Differences in Gene Expression Profiles between Early and Late Isolates in Monospecies Achromobacter Biofilm
Abstract
:1. Introduction
2. Results and Discussion
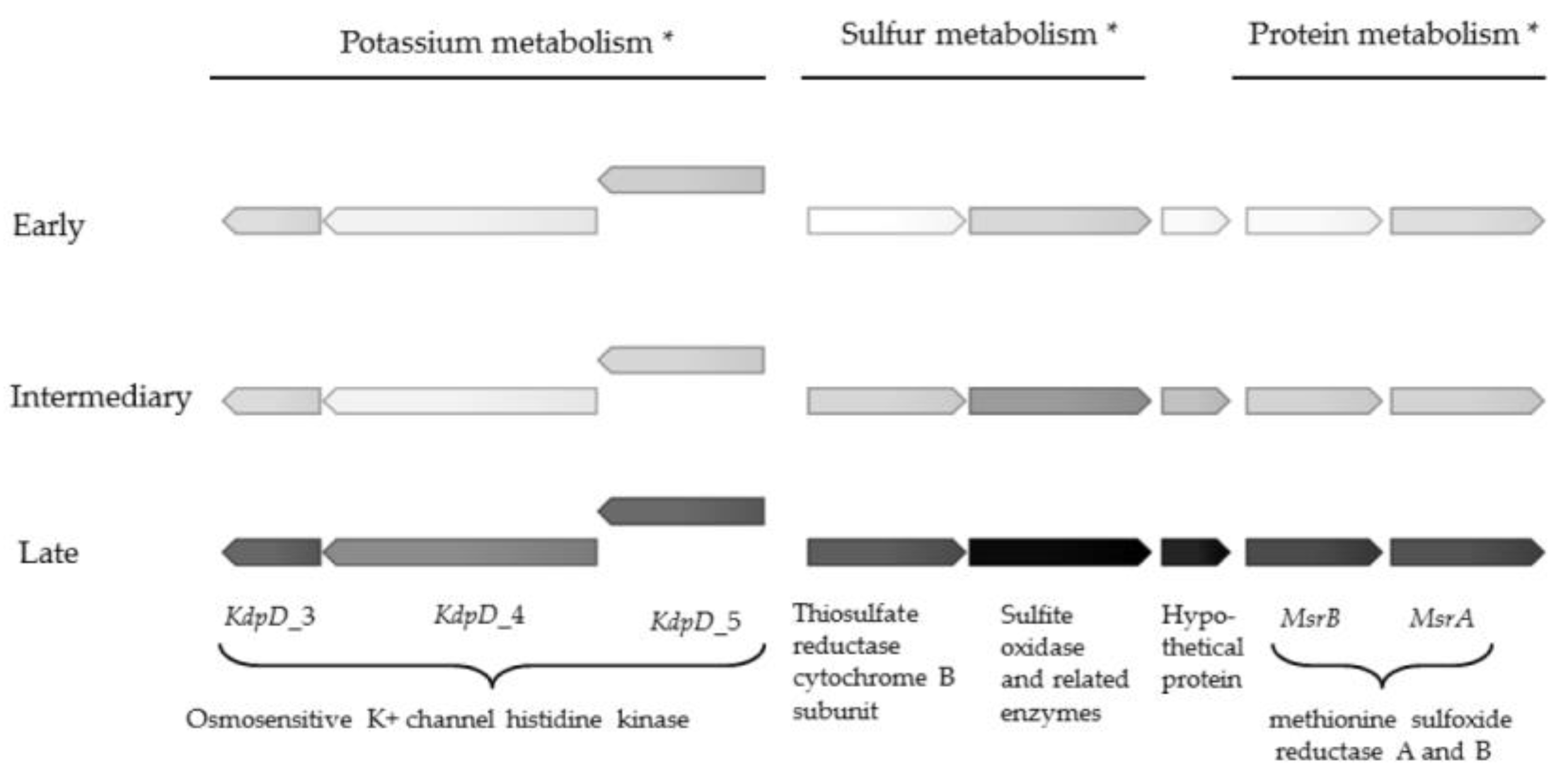
2.1. Gene Expression Profiles
2.2. Antimicrobial Resistance
3. Materials and Methods
3.1. Strains and Growth Conditions
3.2. Preparation of Biofilms and RNA Extraction
3.3. Sequencing and Transcriptomic Data Processing
3.4. Antimicrobial Susceptibility Measurements
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Davies, J.C.; Rubin, B.K. Emerging and unusual gram-negative infections in cystic fibrosis. Semin. Respir. Crit. Care Med. 2007, 28, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Ridderberg, W.; Bendstrup, K.E.; Olesen, H.V.; Jensen-Fangel, S.; Norskov-Lauritsen, N. Marked increase in incidence of Achromobacter xylosoxidans infections caused by sporadic acquisition from the environment. J. Cyst. Fibros. 2011, 10, 466–469. [Google Scholar] [CrossRef] [PubMed]
- Hansen, L.B.; Ren, D.; Burmolle, M.; Sorensen, S.J. Distinct gene expression profile of Xanthomonas retroflexus engaged in synergistic multispecies biofilm formation. ISME J. 2017, 11, 300–303. [Google Scholar] [CrossRef] [PubMed]
- Hogardt, M.; Heesemann, J. Microevolution of Pseudomonas aeruginosa to a chronic pathogen of the cystic fibrosis lung. Curr. Top. Microbiol. Immunol. 2013, 358, 91–118. [Google Scholar] [PubMed]
- Silva, I.N.; Santos, P.M.; Santos, M.R.; Zlosnik, J.E.; Speert, D.P.; Buskirk, S.W.; Bruger, E.L.; Waters, C.M.; Cooper, V.S.; Moreira, L.M. Long-Term Evolution of Burkholderia multivorans during a Chronic Cystic Fibrosis Infection Reveals Shifting Forces of Selection. mSystems 2016, 1, e00029-16. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.M.; Pereira, M.O. Pseudomonas aeruginosa Diversification during Infection Development in Cystic Fibrosis Lungs—A Review. Pathogens 2014, 3, 680–703. [Google Scholar] [CrossRef] [PubMed]
