Identification of Agents Active against Methicillin-Resistant Staphylococcus aureus USA300 from a Clinical Compound Library
Abstract
:1. Introduction
2. Results and Discussion
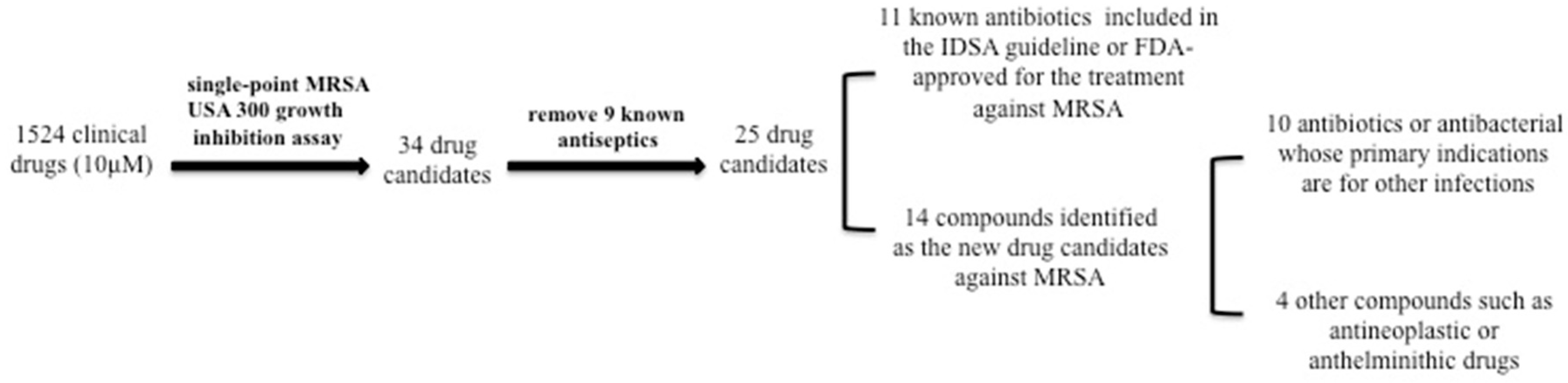
2.1. Identification of active hits from the clinical compound library that had good activity against MRSA strain USA300
2.2. Ranking the Activity of the Active Hits by MIC Testing
3. Materials and Methods
3.1. Bacterial Strain
3.2. The Clinical Compound Library
3.3. Log-phase Inhibition Screens
3.4. Minimum Inhibitory Concentration (MIC) Assay
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lowy, F.D. Staphylococcus aureus infections. N. Eng. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Ferri, M.; Ranucci, E.; Romagnoli, P.; Giaccone, V. Antimicrobial resistance: A global emerging threat to public health systems. Crit. Rev. Food Sci. Nutr. 2015, 57, 2857–2876. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance 2014. Avaiable online: http://www.who.int/drugresistance/documents/surveillancereport/en/ (accessed on 28 July 2017).
- Liu, C.; Bayer, A.; Cosgrove, S.E.; Daum, R.S.; Fridkin, S.K.; Gorwitz, R.J.; Kaplan, S.L.; Karchmer, A.W.; Levine, D.P.; Murray, B.E.; et al. Clinical practice guidelines by the infectious diseases society of america for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children. Clin. Infect. Dis. 2011, 52, e18–e55. [Google Scholar] [CrossRef] [PubMed]
- Anstead, G.M.; Cadena, J.; Javeri, H. Treatment of infections due to resistant Staphylococcus aureus. Methods Mol. Biol. 2014, 1085, 259–309. [Google Scholar] [PubMed]
- Rello, J.; Torres, A.; Ricart, M.; Valles, J.; Gonzalez, J.; Artigas, A.; Rodriguez-Roisin, R. Ventilator-associated pneumonia by Staphylococcus aureus. Comparison of methicillin-resistant and methicillin-sensitive episodes. Am. J. Respir. Crit. Care Med. 1994, 150, 1545–1549. [Google Scholar] [CrossRef] [PubMed]
- Sader, H.S.; Fey, P.D.; Limaye, A.P.; Madinger, N.; Pankey, G.; Rahal, J.; Rybak, M.J.; Snydman, D.R.; Steed, L.L.; Waites, K.; et al. Evaluation of vancomycin and daptomycin potency trends (mic creep) against methicillin-resistant Staphylococcus aureus isolates collected in nine U.S. Medical centers from 2002 to 2006. Antimicrob. Agents Chemother. 2009, 53, 4127–4132. [Google Scholar] [CrossRef] [PubMed]
- Rajamuthiah, R.; Fuchs, B.B.; Jayamani, E.; Kim, Y.; Larkins-Ford, J.; Conery, A.; Ausubel, F.M.; Mylonakis, E. Whole animal automated platform for drug discovery against multi-drug resistant Staphylococcus aureus. PLoS ONE 2014, 9, e89189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y. Persisters, persistent infections and the yin-yang model. Emerg. Microb. Infect. 2014, 3, e3. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Cui, P.; Yee, R.; Shi, W.; Zhang, S.; Feng, J.; Sullivan, D.; Zhang, W.; Zhu, B.; Zhang, Y. A clinical drug library screen identifies tosufloxacin as being highly active against Staphylococcus aureus persisters. Antibiotics 2015, 4, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Cheng, K.C.; Johnson, R.L.; Huang, R.; Pattaradilokrat, S.; Liu, A.; Guha, R.; Fidock, D.A.; Inglese, J.; Wellems, T.E.; et al. Chemical genomic profiling for antimalarial therapies, response signatures, and molecular targets. Science 2011, 333, 724–729. [Google Scholar] [CrossRef] [PubMed]
