bartoli, K.M.; Gilbert, W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014, 515, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Yu, Y.T. RNA pseudouridylation: New insights into an old modification. Trends Biochem. Sci. 2013, 38, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Harcourt, E.M.; Kietrys, A.M.; Kool, E.T. Chemical and structural effects of base modifications in messenger RNA. Nature 2017, 541, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xiong, X.; Yi, C. Epitranscriptome sequencing technologies: Decoding RNA modifications. Nat. Methods 2016, 14, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.; Agarwala, S.D.; Mumbach, M.R.; Jovanovic, M.; Mertins, P.; Shishkin, A.; Tabach, Y.; Mikkelsen, T.S.; Satija, R.; Ruvkun, G.; et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 2013, 155, 1409–1421. [Google Scholar] [CrossRef] [PubMed]
- Widagdo, J.; Zhao, Q.Y.; Kempen, M.J.; Tan, M.C.; Ratnu, V.S.; Wei, W.; Leighton, L.; Spadaro, P.A.; Edson, J.; Anggono, V.; et al. Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J. Neurosci. 2016, 36, 6771–6777. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Iles, M.M.; Law, M.H.; Stacey, S.N.; Han, J.; Fang, S.; Pfeiffer, R.; Harland, M.; Macgregor, S.; Taylor, J.C.; Aben, K.K.; et al. A variant in FTO shows association with melanoma risk not due to BMI. Nat. Genet. 2013, 45, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Tung, Y.C.; Yeo, G.S. From GWAS to biology: Lessons from FTO. Ann. N. Y. Acad. Sci. 2011, 1220, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Choudhry, Z.; Sengupta, S.M.; Grizenko, N.; Thakur, G.A.; Fortier, M.E.; Schmitz, N.; Joober, R. Association between obesity-related gene FTO and ADHD. Obesity (Silver Spring) 2013, 21, E738–E744. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.; Xu, W.; Wang, H.X.; Winblad, B.; Fratiglioni, L.; Graff, C. The obesity related gene, FTO, interacts with APOE, and is associated with alzheimer’s disease risk: A prospective cohort study. J. Alzheimer’s Dis. 2011, 23, 461–469. [Google Scholar]
- Reitz, C.; Tosto, G.; Mayeux, R.; Luchsinger, J.A. Genetic variants in the fat and obesity associated (FTO) gene and risk of alzheimer’s disease. PLoS ONE 2012, 7, e50354. [Google Scholar] [CrossRef] [PubMed]
- Hess, M.E.; Hess, S.; Meyer, K.D.; Verhagen, L.A.; Koch, L.; Bronneke, H.S.; Dietrich, M.O.; Jordan, S.D.; Saletore, Y.; Elemento, O.; et al. The fat mass and obesity associated gene (FTO) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 2013, 16, 1042–1048. [Google Scholar] [CrossRef] [PubMed]
- Amort, T.; Souliere, M.F.; Wille, A.; Jia, X.Y.; Fiegl, H.; Worle, H.; Micura, R.; Lusser, A. Long non-coding RNAs as targets for cytosine methylation. RNA Biol. 2013, 10, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Edelheit, S.; Schwartz, S.; Mumbach, M.R.; Wurtzel, O.; Sorek, R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in Bacteria, Archaea, and yeast reveals m5C within Archaeal mRNAs. PLoS Genet. 2013, 9, e1003602. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, A.; Chernyakov, I.; Gu, W.; Hiley, S.L.; Hughes, T.R.; Grayhack, E.J.; Phizicky, E.M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 2006, 21, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.T.; Pang, Y.L.; Deng, W.; Babu, I.R.; Dyavaiah, M.; Begley, T.J.; Dedon, P.C. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 2012, 3, 937. [Google Scholar] [CrossRef] [PubMed]