- Hansen, C.R.; Pressler, T.; Nielsen, K.G.; Jensen, P.O.; Bjarnsholt, T.; Hoiby, N. Inflammation in Achromobacter xylosoxidans infected cystic fibrosis patients. J. Cyst. Fibros. 2010, 9, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, T.H.; Hansen, M.A.; Jensen, P.O.; Hansen, L.; Riber, L.; Cockburn, A.; Kolpen, M.; Ronne Hansen, C.; Ridderberg, W.; Eickhardt, S.; et al. Complete genome sequence of the cystic fibrosis pathogen Achromobacter xylosoxidans NH44784–1996 complies with important pathogenic phenotypes. PLoS ONE 2013, 8, e68484. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.M.; Norskov-Lauritsen, N.; Bjarnsholt, T.; Meyer, R.L. Achromobacter Species Isolated from Cystic Fibrosis Patients Reveal Distinctly Different Biofilm Morphotypes. Microorganisms 2016, 4, 33. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed]
- Dotsch, A.; Eckweiler, D.; Schniederjans, M.; Zimmermann, A.; Jensen, V.; Scharfe, M.; Geffers, R.; Haussler, S. The Pseudomonas aeruginosa transcriptome in planktonic cultures and static biofilms using RNA sequencing. PLoS ONE 2012, 7, e31092. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ridderberg, W.; Hansen, C.R.; Hoiby, N.; Jensen-Fangel, S.; Olesen, H.V.; Skov, M.; Lemming, L.E.; Pressler, T.; Johansen, H.K.; et al. Early treatment with inhaled antibiotics postpones next occurrence of Achromobacter in cystic fibrosis. J. Cyst. Fibros. 2013, 12, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Hoiby, N.; Frederiksen, B.; Pressler, T. Eradication of early Pseudomonas aeruginosa infection. J. Cyst. Fibros. 2005, 4 (Suppl. 2), 49–54. [Google Scholar] [CrossRef] [PubMed]