- Duthie, E.S.; Lorenz, L.L. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 1952, 6, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Barry, V.C.; Belton, J.G.; Conalty, M.L.; Denneny, J.M.; Edward, D.W.; O’Sullivan, J.F.; Twomey, D.; Winder, F. A new series of phenazines (rimino-compounds) with high antituberculosis activity. Nature 1957, 179, 1013–1015. [Google Scholar] [CrossRef] [PubMed]
- Gopal, M.; Padayatchi, N.; Metcalfe, J.Z.; O’Donnell, M.R. Systematic review of clofazimine for the treatment of drug-resistant tuberculosis. Int. J. Tuberc. Lung Dis. 2013, 17, 1001–1007. [Google Scholar] [CrossRef] [PubMed]
- Oliva, B.; O’Neill, A.J.; Miller, K.; Stubbings, W.; Chopra, I. Anti-staphylococcal activity and mode of action of clofazimine. J. Antimicrob. Chemother. 2004, 53, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Koval, A.V.; Vlasov, P.; Shichkova, P.; Khunderyakova, S.; Markov, Y.; Panchenko, J.; Volodina, A.; Kondrashov, F.A.; Katanaev, V.L. Anti-leprosy drug clofazimine inhibits growth of triple-negative breast cancer cells via inhibition of canonical wnt signaling. Biochem. Pharmacol. 2014, 87, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Mahon, R.N.; Hafner, R. Immune cell regulatory pathways unexplored as host-directed therapeutic targets for mycobacterium tuberculosis: An opportunity to apply precision medicine innovations to infectious diseases. Clin. Infect. Dis. 2015, 61 (Suppl. 3), S200–S216. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.D.; Hunter, M.; Cobb, M.; Traeger, G.; Spiegel, P.C. Thiostrepton inhibits stable 70s ribosome binding and ribosome-dependent gtpase activation of elongation factor g and elongation factor 4. Nucleic Acids Res. 2012, 40, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Macielag, M.J.; Demers, J.P.; Fraga-Spano, S.A.; Hlasta, D.J.; Johnson, S.G.; Kanojia, R.M.; Russell, R.K.; Sui, Z.; Weidner-Wells, M.A.; Werblood, H.; et al. Substituted salicylanilides as inhibitors of two-component regulatory systems in bacteria. J. Med. Chem. 1998, 41, 2939–2945. [Google Scholar] [CrossRef] [PubMed]
- Downey, A.S.; Chong, C.R.; Graczyk, T.K.; Sullivan, D.J. Efficacy of pyrvinium pamoate against cryptosporidium parvum infection in vitro and in a neonatal mouse model. Antimicrob. Agents Chemother. 2008, 52, 3106–3112. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Patel, T.R.; Sirianni, R.W.; Strohbehn, G.; Zheng, M.Q.; Duong, N.; Schafbauer, T.; Huttner, A.J.; Huang, Y.; Carson, R.E.; et al. Highly penetrative, drug-loaded nanocarriers improve treatment of glioblastoma. Proc. Natl. Acad. Sci. USA 2013, 110, 11751–11756. [Google Scholar] [CrossRef] [PubMed]
- Lau, Q.Y.; Tan, Y.Y.F.; Goh, V.C.Y.; Lee, D.J.Q.; Ng, F.M.; Ong, E.H.Q.; Hill, E.; Chia, C.S.B. An fda-drug library screen for compounds with bioactivities against meticillin-resistant Staphylococcus aureus (MRSA). Antibiotics 2015, 4, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Cui, P.; Shi, W.; Zhang, S.; Feng, J.; Wang, Y.; Sullivan, D.; Zhang, W.; Zhu, B.; Zhang, Y. Identification of anti-persister activity against uropathogenic escherichia coli from a clinical drug library. Antibiotics 2015, 4, 179–187. [Google Scholar]
