3D-Printed Bioreceptive Tiles of Reaction–Diffusion (Gierer–Meinhardt Model) for Multi-Scale Algal Strains’ Passive Immobilization
Abstract
:1. Introduction
2. Results
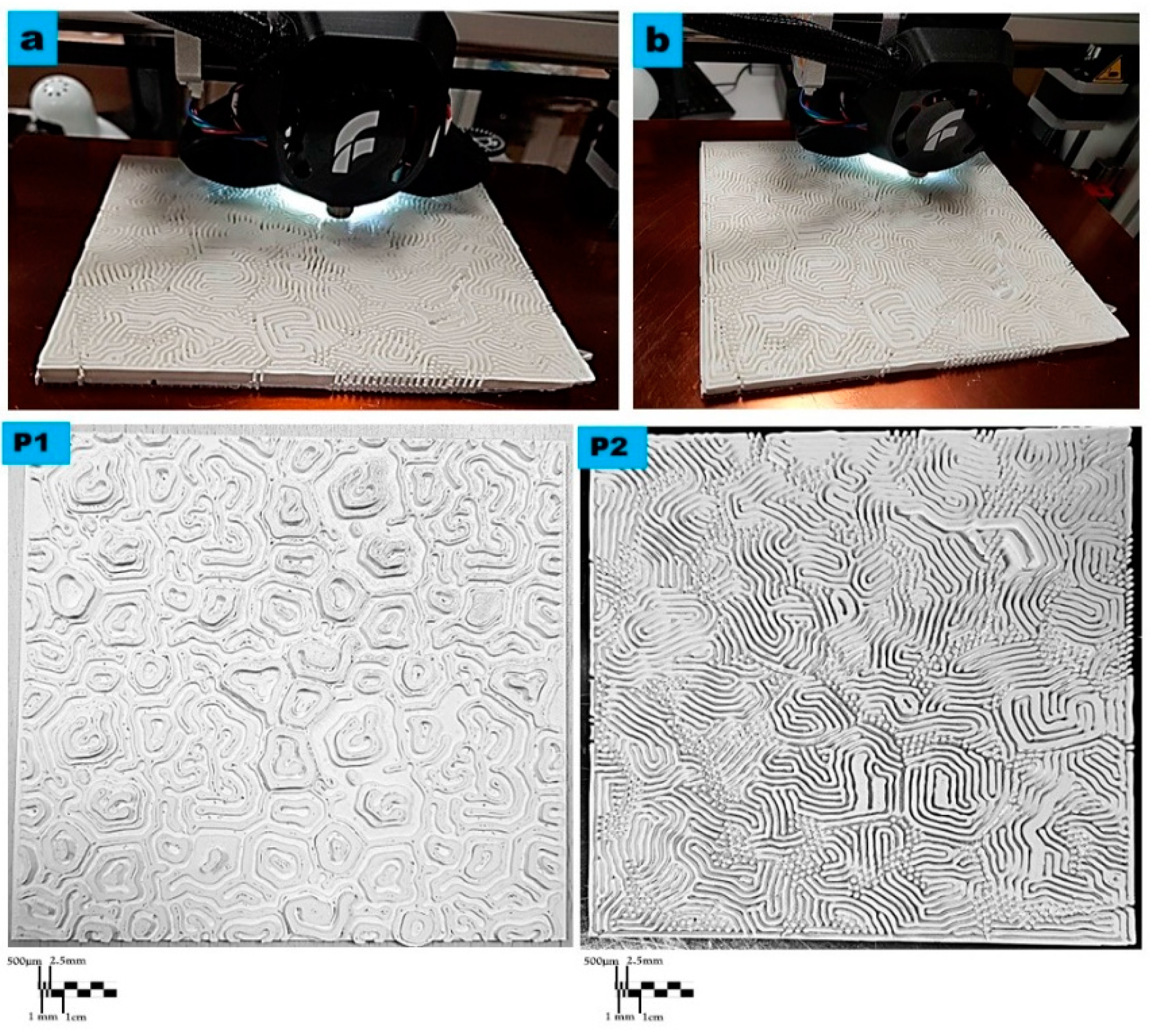
2.1. 3D-Printed Bioreceptive Tiles Following the Reaction–Diffusion Gierer–Meinhardt Model: Pattern 1 and 2
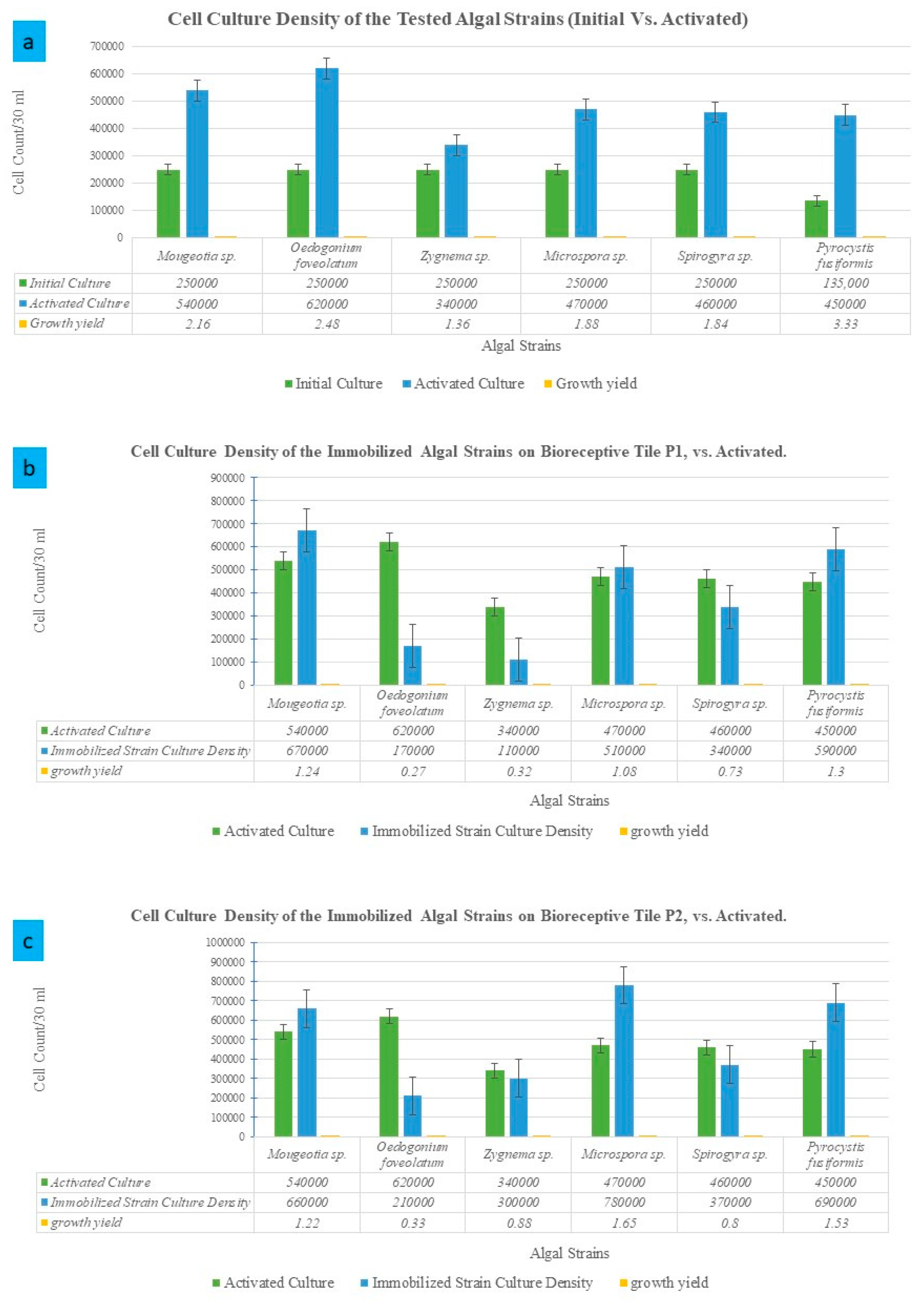
2.2. Passive Immobilization of the Multi-Scale Lengths Strains of a Mixed Algal Culture on the 3D-Printed Bioreceptive Tiles: P1 vs. P2
2.3. Relation between Scale and Morphology of the Bioreceptive Tile Pattern and the Scale and Morphology of the Immobilized Algae Strain
3. Discussion
3.1. 3D-Printed Bioreceptive Tiles from The Reaction–Diffusion Gierer–Meinhardt Model: Pattern 1 and 2
3.2. Multi-Scale-Lengths Algal Strains’ Cell Immobilization on the Bioreceptive Tiles P1 and P2
4. Conclusions
5. Materials and Methods
5.1. Designing Bioreceptive Surfaces from the Activator-Inhibitor Gierer–Meinhardt Model
5.1.1. Two Biopatterns from Gierer–Meinhardt Model: P1: Polar/Periodic and P2: Strip/Labyrinth Patterns
5.1.2. 3D Translation and 3D Printing of the Bioreceptive Tiles: P1, and P2: Z-Offsetting, and Fractal Dimension
5.1.3. Multi-Scale Mixed Algal Culture Medium and Inoculation
5.1.4. Culture Density and Microscopy Study
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Marcos Cruz Architect. Available online: http://marcoscruzarchitect.blogspot.com/2017/10/bioreceptive-concrete-facades-design.html (accessed on 15 May 2023).