- Chmiel, J.F.; Aksamit, T.R.; Chotirmall, S.H.; Dasenbrook, E.C.; Elborn, J.S.; LiPuma, J.J.; Ranganathan, S.C.; Waters, V.J.; Ratjen, F.A. Antibiotic management of lung infections in cystic fibrosis. I. The microbiome, methicillin-resistant Staphylococcus aureus, gram-negative bacteria, and multiple infections. Ann. Am. Thorac. Soc. 2014, 11, 1120–1129. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Sadikot, R.T.; Blackwell, T.S.; Christman, J.W.; Prince, A.S. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 2005, 171, 1209–1223. [Google Scholar] [CrossRef] [PubMed]
- Sommer, L.M.; Marvig, R.L.; Lujan, A.; Koza, A.; Pressler, T.; Molin, S.; Johansen, H.K. Is genotyping of single isolates sufficient for population structure analysis of Pseudomonas aeruginosa in cystic fibrosis airways? BMC Genom. 2016, 17, 589. [Google Scholar] [CrossRef] [PubMed]
- Lee, V.T.; Smith, R.S.; Tummler, B.; Lory, S. Activities of Pseudomonas aeruginosa effectors secreted by the Type III secretion system in vitro and during infection. Infect. Immun. 2005, 73, 1695–1705. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Ramirez, D.; Seshadri, R.; Cullina, J.F.; Powers, C.A.; Schulert, G.S.; Bar-Meir, M.; Sullivan, C.L.; McColley, S.A.; Hauser, A.R. Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. J. Clin. Microbiol. 2004, 42, 5229–5237. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.; Singh, P.K. Evolving stealth: Genetic adaptation of Pseudomonas aeruginosa during cystic fibrosis infections. Proc. Natl. Acad. Sci. USA 2006, 103, 8305–8306. [Google Scholar] [CrossRef] [PubMed]
- Haiko, J.; Westerlund-Wikstrom, B. The role of the bacterial flagellum in adhesion and virulence. Biology 2013, 2, 1242–1267. [Google Scholar] [CrossRef] [PubMed]
- Hogardt, M.; Heesemann, J. Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int. J. Med. Microbiol. 2010, 300, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Ridderberg, W.; Handberg, K.J.; Nørskov-Lauritsen, N. Prevalence of hypermutator isolates of Achromobacter sp. from cystic fibrosis patients. Unpublished, manuscript in preparation.