- Chernyakov, I.; Whipple, J.M.; Kotelawala, L.; Grayhack, E.J.; Phizicky, E.M. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes Dev. 2008, 22, 1369–1380. [Google Scholar] [CrossRef]
- Chow, C.S.; Lamichhane, T.N.; Mahto, S.K. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem. Biol. 2007, 2, 610–619. [Google Scholar] [CrossRef]
- Tuorto, F.; Liebers, R.; Musch, T.; Schaefer, M.; Hofmann, S.; Kellner, S.; Frye, M.; Helm, M.; Stoecklin, G.; Lyko, F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012, 19, 900–905. [Google Scholar] [CrossRef]
- Metodiev, M.D.; Spahr, H.; Loguercio Polosa, P.; Meharg, C.; Becker, C.; Altmueller, J.; Habermann, B.; Larsson, N.G.; Ruzzenente, B. NSun4 is a dual function mitochondrial protein required for both methylation of 12s rRNA and coordination of mitoribosomal assembly. PLoS Genet. 2014, 10, e1004110. [Google Scholar] [CrossRef] [Green Version]
- Hussain, S.; Aleksic, J.; Blanco, S.; Dietmann, S.; Frye, M. Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol. 2013, 14, 215. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Z.; Yi, J.; Tang, H.; Xing, J.; Yu, M.; Tong, T.; Shang, Y.; Gorospe, M.; Wang, W. The tRNA methyltransferase Nsun2 stabilizes p16INK(4) mRNA by methylating the 3′-untranslated region of p16. Nat. Commun. 2012, 3, 712. [Google Scholar] [CrossRef]
- Brzezicha, B.; Schmidt, M.; Makalowska, I.; Jarmolowski, A.; Pienkowska, J.; Szweykowska-Kulinska, Z. Identification of human tRNA:M5c methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA). Nucleic Acids Res. 2006, 34, 6034–6043. [Google Scholar] [CrossRef]
- Frye, M.; Watt, F.M. The RNA methyltransferase MIsu (NSun2) mediates myc-induced proliferation and is upregulated in tumors. Curr. Biol. 2006, 16, 971–981. [Google Scholar] [CrossRef]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of trnaasp by the DNA methyltransferase homolog Dnmt2. Science 2006, 311, 395–398. [Google Scholar] [CrossRef]
- Sakita-Suto, S.; Kanda, A.; Suzuki, F.; Sato, S.; Takata, T.; Tatsuka, M. Aurora-b regulates RNA methyltransferase NSun2. Mol. Biol. Cell 2007, 18, 1107–1117. [Google Scholar] [CrossRef]
- Chi, L.; Delgado-Olguin, P. Expression of NOL1/NOP2/sun domain (NSun) RNA methyltransferase family genes in early mouse embryogenesis. Gene Expr. Patterns 2013, 13, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Blanco, S.; Kurowski, A.; Nichols, J.; Watt, F.M.; Benitah, S.A.; Frye, M. The RNA-methyltransferase MIsu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 2011, 7, e1002403. [Google Scholar] [CrossRef] [PubMed]