- Mustafa, K.F.; Prieto, A.; Ottele, M. The Role of Geometry on a Self-Sustaining Bio-Receptive Concrete Panel for Facade Application. Sustainability 2021, 13, 7453. [Google Scholar] [CrossRef]
- Castillo, J.G.; Gennett, A.; Estévez, A.T.; Abdallah, Y.K. Employing Columba livia Swarmal Patterns in Designing Self-Sufficient Photo Bioreactor of Chlorella spp. Cultivation in Plaça de Catalunya. In Renewable Energy for Mitigating Climate Change; CRC Press: Boca Raton, FL, USA, 2021; pp. 153–168. [Google Scholar] [CrossRef]
- Veeger, M.; Ottelé, M.; Prieto, A. Making bioreceptive concrete: Formulation and testing of bioreceptive concrete mixtures. J. Build. Eng. 2021, 44, 102545. [Google Scholar] [CrossRef]
- Mal, N.; Satpati, G.; Raghunathan, S.; Davoodbasha, M. Current strategies on algae-based biopolymer production and scale-up. Chemosphere 2022, 289, 133178. [Google Scholar] [CrossRef]
- Onen Cinar, S.; Chong, Z.K.; Kucuker, M.A.; Wieczorek, N.; Cengiz, U.; Kuchta, K. Bioplastic Production from Microalgae: A Review. Int. J. Environ. Res. Public Health 2020, 17, 3842. [Google Scholar] [CrossRef]
- El Semary, N.; Alsuhail, M.; Al Amer, K.; AlNaim, A. Applications of algae for environmental sustainability: Novel bioplastic formulation method from marine green alga. Front. Mar. Sci. 2022, 9, 1047284. [Google Scholar] [CrossRef]
- López-Pacheco, I.Y.; Rodas-Zuluaga, L.I.; Cuellar-Bermudez, S.P.; Hidalgo-Vázquez, E.; Molina-Vazquez, A.; Araújo, R.G.; Martínez-Ruiz, M.; Varjani, S.; Barceló, D.; Iqbal, H.M.N.; et al. Revalorization of Microalgae Biomass for Synergistic Interaction and Sustainable Applications: Bioplastic Generation. Mar. Drugs 2022, 20, 601. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.K.; Mak, A.L.; Trevan, M.D. Immobilized algae: A review. Process Biochem. 1986, 21, 122–127. [Google Scholar]