- Worlitzsch, D.; Tarran, R.; Ulrich, M.; Schwab, U.; Cekici, A.; Meyer, K.C.; Birrer, P.; Bellon, G.; Berger, J.; Weiss, T.; et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Investig. 2002, 109, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Soutourina, O.; Poupel, O.; Coppee, J.Y.; Danchin, A.; Msadek, T.; Martin-Verstraete, I. CymR, the master regulator of cysteine metabolism in Staphylococcus aureus, controls host sulphur source utilization and plays a role in biofilm formation. Mol. Microbiol. 2009, 73, 194–211. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.; Motta, S.; Mauri, P.; Landini, P. Sulfate assimilation pathway intermediate phosphoadenosine 59-phosphosulfate acts as a signal molecule affecting production of curli fibres in Escherichia coli. Microbiology 2014, 160, 1832–1844. [Google Scholar] [CrossRef] [PubMed]
- Imperi, F.; Tiburzi, F.; Fimia, G.M.; Visca, P. Transcriptional control of the pvdS iron starvation sigma factor gene by the master regulator of sulfur metabolism CysB in Pseudomonas aeruginosa. Environ. Microbiol. 2010, 12, 1630–1642. [Google Scholar] [PubMed]
- Lundberg, M.E.; Becker, E.C.; Choe, S. MstX and a putative potassium channel facilitate biofilm formation in Bacillus subtilis. PLoS ONE 2013, 8, e60993. [Google Scholar] [CrossRef] [PubMed]
- Prindle, A.; Liu, J.; Asally, M.; Ly, S.; Garcia-Ojalvo, J.; Suel, G.M. Ion channels enable electrical communication in bacterial communities. Nature 2015, 527, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Prindle, A.; Humphries, J.; Gabalda-Sagarra, M.; Asally, M.; Lee, D.Y.; Ly, S.; Garcia-Ojalvo, J.; Suel, G.M. Metabolic co-dependence gives rise to collective oscillations within biofilms. Nature 2015, 523, 550–554. [Google Scholar] [CrossRef] [PubMed]
- Humphries, J.; Xiong, L.; Liu, J.; Prindle, A.; Yuan, F.; Arjes, H.A.; Tsimring, L.; Suel, G.M. Species-Independent Attraction to Biofilms through Electrical Signaling. Cell 2017, 168, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Ridderberg, W.; Nielsen, S.M.; Norskov-Lauritsen, N. Genetic Adaptation of Achromobacter sp. during Persistence in the Lungs of Cystic Fibrosis Patients. PLoS ONE 2015, 10, e0136790. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y.; Poirel, L.; Paterson, D.L.; Nordmann, P. Characterization of a naturally occurring class D beta-lactamase from Achromobacter xylosoxidans. Antimicrob. Agents Chemother. 2008, 52, 1952–1956. [Google Scholar] [CrossRef] [PubMed]
- Bador, J.; Amoureux, L.; Duez, J.M.; Drabowicz, A.; Siebor, E.; Llanes, C.; Neuwirth, C. First description of an RND-type multidrug efflux pump in Achromobacter xylosoxidans, AxyABM. Antimicrob. Agents Chemother. 2011, 55, 4912–4914. [Google Scholar] [CrossRef] [PubMed]
- Bernier, S.P.; Lebeaux, D.; DeFrancesco, A.S.; Valomon, A.; Soubigou, G.; Coppee, J.Y.; Ghigo, J.M.; Beloin, C. Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet. 2013, 9, e1003144. [Google Scholar] [CrossRef] [PubMed]
- Waters, V.; Ratjen, F. Standard versus biofilm antimicrobial susceptibility testing to guide antibiotic therapy in cystic fibrosis. Cochrane Database Syst. Rev. 2015, 3, CD009528. [Google Scholar] [CrossRef]
- Baugh, S.; Ekanayaka, A.S.; Piddock, L.J.; Webber, M.A. Loss of or inhibition of all multidrug resistance efflux pumps of Salmonella enterica serovar Typhimurium results in impaired ability to form a biofilm. J. Antimicrob. Chemother. 2012, 67, 2409–2417. [Google Scholar] [CrossRef] [PubMed]
- Kvist, M.; Hancock, V.; Klemm, P. Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl. Environ. Microbiol. 2008, 74, 7376–7382. [Google Scholar] [CrossRef] [PubMed]
- Lamers, R.P.; Cavallari, J.F.; Burrows, L.L. The efflux inhibitor phenylalanine-arginine beta-naphthylamide (PAβN) permeabilizes the outer membrane of gram-negative bacteria. PLoS ONE 2013, 8, e60666. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.F.; Lin, Y.Y.; Lan, C.Y. Contribution of EmrAB efflux pumps to colistin resistance in Acinetobacter baumannii. J. Microbiol. 2017, 55, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Pamp, S.J.; Gjermansen, M.; Johansen, H.K.; Tolker-Nielsen, T. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 2008, 68, 223–240. [Google Scholar] [CrossRef] [PubMed]
- Bador, J.; Amoureux, L.; Blanc, E.; Neuwirth, C. Innate aminoglycoside resistance of Achromobacter. xylosoxidans is due to AxyXY-OprZ, an RND-type multidrug efflux pump. Antimicrob. Agents Chemother. 2013, 57, 603–605. [Google Scholar] [CrossRef] [PubMed]
- Spengler, G.; Kincses, A.; Gajdacs, M.; Amaral, L. New Roads Leading to Old Destinations: Efflux Pumps as Targets to Reverse Multidrug Resistance in Bacteria. Molecules 2017, 22, 448. [Google Scholar] [CrossRef] [PubMed]
- Bjarnsholt, T.; Alhede, M.; Alhede, M.; Eickhardt-Sorensen, S.R.; Moser, C.; Kuhl, M.; Jensen, P.O.; Hoiby, N. The in vivo biofilm. Trends Microbiol. 2013, 21, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Palmer, K.L.; Aye, L.M.; Whiteley, M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 2007, 189, 8079–8087. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 9 May 2016).