- Abbasi-Moheb, L.; Mertel, S.; Gonsior, M.; Nouri-Vahid, L.; Kahrizi, K.; Cirak, S.; Wieczorek, D.; Motazacker, M.M.; Esmaeeli-Nieh, S.; Cremer, K.; et al. Mutations in NSun2 cause autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2012, 90, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Blanco, S.; Bandiera, R.; Popis, M.; Hussain, S.; Lombard, P.; Aleksic, J.; Sajini, A.; Tanna, H.; Cortes-Garrido, R.; Gkatza, N.; et al. Stem cell function and stress response are controlled by protein synthesis. Nature 2016, 534, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Frye, M.; Dragoni, I.; Chin, S.F.; Spiteri, I.; Kurowski, A.; Provenzano, E.; Green, A.; Ellis, I.O.; Grimmer, D.; Teschendorff, A.; et al. Genomic gain of 5p15 leads to over-expression of MIsu (NSun2) in breast cancer. Cancer Lett. 2010, 289, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Benavente, S.B.; Nascimento, E.; Dragoni, I.; Kurowski, A.; Gillich, A.; Humphreys, P.; Frye, M. The nucleolar RNA methyltransferase MIsu (NSun2) is required for mitotic spindle stability. J. Cell Biol. 2009, 186, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Tuorto, F.; Menon, S.; Blanco, S.; Cox, C.; Flores, J.V.; Watt, S.; Kudo, N.R.; Lyko, F.; Frye, M. The mouse cytosine-5 RNA methyltransferase NSun2 is a component of the chromatoid body and required for testis differentiation. Mol. Cell Biol. 2013, 33, 1561–1570. [Google Scholar] [CrossRef] [PubMed]
- Satterlee, J.S.; Basanta-Sanchez, M.; Blanco, S.; Li, J.B.; Meyer, K.; Pollock, J.; Sadri-Vakili, G.; Rybak-Wolf, A. Novel RNA modifications in the nervous system: Form and function. J. Neurosci. 2014, 34, 15170–15177. [Google Scholar] [CrossRef] [PubMed]
- Fahiminiya, S.; Almuriekhi, M.; Nawaz, Z.; Staffa, A.; Lepage, P.; Ali, R.; Hashim, L.; Schwartzentruber, J.; Abu Khadija, K.; Zaineddin, S.; et al. Whole exome sequencing unravels disease-causing genes in consanguineous families in Qatar. Clin. Genet. 2014, 86, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Rafiq, M.A.; Noor, A.; Hussain, S.; Flores, J.V.; Rupp, V.; Vincent, A.K.; Malli, R.; Ali, G.; Khan, F.S.; et al. Mutation in NSun2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2012, 90, 856–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, F.J.; Lee, J.H.; Lee, J.E.; Blanco, S.; Nickerson, E.; Gabriel, S.; Frye, M.; Al-Gazali, L.; Gleeson, J.G. Whole exome sequencing identifies a splicing mutation in NSun2 as a cause of a dubowitz-like syndrome. J. Med. Genet. 2012, 49, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.; Chidester, S.; Zavala, C.V.; Manos, E.J.; James, S.R.; Karpf, A.R.; Jones, D.A.; Cairns, B.R. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 2007, 21, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Pollex, T.; Hanna, K.; Tuorto, F.; Meusburger, M.; Helm, M.; Lyko, F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010, 24, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Emara, M.M.; Ivanov, P.; Hickman, T.; Dawra, N.; Tisdale, S.; Kedersha, N.; Hu, G.F.; Anderson, P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010, 285, 10959–10968. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Feng, J.; Liu, Q.; Sun, F.