- Moreno-Garrido, I. Microalgae immobilization: Current techniques and uses. Bioresour. Technol. 2008, 99, 3949–3964. [Google Scholar] [CrossRef]
- Codd, G.A. Immobilized micro-algae and cyanobacteria. Br. Phycol. Soc. Newslett. 1987, 24, 1–5. [Google Scholar]
- Chen, Z.; Osman, A.I.; Rooney, D.W.; Oh, W.-D.; Yap, P.-S. Remediation of Heavy Metals in Polluted Water by Immobilized Algae: Current Applications and Future Perspectives. Sustainability 2023, 15, 5128. [Google Scholar] [CrossRef]
- Akhtar, N. Removal and recovery of nickel(II) from aqueous solution by loofa sponge-immobilized biomass of Chlorella sorokiniana: Characterization studies. J. Hazard. Mater. 2004, 108, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Danilov, R.A.; Ekelund, N.G.A. Comparison of usefulness of threetypes of artificial substrata (glass, wood and plastic) when studyingsettlement patterns of periphyton in lakes of different trophic status. J. Microbiol. Methods 2001, 45, 167–170. [Google Scholar] [CrossRef]
- Ghosh, M.; Gaur, J.P. Current velocity and the establishment ofstream algal periphyton communities. Aquat. Bot. 1998, 60, 1–10. [Google Scholar] [CrossRef]
- Kuehner, M. Bioplastics—Study: Market, Analysis, Trends. Ceresana. Archive.org. 2016. Available online: https://web.archive.org/web/20171104212623/http://www.ceresana.com/en/market-studies/plastics/bioplastics (accessed on 25 May 2023).