| Antibiotic (µg/mL) | CF2-a (Early) | CF2-b (Intermediate) | CF2-d (Late) | |||
|---|---|---|---|---|---|---|
| MIC | Categorisation * | MIC | Categorisation * | MIC | Categorisation * | |
| Amikacin | >32 | R | >32 | R | >32 | R |
| Aztreonam | >16 | R | >16 | R | >16 | R |
| Cefepime | 16 | R | 8 | S | >16 | R |
| Cefotaxime | 32 | NI | >32 | NI | >32 | NI |
| Ceftazidime | 4 | S | 4 | S | 8 | S |
| Ciprofloxacin | 2 | R | 2 | R | >2 | R |
| Colistin | 1 | S | 1 | S | > 8 | R |
| Doripenem | 0.25 | S | 0.25 | S | 2 | I |
| Doxycycline | 8 | NI | 8 | NI | 8 | NI |
| Ertapenem | ≤0.25 | NI | 1 | NI | >4 | NI |
| Gentamicin | >8 | R | >8 | R | >8 | R |
| Imipenem | ≤1 | S | 2 | S | ≤1 | S |
| Levofloxacin | 2 | R | 2 | R | > 8 | R |
| Meropenem | ≤1 | S | ≤1 | S | 4 | I |
| Minocycline | ≤2 | NI | ≤2 | NI | 4 | NI |
| Piperacillin | 8 | S | 8 | S | 8 | S |
| Polymyxin B | 1 | NI | 1 | NI | 4 | NI |
| Ticarcillin/Clavulanic Acid | ≤16 | S | ≤16 | S | ≤16 | S |
| Tigecycline | ≤0.25 | NI | 0.5 | NI | 0.5 | NI |
| Tobramycin | > 8 | R | > 8 | R | > 8 | R |
| Trimethoprim/ Sulfamethoxazole | ≤0.5 | NI | ≤0.5 | NI | ≤0.5 | NI |
| Gene Function | Downregulated Genes in the Late Isolate | Upregulated Genes in the Late Isolate | Fold Change * |
|---|---|---|---|
| Virulence | |||
| Arsenic resistance ArsH | 1 | 8.2 | |
| Zinc resistance | 2 | 14.1 to 20.5 | |
| Type III secretion system | 10 | −29.0 to −7.8 | |
| Motility | |||
| Flagellar motility | 3 | −11.3 to −5.4 | |
| Antimicrobial susceptibility | |||
| Beta-lactamase | 1 | 23.3 | |
| Multidrug resistance efflux pumps | 4 | 6.1 to 23.5 | |
| Cell Wall and Capsule | |||
| Capsular and extracellular polysaccharides | 2 | 2 | −9.4 to 5.2 |
| Lipopolysaccharide assembly YrbC | 1 | 8.8 | |
| Bacterial peptidoglycan hydrolases | 1 | 9.2 | |
| EPS biosynthesis EpsF | 1 | 5.9 | |
| Capsular polysaccharide ABC transporter KpsT | 1 | 6.6 | |
| Respiration | |||
| Anaerobic respiratory reductases | 2 | 6.1 to 9.0 | |
| Formate dehydrogenase | 2 | 5.3 to 6.7 | |
| Soluble cytochromes | 1 | 9.6 | |
| Fermentation | 1 | 7.3 | |
| Nitrogen Metabolism | 1 | 7.2 | |
| Dentrification | 1 | 46.0 | |