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009, 583, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Gebetsberger, J.; Zywicki, M.; Kunzi, A.; Polacek, N. Trna-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012, 2012, 260909. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, P.; Emara, M.M.; Villen, J.; Gygi, S.P.; Anderson, P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 2011, 43, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Sobala, A.; Hutvagner, G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013, 10, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Spriggs, K.A.; Bushell, M.; Willis, A.E. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010, 40, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Lu, C.; Green, P.J.; Parker, R. Trna cleavage is a conserved response to oxidative stress in eukaryotes. RNA 2008, 14, 2095–2103. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, S.; Ivanov, P.; Hu, G.F.; Anderson, P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009, 185, 35–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karaca, E.; Weitzer, S.; Pehlivan, D.; Shiraishi, H.; Gogakos, T.; Hanada, T.; Jhangiani, S.N.; Wiszniewski, W.; Withers, M.; Campbell, I.M.; et al. Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell 2014, 157, 636–650. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, A.E.; Eggens, V.R.; Caglayan, A.O.; Reuter, M.S.; Scott, E.; Coufal, N.G.; Silhavy, J.L.; Xue, Y.; Kayserili, H.; Yasuno, K.; et al. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell 2014, 157, 651–663. [Google Scholar] [CrossRef] [PubMed]
- De Felice, C.; Signorini, C.; Leoncini, S.; Pecorelli, A.; Durand, T.; Valacchi, G.; Ciccoli, L.; Hayek, J. The role of oxidative stress in rett syndrome: An overview. Ann. N. Y. Acad. Sci. 2012, 1259, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Lintas, C.; Sacco, R.; Persico, A.M. Genome-wide expression studies in autism spectrum disorder, rett syndrome, and down syndrome. Neurobiol. Dis. 2012, 45, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Nishikura, K. Functions and regulation of RNA editing by adar deaminases. Annu. Rev. Biochem. 2010, 79, 321–349. [Google Scholar] [CrossRef] [PubMed]
- Bass, B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002, 71, 817–846. [Google Scholar] [CrossRef] [PubMed]
- Gerber, A.P.; Keller, W. RNA editing by base deamination: More enzymes, more targets, new mysteries. Trends Biochem. Sci. 2001, 26, 376–384. [Google Scholar] [CrossRef]
- Valente, L.; Nishikura, K. ADAR gene family and A-to-I RNA editing: Diverse roles in posttranscriptional gene regulation. Prog. Nucleic Acid Res. Mol. Biol. 2005, 79, 299–338. [Google Scholar] [PubMed]
- Slotkin, W.; Nishikura, K. Adenosine-to-inosine RNA editing and human disease. Genome Med. 2013, 5, 105. [Google Scholar] [CrossRef] [PubMed]
- Tariq, A.; Jantsch, M.F. Transcript diversification in the nervous system: A to I RNA editing in CNS function and disease development. Front. Neurosci. 2012, 6, 99. [Google Scholar] [CrossRef] [PubMed]
- Brusa, R.; Zimmermann, F.; Koh, D.S.; Feldmeyer, D.; Gass, P.; Seeburg, P.H.; Sprengel, R. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science 1995, 270, 1677–1680. [Google Scholar] [CrossRef] [PubMed]