- Erickson, L.E. Bioreactors. In Encyclopedia of Microbiology; Academic Press: Cambridge, MA, USA, 2009; pp. 206–211. [Google Scholar] [CrossRef]
- Bioreactor. The IUPAC Compendium of Chemical Terminology; Blackwell Scientific Publications: Oxford, UK, 2014. [Google Scholar] [CrossRef]
- Popović, M.K.; Pörtner, R. Bioreactors and Cultivation Systems for Cell and Tissue Culture. Biotechnology—Bioreactoes and Cultivation Systems for Cell and Tissue Culture—M.K. Available online: http://www.eolss.net/Sample-Chapters/C17/E6-58-04-15.pdf (accessed on 25 May 2023).
- López, A.; Lázaro, N.; Marqués, A.M. The interphase technique: A simple method of cell immobilization in gel-beads. J. Microbiol. Methods 1997, 30, 231–234. [Google Scholar] [CrossRef]
- Kowalczyk, T.; Sitarek, P.; Toma, M.; Rijo, P.; Domínguez-Martín, E.; Falcó, I.; Sánchez, G.; Śliwiński, T. Enhanced Accumulation of Betulinic Acid in Transgenic Hairy Roots of Senna obtusifolia Growing in the Sprinkle Bioreactor and Evaluation of Their Biological Properties in Various Biological Models. Chem. Biodivers. 2021, 18, e2100455. [Google Scholar] [CrossRef] [PubMed]
- Peinado, R.A.; Moreno, J.J.; Villalba, J.M.; González-Reyes, J.A.; Ortega, J.M.; Mauricio, J.C. Yeast biocapsules: A new immobilization method and their applications. Enzym. Microb. Technol. 2006, 40, 79–84. [Google Scholar] [CrossRef]
- Prosenc, F.; Piechocka, J.; Škufca, D.; Heath, E.; Griessler Bulc, T.; Istenič, D.; Buttiglieri, G. Microalgae-based removal of contaminants of emerging concern: Mechanisms in Chlorella vulgaris and mixed algal-bacterial cultures. J. Hazard. Mater. 2021, 418, 126284. [Google Scholar] [CrossRef]
- Satpati, G.G.; Pal, R. Co-Cultivation of Leptolyngbya tenuis (Cyanobacteria) and Chlorella ellipsoidea (Green alga) for Biodiesel Production, Carbon Sequestration, and Cadmium Accumulation. Curr. Microbiol. 2021, 78, 1466–1481. [Google Scholar] [CrossRef]
- Estévez, A.T.; Navarro, D. Biomanufacturing the Future: Biodigital Architecture & Genetics. Procedia Manuf. 2017, 12, 7–16. [Google Scholar] [CrossRef]
- ecoLogicStudio. Available online: https://www.ecologicstudio.com/ (accessed on 23 June 2023).