| Cytochrome O ubiquinol oxidase subunit I–IV | 4 | −20.7 to −7.3 | |
| Succinate dehydrogenase | 1 | −9.4 | |
| Stress response | |||
| Cold shock CspA/CspG | 2 | 5.5 to 10.7 | |
| Detoxification | 1 | 10.4 | |
| Osmotic stress | 3 | 5.1 to 8.6 | |
| Oxidative stress | 2 | 9.5 to 11.8 | |
| Heat shock | 1 | −5.4 | |
| DNA metabolism | |||
| DNA recombination RuvA/RuvC | 2 | −6.5 to −5.9 | |
| DNA repair RecO | 1 | −6.7 | |
| Sulfur metabolism | |||
| Inorganic sulfur assimilation | 4 | 10.1 | |
| Organic sulfur assimilation | 1 | 6 | −5.7 to 22.7 |
| Sulfur metabolism | 2 | 313.8; 1516.2 | |
| Protein metabolism | |||
| Protein degradation | 3 | 7.9 to 24.7 | |
| Protein biosynthesis | 1 | 6.3 | |
| Protein processing and modification MsrA and MrsB | 2 | 201.2 to 756.1 | |
| Potassium metabolism | |||
| Osmosensitive K+ channel histidine kinase KdpD II-V | 4 | 8.7 to 128.0 |
| Antibiotic (µg/mL) | CF2-a (Early) | CF2-b (Intermediate) | CF2-d (Late) | |||
|---|---|---|---|---|---|---|
| MIC | MBEC | MIC | MBEC | MIC | MBEC | |
| Colistin | 1 | 128 | 1 | 128 | >8 | 2048 |
| +PAβN (100 µg/mL) | 1 | ≤32 | 1 | 64 | >8 | 2048 |
| Levofloxacin | 2 | 128 | 2 | 128 | >8 | 256 |
| +PAβN (100 µg/mL) | 2 | 128 | 2 | 128 | 4 | 256 |
| Tobramycin | >8 | 512 | >8 | >1024 | >8 | >1024 |
| +PAβN (100 µg/mL) | >8 | 512 | >8 | >1024 | >8 | >1024 |
| Piperacillin | 8 | 2048 | 8 | 2048 | 8 | >2048 |
| +PAβN (100 µg/mL) | 8 | 2048 | 8 | 2048 | 8 | >2048 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nielsen, S.M.; Meyer, R.L.; Nørskov-Lauritsen, N. Differences in Gene Expression Profiles between Early and Late Isolates in Monospecies Achromobacter Biofilm. Pathogens 2017, 6, 20. https://doi.org/10.3390/pathogens6020020
Nielsen SM, Meyer RL, Nørskov-Lauritsen N. Differences in Gene Expression Profiles between Early and Late Isolates in Monospecies Achromobacter Biofilm. Pathogens. 2017; 6(2):20. https://doi.org/10.3390/pathogens6020020
Chicago/Turabian StyleNielsen, Signe M., Rikke L. Meyer, and Niels Nørskov-Lauritsen. 2017. "Differences in Gene Expression Profiles between Early and Late Isolates in Monospecies Achromobacter Biofilm" Pathogens 6, no. 2: 20. https://doi.org/10.3390/pathogens6020020