- Paez-Colasante, X.; Figueroa-Romero, C.; Sakowski, S.A.; Goutman, S.A.; Feldman, E.L. Amyotrophic lateral sclerosis: Mechanisms and therapeutics in the epigenomic era. Nat. Rev. Neurol. 2015, 11, 266–279. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Chai, H.L.; Teramoto, S.; Tsuji, S.; Shimazaki, K.; Muramatsu, S.; Kwak, S. Rescue of amyotrophic lateral sclerosis phenotype in a mouse model by intravenous AAV9-ADAR2 delivery to motor neurons. EMBO Mol. Med. 2013, 5, 1710–1719. [Google Scholar] [CrossRef] [PubMed]
- Bodian, D. A suggestive relationship of nerve cell RNA with specific synaptic sites. Proc. Natl. Acad. Sci. USA 1965, 53, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Steward, O.; Levy, W.B. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J. Neurosci. 1982, 2, 284–291. [Google Scholar] [PubMed]
- Torre, E.R.; Steward, O. Demonstration of local protein synthesis within dendrites using a new cell culture system that permits the isolation of living axons and dendrites from their cell bodies. J. Neurosci. 1992, 12, 762–772. [Google Scholar] [PubMed]
- Aakalu, G.; Smith, W.B.; Nguyen, N.; Jiang, C.; Schuman, E.M. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron 2001, 30, 489–502. [Google Scholar] [CrossRef]
- Feig, S.; Lipton, P. Pairing the cholinergic agonist carbachol with patterned schaffer collateral stimulation initiates protein synthesis in hippocampal CA1 pyramidal cell dendrites via a muscarinic, NMDA-dependent mechanism. J. Neurosci. 1993, 13, 1010–1021. [Google Scholar] [PubMed]
- Weiler, I.J.; Greenough, W.T. Potassium ion stimulation triggers protein translation in synaptoneurosomal polyribosomes. Mol. Cell Neurosci. 1991, 2, 305–314. [Google Scholar] [CrossRef]
- Weiler, I.J.; Greenough, W.T. Metabotropic glutamate receptors trigger postsynaptic protein synthesis. Proc. Natl. Acad. Sci. USA 1993, 90, 7168–7171. [Google Scholar] [CrossRef] [PubMed]
- Antar, L.N.; Afroz, R.; Dictenberg, J.B.; Carroll, R.C.; Bassell, G.J. Metabotropic glutamate receptor activation regulates fragile X mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J. Neurosci. 2004, 24, 2648–2655. [Google Scholar] [CrossRef] [PubMed]
- Tiruchinapalli, D.M.; Oleynikov, Y.; Kelic, S.; Shenoy, S.M.; Hartley, A.; Stanton, P.K.; Singer, R.H.; Bassell, G.J. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J. Neurosci. 2003, 23, 3251–3261. [Google Scholar] [PubMed]
- Tongiorgi, E.; Righi, M.; Cattaneo, A. Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. J. Neurosci. 1997, 17, 9492–9505. [Google Scholar] [PubMed]
- Gong, R.; Park, C.S.; Abbassi, N.R.; Tang, S.J. Roles of glutamate receptors and the mammalian target of rapamycin (mTOR) signaling pathway in activity-dependent dendritic protein synthesis in hippocampal neurons. J. Biol. Chem. 2006, 281, 18802–18815. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.N.; Smith, L.T.; Logan, S.M.; Simpkins, J.W. Estrogen-induced activation of extracellular signal-regulated kinase signaling triggers dendritic resident mRNA translation. Neuroscience 2010, 170, 1080–1085. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.O.; Kim, S.M.; Zhao, Y.; Hwang, H.; Miura, S.K.; Sossin, W.S.; Martin, K.C. Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science 2009, 324, 1536–1540. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Martin, A.; Crespo, M.; Portera-Cailliau, C. Delayed stabilization of dendritic spines in fragile X mice. J. Neurosci. 2010, 30, 7793–7803. [Google Scholar] [CrossRef] [PubMed]