- Abdallah, Y.K.; Estevez, A.T.; Tantawy, D.E.D.M.; Ibraheem, A.M.; Khalil, N.M. Employing Laccase-Producing Aspergillus sydowii NYKA 510 as a Cathodic Biocatalyst in Self-Sufficient Lighting Microbial Fuel Cell. J. Microbiol. Biotechnol. 2019, 29, 1861–1872. [Google Scholar] [CrossRef]
- Jaafari, A.A.Q.; Roznowski, V.; Estévez, A.T.; Abdullah, Y.K. Self-Sufficient Bioelectricity Systems in Architecture: Employing Spirulina Platensis in Photosynthetic Microbial Fuel Cells for the Generation of Domestic and Urban Bio-Electricity through a Diffusion-Limited Aggregation Pattern. Sustain. Eng. Technol. Archit. 2021, 29, 1–18. [Google Scholar] [CrossRef]
- Abdallah, Y.K.; Estevez, A.T. Bioactive Devices as Self-Sufficient Systems for Energy Production in Architecture. J. Green Build. 2021, 16, 3–22. [Google Scholar] [CrossRef]
- Abdallah, Y.K.; Estevez, A.T. Methodology of Implementing Transformative Bioactive Hybrids in Built Environment to Achieve Sustainability. In Proceedings of the Blucher Design Proceedings, Medellín, Colombia, 18–20 November 2020. [Google Scholar] [CrossRef]
- Huesemann, M.; Williams, P.; Edmundson, S.; Chen, P.; Kruk, R.; Cullinan, V.; Crowe, B.; Lundquist, T. The laboratory environmental algae pond simulator (LEAPS) photobioreactor: Validation using outdoor pond cultures of Chlorella sorokiniana and Nannochloropsis salina. Algal Res. 2017, 26, 39–46. [Google Scholar] [CrossRef]
- Ozkan, A.; Berberoglu, H. Adhesion of algal cells to surfaces. Biofouling 2013, 29, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Sedighi, M.; Pourmoghaddam Qhazvini, P.; Amidpour, M. Algae-Powered Buildings: A Review of an Innovative, Sustainable Approach in the Built Environment. Sustainability 2023, 15, 3729. [Google Scholar] [CrossRef]
- Vecchi, V.; Barera, S.; Bassi, R.; Dall’Osto, L. Potential and Challenges of Improving Photosynthesis in Algae. Plants 2020, 9, 67. [Google Scholar] [CrossRef] [Green Version]
- Brickley, M.R.; Weise, V.; Hawes, C.; Cobb, A.H. Morphology and dynamics of mitochondria in Mougeotia sp. Eur. J. Phycol. 2010, 45, 258–266. [Google Scholar] [CrossRef]
- Abdallah, Y.K.; Estevez, A.T. Integrating Chlorella vulgaris and Monoraphidium contortum in Architectural Systems for the Biodegradation of Sulfamethoxazole from Wastewater. In Nourishing Tomorrow; Ting, D.S.-K., Stagner, J.A., Eds.; World Scientific: Singapore, 2023. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M. AlgaeBase; algalwebofc.github.io; World-Wide Electronic Publication, National University of Ireland: Galway, Ireland, 2008; Available online: http://www.algaebase.org (accessed on 13 February 2019).