- Chong, C.R.; Chen, X.C.; Shi, L.R.; Liu, J.O.; Sullivan, D.J. A clinical drug library screen identifies astemizole as an antimalarial agent. Nat. Chem. Biol. 2006, 2, 415–416. [Google Scholar] [CrossRef] [PubMed]
| Drugs | MIC (μM) | MIC (μg/mL) | Original Indication | Cmax * (μg/mL) |
|---|---|---|---|---|
| Quinaldine Blue | 1.2 | 0.57 | Antimalarial | - |
| Thiostrepton | 2.5 | 4.16 | Antibiotic | - |
| Nafcillin | 2.5 | 1.13 | Antibiotic | 14.34 |
| Carbomycin | 5.0 | 4.20 | Antibiotic | - |
| Moxalactam | 5.0 | 2.82 | Antibiotic | 49 |
| Cefotiam | 5.0 | 2.62 | Antibiotic | 20 |
| Closantel | 5.0 | 3.31 | Anthelminthic | - |
| Dithiazanine Iodide | 10.0 | 5.18 | Anthelminthic | - |
| Clofazimine | 10.0 | 4.73 | Antibacterial | 0.7–1.0 |
| Pyrvinium Pamoate | 10.0 | 11.51 | Anthelminthic | - |
| Cefmenoxime | 10.0 | 5.25 | Antibiotic | 10800 |
| Cefdinir | 10.0 | 3.95 | Antibiotic | 0.64–1.74 |
| Chloroxine | 10.0 | 2.14 | Antibacterial | - |
| Spiramycin | 10.0 | 8.43 | Antibiotic | 1000 |
| Drugs | MIC (μM) | MIC (μg/mL) | Original Indication |
|---|---|---|---|
| Brilliant green | 1.2 | 0.58 | Antiseptic |
| Gentian Violet | 1.2 | 0.49 | Antiseptic |
| Bithionol | 5.0 | 1.78 | Antiseptic |
| Trilocarban | 10.0 | 1.56 | Antiseptic |
| Benzododecinium | 10.0 | 12.27 | Antiseptic |
| Chlorquinaldol | 10.0 | 2.28 | Antiseptic |
| Methylbenzethonium | 10.0 | 4.62 | Antiseptic |
| Thonzonium | 10.0 | 5.9 | Antiseptic |
| Cetylpyridinium | 10.0 | 3.58 | Antiseptic |
| Drugs | MIC (μM) | MIC (μg/mL) | Original Indication |
|---|---|---|---|
| Minocycline | <0.3 | <0.14 | Antibiotic |
| Meclocycline | <0.3 | <0.20 | Antibacterial |
| Rifampin | <0.3 | <0.24 | Antibiotic |
| Clindamycin | 0.3 | 0.14 | Antibiotic |
| Vancomycin | 0.6 | 0.89 | Antibiotic |
| Formylrifamycin | 0.6 | 0.43 | Antibacterial |
| Doxycycline | 0.6 | 0.30 | Antibiotic |
| Chlortetracycline | 1.2 | 0.62 | Antibiotic |
| Tetracycline | 2.5 | 1.11 | Antibiotic |
| Daptomycin | 10.0 | 16 | Antibiotic |
| Linezolid | 10.0 | 3.47 | Antibiotic |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, H.; Yee, R.; Cui, P.; Tian, L.; Zhang, S.; Shi, W.; Sullivan, D.; Zhu, B.; Zhang, W.; Zhang, Y. Identification of Agents Active against Methicillin-Resistant Staphylococcus aureus USA300 from a Clinical Compound Library. Pathogens 2017, 6, 44. https://doi.org/10.3390/pathogens6030044
Niu H, Yee R, Cui P, Tian L, Zhang S, Shi W, Sullivan D, Zhu B, Zhang W, Zhang Y. Identification of Agents Active against Methicillin-Resistant Staphylococcus aureus USA300 from a Clinical Compound Library. Pathogens. 2017; 6(3):44. https://doi.org/10.3390/pathogens6030044
Chicago/Turabian StyleNiu, Hongxia, Rebecca Yee, Peng Cui, Lili Tian, Shuo Zhang, Wanliang Shi, David Sullivan, Bingdong Zhu, Wenhong Zhang, and Ying Zhang. 2017. "Identification of Agents Active against Methicillin-Resistant Staphylococcus aureus USA300 from a Clinical Compound Library" Pathogens 6, no. 3: 44. https://doi.org/10.3390/pathogens6030044