- Engert, F.; Bonhoeffer, T. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature 1999, 399, 66–70. [Google Scholar] [PubMed]
- Huber, K.M.; Kayser, M.S.; Bear, M.F. Role for rapid dendritic protein synthesis in hippocampal mglur-dependent long-term depression. Science 2000, 288, 1254–1257. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Schuman, E.M. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science 1996, 273, 1402–1406. [Google Scholar] [CrossRef] [PubMed]
- Raymond, C.R.; Thompson, V.L.; Tate, W.P.; Abraham, W.C. Metabotropic glutamate receptors trigger homosynaptic protein synthesis to prolong long-term potentiation. J. Neurosci. 2000, 20, 969–976. [Google Scholar] [PubMed]
- Sidorov, M.S.; Auerbach, B.D.; Bear, M.F. Fragile X mental retardation protein and synaptic plasticity. Mol. Brain 2013, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.; Joseph, S. Fragile X mental retardation protein: A paradigm for translational control by RNA-binding proteins. Biochimie 2015, 114, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Menon, L.; Mader, S.A.; Mihailescu, M.R. Fragile X mental retardation protein interactions with the microtubule associated protein 1B RNA. RNA 2008, 14, 1644–1655. [Google Scholar] [CrossRef] [PubMed]
- Vasilyev, N.; Polonskaia, A.; Darnell, J.C.; Darnell, R.B.; Patel, D.J.; Serganov, A. Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP. Proc. Natl. Acad. Sci. USA 2015, 112, E5391–E5400. [Google Scholar] [CrossRef] [PubMed]
- Buffington, S.A.; Huang, W.; Costa-Mattioli, M. Translational control in synaptic plasticity and cognitive dysfunction. Annu. Rev. Neurosci. 2014, 37, 17–38. [Google Scholar] [CrossRef] [PubMed]
- Osterweil, E.K.; Krueger, D.D.; Reinhold, K.; Bear, M.F. Hypersensitivity to mGLUR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile X syndrome. J. Neurosci. 2010, 30, 15616–15627. [Google Scholar] [CrossRef] [PubMed]
- Majumder, P.; Chu, J.F.; Chatterjee, B.; Swamy, K.B.; Shen, C.J. Co-regulation of mRNA translation by TDP-43 and fragile X syndrome protein fmrp. Acta Neuropathol. 2016, 132, 721–738. [Google Scholar] [CrossRef] [PubMed]
- Majumder, P.; Chen, Y.T.; Bose, J.K.; Wu, C.C.; Cheng, W.C.; Cheng, S.J.; Fang, Y.H.; Chen, Y.L.; Tsai, K.J.; Lien, C.C.; et al. TDP-43 regulates the mammalian spinogenesis through translational repression of Rac1. Acta Neuropathol. 2012, 124, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Coyne, A.N.; Siddegowda, B.B.; Estes, P.S.; Johannesmeyer, J.; Kovalik, T.; Daniel, S.G.; Pearson, A.; Bowser, R.; Zarnescu, D.C. Futsch/MAP1B mRNA is a translational target of TDP-43 and is neuroprotective in a drosophila model of amyotrophic lateral sclerosis. J. Neurosci. 2014, 34, 15962–15974. [Google Scholar] [CrossRef] [PubMed]