- Herburger, K.; Lewis, L.A.; Holzinger, A. Photosynthetic efficiency, desiccation tolerance and ultrastructure in two phylogenetically distinct strains of alpine Zygnema sp. (Zygnematophyceae 2014, Streptophyta): Role of pre-akinete formation. Protoplasma 2015, 252, 571–589. [Google Scholar] [CrossRef] [Green Version]
- Memon, A.A.S.; Pathan, A.A.; Kandhar, I.A.; Mahar, R.B.; Brohi, R.-Z.; Balouch, A. Microspora Floccosa: A Potential Biofuel Producer. Pak. J. Anal. Environ. Chem. 2016, 17, 106. [Google Scholar] [CrossRef]
- Frost, O.; Nazanin Owji Thorogate, R.; Christos Kyriakidis Prasad Sawadkar Mordan, N.; Knowles, J.C.; Lali, F.V.; García-Gareta, E. Cell morphology as a design parameter in the bioengineering of cell–biomaterial surface interactions. Biomater. Sci. 2021, 9, 8032–8050. [Google Scholar] [CrossRef]
- Bhovichitra, M.; Swift, E. Light and dark uptake of nitrate and ammonium by large oceanic dinoflagellates: Pyrocystis noctiluca, Pyrocystis fusiformis, and Dissodinium lunula1. Limnol. Oceanogr. 1977, 22, 73–83. [Google Scholar] [CrossRef]
- Guo Luyao Shi, X.; Cao, J. Turing patterns of Gierer–Meinhardt model on complex networks. Nonlinear Dyn. 2021, 105, 899–909. [Google Scholar] [CrossRef]
- Wongsawad, P.; Peerapornpisal, Y. Morphological and molecular profiling of Spirogyra from northeastern and northern Thailand using inter simple sequence repeat (ISSR) markers. Saudi J. Biol. Sci. 2015, 22, 382–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.-L.; Hou, X.; Jing, Z. Stripe and Spot Patterns in a Gierer–Meinhardt Activator–Inhibitor Model with Different Sources. Int. J. Bifurc. Chaos 2015, 25, 1550108. [Google Scholar] [CrossRef]
- Stephen, W. Statistical Mechanics of Cellular Automata. Rev. Mod. Phys. 1983, 55, 601–644. [Google Scholar] [CrossRef]
- Tommaso, T.; Norman, M. Cellular Automata Machines: A New Environment for Modeling; MIT Press: Cambridge, MA, USA, 1987; p. 27. ISBN 9780262200608. [Google Scholar]
- Gierer, A.; Meinhardt, H. A theory of biological pattern formation. Kybernetik 1972, 12, 30–39. [Google Scholar] [CrossRef] [Green Version]
- Gierer, A. Generation of biological patterns and form: Some physical, mathematical, and logical aspects. Prog. Biophys. Mol. Biol. 1981, 37, 1–47. [Google Scholar] [CrossRef] [Green Version]
- Meinhardt, H. Models of Biological Pattern Formation; Academic Press: Cambridge, MA, USA, 1982. [Google Scholar]
- Meinhardt, H. Gierer-Meinhardt model. Scholarpedia 2006, 1, 1418. [Google Scholar] [CrossRef]
- Doelman, A. Pattern Formation in Reaction-Diffusion Systems—An Explicit Approach. 2018. Available online: https://www.math.leidenuniv.nl/~doelman/RDSPF-web.pdf (accessed on 10 July 2022).
- Schnörr, D.; Schnörr, C. Learning System Parameters from Turing Patterns. Available online: https://ipa.math.uni-heidelberg.de/dokuwiki/Papers/Schnorr2021vr.pdf (accessed on 10 July 2022).
- Jiang, X.; Bruzewicz, D.A.; Wong, A.E.; Piel, M.; Whitesides, G.M. Directing cell migration with asymmetric micropatterns. Proc. Natl. Acad. Sci. USA 2005, 102, 975–978. [Google Scholar] [CrossRef]
- Spalding, M.H. Photosynthesis and photorespiration in freshwater green algae. Aquat. Bot. 1989, 34, 181–209. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Z.; Li, X.; Wu, X.; Chen, L.; Wang, G. Photosynthesis Responses of Tibetan Freshwater Algae Chlorella vulgaris to Herbicide Glyphosate. Int. J. Environ. Res. Public Health 2022, 20, 386. [Google Scholar] [CrossRef] [PubMed]
- Valiadi, M.; Iglesias-Rodriguez, D. Understanding Bioluminescence in Dinoflagellates—How Far Have We Come? Microorganisms 2013, 1, 3–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanley, K.A.; Widder, E.A. Bioluminescence in Dinoflagellates: Evidence that the Adaptive Value of Bioluminescence in Dinoflagellates is Concentration Dependent. Photochem. Photobiol. 2017, 93, 519–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woelkerling, W.J.; Kowal, R.R.; Gough, S.B. Sedgwick-rafter cell counts: A procedural analysis. Hydrobiologia 1976, 48, 95–107. [Google Scholar] [CrossRef]
- Steinberg, M.K.; First, M.R.; Lemieux, E.J.; Drake, L.A.; Nelson, B.N.; Kulis, D.M.; Anderson, D.M.; Welschmeyer, N.A.; Herring, P.R. Comparison of techniques used to count single-celled viable phytoplankton. J. Appl. Phycol. 2011, 24, 751–758. [Google Scholar] [CrossRef]
- Latz, M.I. Bioluminescence response of four species of dinoflagellates to fully developed pipe flow. J. Plankton Res. 2004, 26, 1529–1546. [Google Scholar] [CrossRef] [Green Version]
- Gross, M.; Zhao, X.; Mascarenhas, V.; Wen, Z. Effects of the surface physico-chemical properties and the surface textures on the initial colonization and the attached growth in algal biofilm. Biotechnol. Biofuels 2016, 9, 38. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Yuan, W.; Cao, J. Effect of surface texturing on microalgal cell attachment to solid carriers. Int. J. Agric. Biol. Eng. 2014, 7, 82–91. [Google Scholar]
- Cao, J.; Yuan, W.; Pei, Z.J.; Davis, T.; Cui, Y.; Beltran, M. A preliminary study of the effect of surface texture on algae cell attachment for a mechanical-biological energy manufacturing system. J. Manuf. Sci. Eng. 2009, 131, 064505. [Google Scholar] [CrossRef]
- Nosonovsky, M.; Ramachandran, R. Geometric Interpretation of Surface Tension Equilibrium in Superhydrophobic Systems. Entropy 2015, 17, 4684–4700. [Google Scholar] [CrossRef] [Green Version]
- Girifalco, L.A.; Good, R.J. A Theory for the Estimation of Surface and Interfacial Energies. I. Derivation and Application to Interfacial Tension. J. Phys. Chem. 1957, 61, 904–909. [Google Scholar] [CrossRef]
- Marmur, A. Soft contact: Measurement and interpretation of contact angles. Soft Matter 2006, 2, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Zhang, C.; Ho, S.-H. Immobilized microalgal system: An achievable idea for upgrading current microalgal wastewater treatment. Environ. Sci. Ecotechnol. 2022, 14, 100227. [Google Scholar] [CrossRef] [PubMed]
- Fofonjka, A.; Milinkovitch, M.C. Reaction-diffusion in a growing 3D domain of skin scales generates a discrete cellular automaton. Nat. Commun. 2021, 12, 2433. [Google Scholar] [CrossRef]
- Landge, A.N.; Jordan, B.M.; Diego, X.; Müller, P. Pattern formation mechanisms of self-organizing reaction-diffusion systems. Dev. Biol. 2020, 460, 2–11. [Google Scholar] [CrossRef]
- Adamatzky, A. (Ed.) Game of Life Cellular Automata; Springer: London, UK, 2010. [Google Scholar] [CrossRef]
- Song, Y.; Yang, R.; Sun, G.-Q. Pattern dynamics in a Gierer–Meinhardt model with a saturating term. Appl. Math. Model. 2017, 46, 476–491. [Google Scholar] [CrossRef]
- Carolina Biological Supply. Alga-Gro® Freshwater Medium, 1 qt. Available online: https://www.carolina.com/dehydrated-media-and-media-ingredients/alga-gro-freshwater-medium-1-qt/153752.pr (accessed on 23 June 2023).
- Kilham, S.S.; Kreeger, D.A.; Lynn, S.G.; Goulden, C.E.; Herrera, L. COMBO: A defined freshwater culture medium for algae and zooplankton. Hydrobiologia 1998, 377, 147–159. [Google Scholar] [CrossRef]
- web.biosci.utexas.edu. Available online: http://web.biosci.utexas.edu/utex/Media%20PDF/soilwater-gr-plus-medium.pdf (accessed on 23 June 2023).
- Carolina Biological Supply. Bioluminescent Dinoflagellate Medium, 1 L. Available online: https://www.carolina.com/dehydrated-media-and-media-ingredients/bioluminescent-dinoflagellate-medium/153757.pr (accessed on 23 June 2023).
- McAlice, B.J. Phytoplankton Sampling with the Sedgwick-Rafter Cell1. Limnol. Oceanogr. 1971, 16, 19–28. [Google Scholar] [CrossRef]
- ANACC Methods and Materials. Cell Counting Using a Sedgwick-Rafter Chamber. Available online: https://research.csiro.au/anaccmethods/phycological-techniques/biomass-estimation/sedgwick-rafter/ (accessed on 25 May 2023).