- Coyne, A.N.; Yamada, S.B.; Siddegowda, B.B.; Estes, P.S.; Zaepfel, B.L.; Johannesmeyer, J.S.; Lockwood, D.B.; Pham, L.T.; Hart, M.P.; Cassel, J.A.; et al. Fragile X protein mitigates TDP-43 toxicity by remodeling RNA granules and restoring translation. Hum. Mol. Genet. 2015, 24, 6886–6898. [Google Scholar] [CrossRef] [PubMed]
- Bernard, P.B.; Castano, A.M.; O’Leary, H.; Simpson, K.; Browning, M.D.; Benke, T.A. Phosphorylation of fmrp and alterations of fmrp complex underlie enhanced mltd in adult rats triggered by early life seizures. Neurobiol. Dis. 2013, 59, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Napoli, I.; Mercaldo, V.; Boyl, P.P.; Eleuteri, B.; Zalfa, F.; De Rubeis, S.; Di Marino, D.; Mohr, E.; Massimi, M.; Falconi, M.; et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 2008, 134, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.R.; Bray, S.M.; Warren, S.T. Molecular mechanisms of fragile X syndrome: A twenty-year perspective. Annu. Rev. Pathol. 2012, 7, 219–245. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.T.; Dyavaiah, M.; DeMott, M.S.; Taghizadeh, K.; Dedon, P.C.; Begley, T.J. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010, 6, e1001247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Y.; Dai, Q.; Zhang, W.; Ren, J.; Pan, T.; He, C. The ALKB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew. Chem. Int. Ed. Engl. 2010, 49, 8885–8888. [Google Scholar] [CrossRef] [PubMed]
- Saikia, M.; Fu, Y.; Pavon-Eternod, M.; He, C.; Pan, T. Genome-wide analysis of N1-methyl-adenosine modification in human tRNAs. RNA 2010, 16, 1317–1327. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, C. Dynamic RNA modifications in posttranscriptional regulation. Mol. Cell 2014, 56, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Komara, M.; Al-Shamsi, A.M.; Ben-Salem, S.; Ali, B.R.; Al-Gazali, L. A novel single-nucleotide deletion (c.1020delA) in NSun2 causes intellectual disability in an emirati child. J. Mol. Neurosci. 2015, 57, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Blanco, S.; Frye, M. Role of RNA methyltransferases in tissue renewal and pathology. Curr. Opin. Cell Biol. 2014, 31, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Flores, J.V.; Cordero-Espinoza, L.; Oeztuerk-Winder, F.; Andersson-Rolf, A.; Selmi, T.; Blanco, S.; Tailor, J.; Dietmann, S.; Frye, M. Cytosine-5 RNA methylation regulates neural stem cell differentiation and motility. Stem Cell Rep. 2017, 8, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Schratt, G. Micrornas at the synapse. Nat. Rev. Neurosci. 2009, 10, 842–849. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Bashir, Z.I. The epitranscriptome in modulating spatiotemporal RNA translation in neuronal post-synaptic function. Front. Cell Neurosci. 2015, 9, 420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceman, S.; O’Donnell, W.T.; Reed, M.; Patton, S.; Pohl, J.; Warren, S.T. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum. Mol. Genet. 2003, 12, 3295–3305. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Pepio, A.M.; Sossin, W.S. Serotonin activates s6 kinase in a rapamycin-sensitive manner in aplysia synaptosomes. J. Neurosci. 2001, 21, 382–391. [Google Scholar] [PubMed]