| Strain Medium Category | Freshwater Green Filamentous Algae | Soil-Water Algae | Marine Water | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain Name | Mougeotia sp. 2nd in P1 3rd in P2 | Oedogonium foveolatum | Zygnema sp. 5th in P1 5th in P2 | Microspora sp. 3rd in P1 1st in P2 | Spirogyra sp. 4th in P1 4th in P2 | Pyrocystis fusiformis 1st in P1 2nd in P2 | |||||||
| Starter Culture Density/30 mL | 250,000 | Growth Ratio 2.16 | 250,000 | Growth Ratio 2.48 | 250,000 | Growth Ratio 1.36 | 250,000 | Growth Ratio 1.88 | 250,000 | Growth Ratio 1.84 | 135,000 | Growth Ratio 3.33 | |
| Activated Culture Density | 540,000 | 620,000 | 340,000 | 470,000 | 460,000 | 450,000 | |||||||
| Culture Density on P1 | 670,000 | Vs. In 2.68 | 170,000 | Vs. In 0.68 | 110,000 | Vs. In 0.44 | 510,000 | Vs. In 2.04 | 340,000 | Vs. In 1.36 | 590,000 | Vs. In 4.8 | |
| Vs. Act 1.24 | Vs. Act 0.27 | Vs. Act 0.32 | Vs. Act 1.08 | Vs. Act 0.73 | Vs. Act 1.3 | ||||||||
| Culture Density on P2 | 660,000 | Vs. In 2.64 | 210,000 | Vs. In 0.84 | 30,0000 | Vs. In 1.2 | 780,000 | Vs. In 3.12 | 370,000 | Vs. In 1.48 | 690,000 | Vs. In 5.1 | |
| Vs. Act 1.22 | Vs. Act 0.33 | Vs. Act 0.88 | Vs. Act 1.65 | Vs. Act 0.8 | Vs. Act 1.53 | ||||||||
| Immobilization Ruling parameters | |||||||||||||
| Morphology | Unbranched intertwining filaments | Unbranched filaments cells wider at one end; occasionally some bulbous cells in between, with rings at the wider end. | Unbranched short cylindrical cells | Unbranched filaments with holdfast cells at the end | Cylindrical cells | Fusiform shaped, elongated with tapered ends | |||||||
| Cell width | 30 µm | 45 µm | 40 µm | 25 µm | 90 µm | 375 µm | |||||||
| Relativity of P1 niches (3000 µm) to cell width of the strain. | 0.01% | 0.015% | 0.013% | 0.008% | 0.03% | 0.12% | |||||||
| P1 Hosting Capacity | 100 cells | 66.6 cells | 75 cells | 120 cells | 33 cells | 8 cells | |||||||
| Relativity of P2 niches (500 µm) to cell width of strain. | 0.06% | 0.09% | 0.08% | 0.05% | 0.18% | 0.75% | |||||||
| P2 Hosting Capacity | 16.6 cells | 11 cells | 12.5 cells | 20 cells | 5 cells | 1.3 Cells | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdallah, Y.K.; Estévez, A.T. 3D-Printed Bioreceptive Tiles of Reaction–Diffusion (Gierer–Meinhardt Model) for Multi-Scale Algal Strains’ Passive Immobilization. Buildings 2023, 13, 1972. https://doi.org/10.3390/buildings13081972
Abdallah YK, Estévez AT. 3D-Printed Bioreceptive Tiles of Reaction–Diffusion (Gierer–Meinhardt Model) for Multi-Scale Algal Strains’ Passive Immobilization. Buildings. 2023; 13(8):1972. https://doi.org/10.3390/buildings13081972
Chicago/Turabian StyleAbdallah, Yomna K., and Alberto T. Estévez. 2023. "3D-Printed Bioreceptive Tiles of Reaction–Diffusion (Gierer–Meinhardt Model) for Multi-Scale Algal Strains’ Passive Immobilization" Buildings 13, no. 8: 1972. https://doi.org/10.3390/buildings13081972