- Colombrita, C.; Onesto, E.; Megiorni, F.; Pizzuti, A.; Baralle, F.E.; Buratti, E.; Silani, V.; Ratti, A. TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J. Biol. Chem. 2012, 287, 15635–15647. [Google Scholar] [CrossRef] [PubMed]
- Xie, P.; Zang, L.Q.; Li, X.K.; Shu, Q. An epigenetic view of developmental diseases: New targets, new therapies. World J. Pediatr. 2016, 12, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Ke, S.; Alemu, E.A.; Mertens, C.; Gantman, E.C.; Fak, J.J.; Mele, A.; Haripal, B.; Zucker-Scharff, I.; Moore, M.J.; Park, C.Y.; et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 2015, 29, 2037–2053. [Google Scholar] [CrossRef] [PubMed]
- Rowles, J.; Wong, M.; Powers, R.; Olsen, M. FTO, RNA epigenetics and epilepsy. Epigenetics 2012, 7, 1094–1097. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.R.; Chopra, P.; Suhl, J.A.; Warren, S.T.; Bassell, G.J. Identification of consensus binding sites clarifies FMRP binding determinants. Nucleic Acids Res. 2016, 44, 6649–6659. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Kwak, S. The molecular link between inefficient GluA2 Q/R site-RNA editing and TDP-43 pathology in motor neurons of sporadic amyotrophic lateral sclerosis patients. Brain Res. 2014, 1584, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; Ito, K.; Sun, H.; Aizawa, H.; Kanazawa, I.; Kwak, S. Glutamate receptors: RNA editing and death of motor neurons. Nature 2004, 427, 801. [Google Scholar] [CrossRef] [PubMed]
- Lacoux, C.; Di Marino, D.; Boyl, P.P.; Zalfa, F.; Yan, B.; Ciotti, M.T.; Falconi, M.; Urlaub, H.; Achsel, T.; Mougin, A.; et al. BC1-FMRP interaction is modulated by 2′-O-methylation: RNA-binding activity of the tudor domain and translational regulation at synapses. Nucleic Acids Res. 2012, 40, 4086–4096. [Google Scholar] [CrossRef] [PubMed]
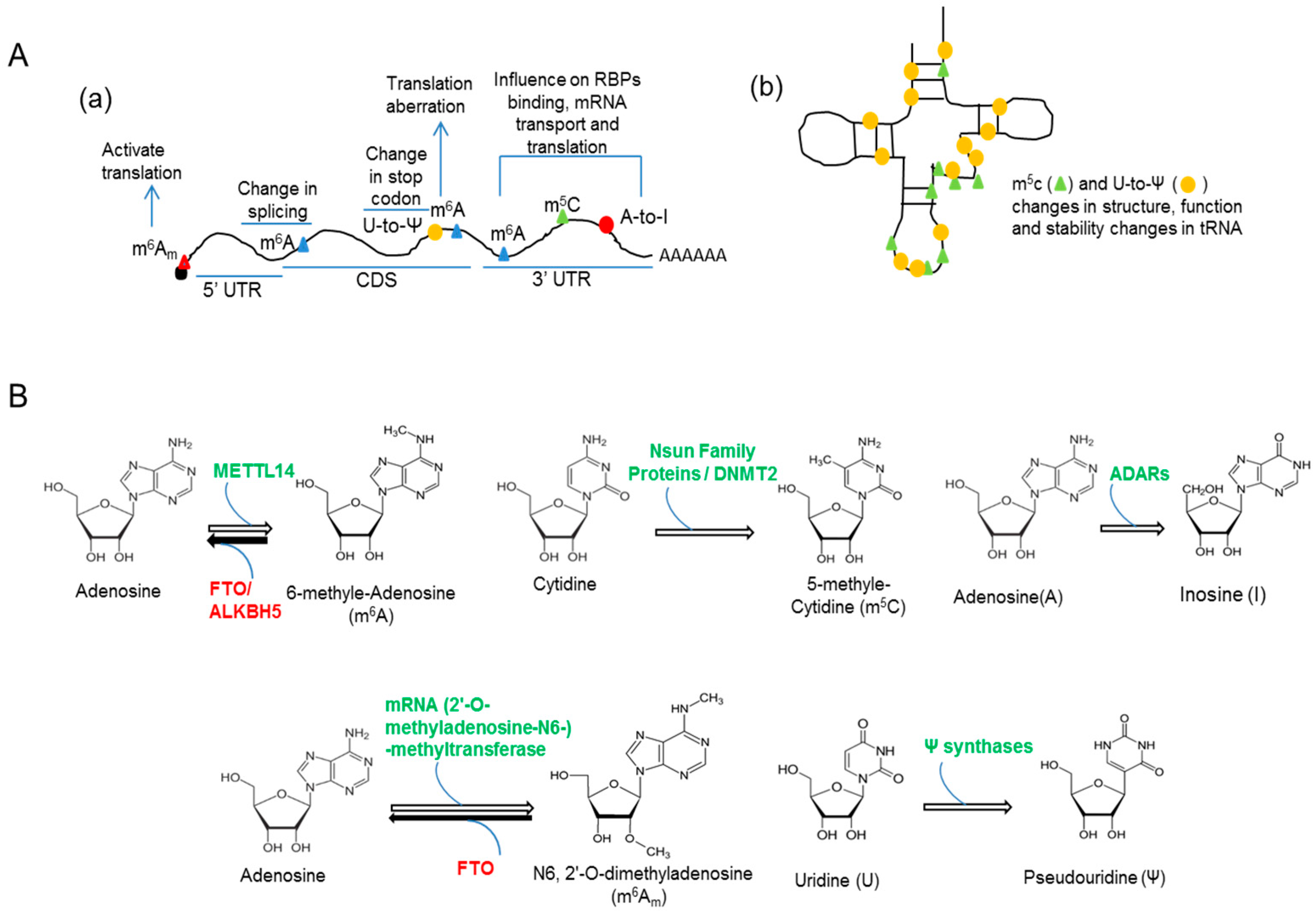
| RNA Modification | Base Involved | Occur in | Modification Enzyme/s |
|---|---|---|---|
| m6A | Adenosine | mRNA, tRNA | METTL3/METTl14 |
| m5C | Cytosine | mRNA, tRNA, ncRNAs | NOP2/NOL1, YebU/Trm4, RsmB and NSun family proteins |
| A-to-I conversion | Adenosine | mRNAs | Adenosine deaminases |
| m6Am | Adenosine | mRNA | mRNA (2′-O-methyladenosine-N6-) methyltransferases |
| Pseudouridylation | Uridine | ncRNA, mRNA | Ψ synthases |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majumder, P.; Chatterjee, B.; Shen, C.-K.J. Epitranscriptome and FMRP Regulated mRNA Translation. Epigenomes 2017, 1, 11. https://doi.org/10.3390/epigenomes1020011
Majumder P, Chatterjee B, Shen C-KJ. Epitranscriptome and FMRP Regulated mRNA Translation. Epigenomes. 2017; 1(2):11. https://doi.org/10.3390/epigenomes1020011
Chicago/Turabian StyleMajumder, Pritha, Biswanath Chatterjee, and C.-K. James Shen. 2017. "Epitranscriptome and FMRP Regulated mRNA Translation" Epigenomes 1, no. 2: 11. https://doi.org/10.3390/epigenomes1020011





