Avian Group D Rotaviruses: Structure, Epidemiology, Diagnosis, and Perspectives on Future Research Challenges
Abstract
:1. Introduction
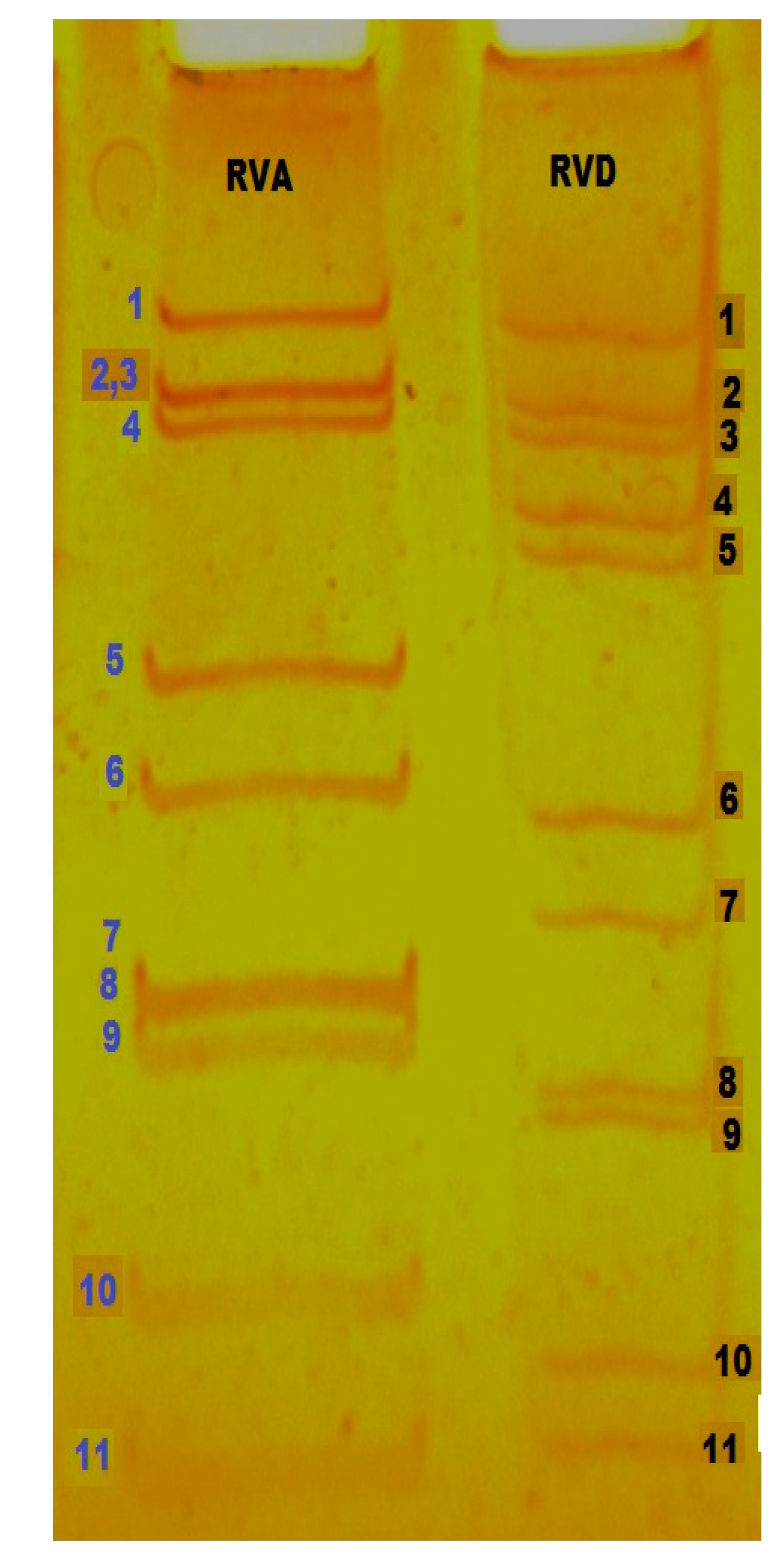
2. Virus Structure and Genome
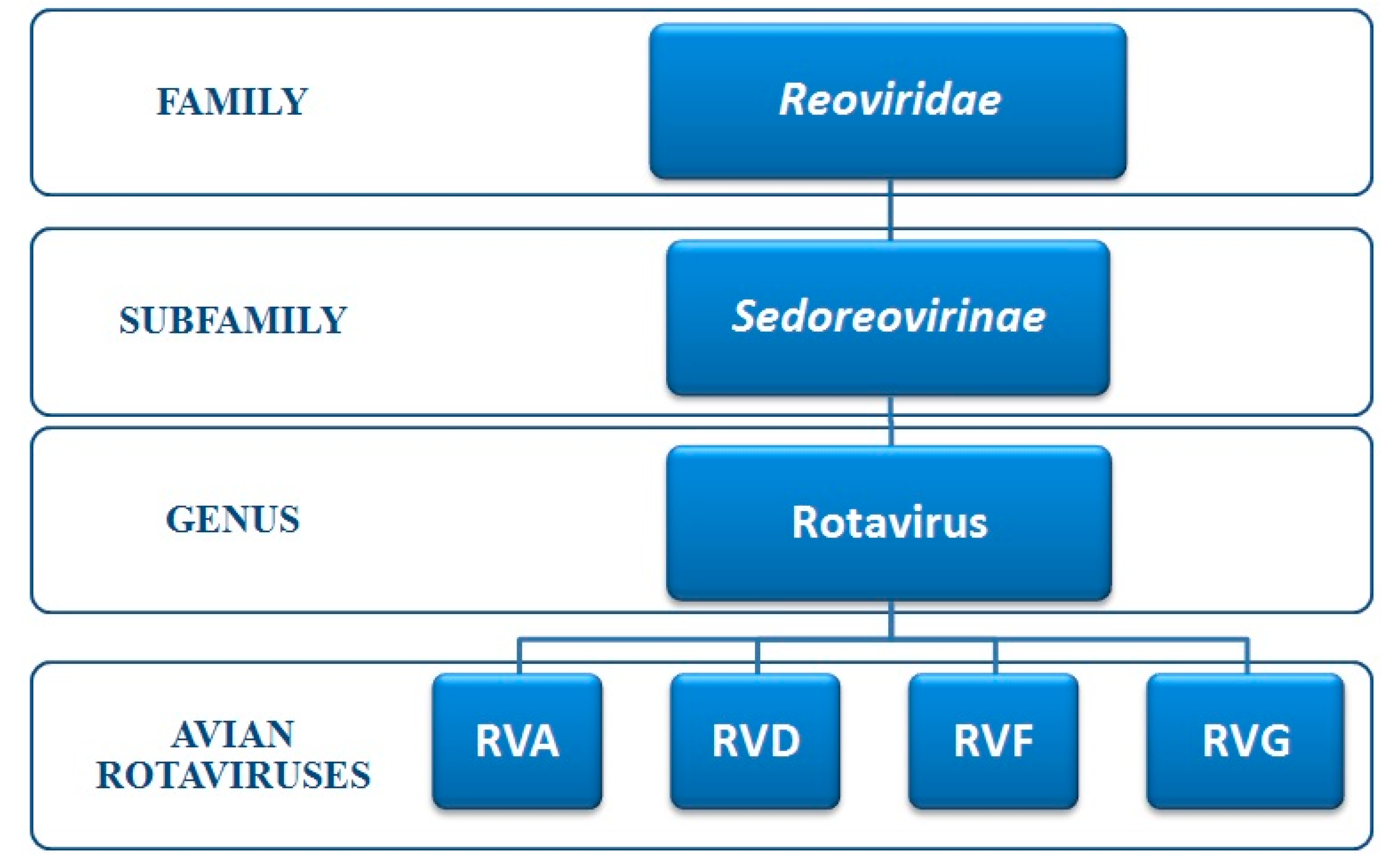
3. Classification
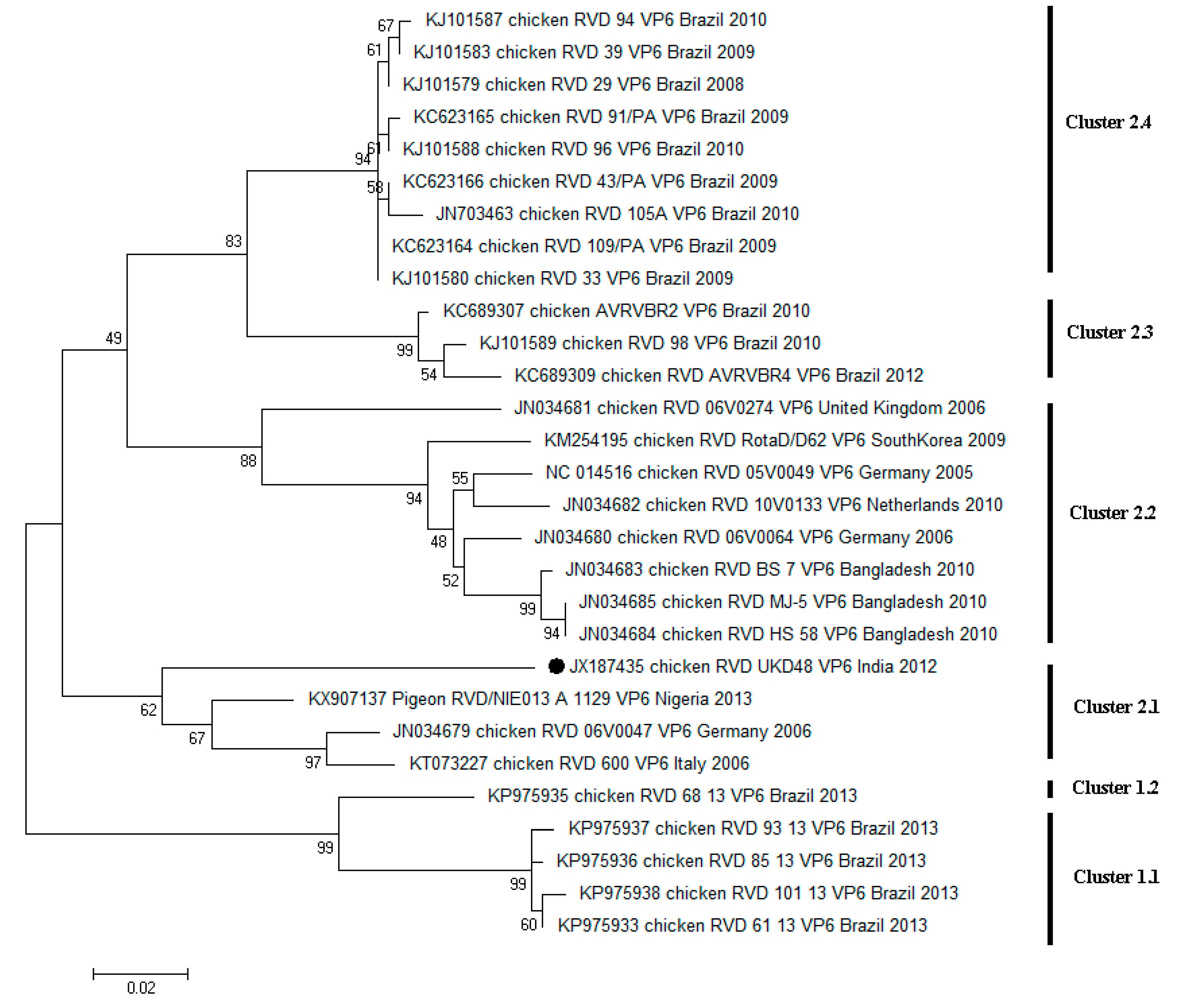
4. Diversity of RVD across the World
5. Identification and Epidemiology
6. Diagnosis
7. Differential Diagnosis
8. Prevention
9. Conclusions and Future Prospects
- ▪
- Studies on host–pathogen interactions, whether they are alike other enteric viruses or not.
- ▪
- As RVD is found in both symptomatic and asymptomatic birds, factors responsible for its virulence and pathogenicity are to be studied.
- ▪
- To date, very few sequences are available, and only for some of the genes of RVD strains. Once enough sequence data are available, a nucleotide sequence-based classification system can be established for RVD, as was achieved for RVAs.
- ▪
- Only one complete genome sequence is available so far, despite the widespread distribution of RVD in chickens.
- ▪
- The function of additional ORF (ORF-2) encoded by the 10th segment of RVD is still not defined.
- ▪
- The development of sensitive and specific diagnostic tests, including the improvement of available ones, is of prime importance.
- ▪
- The development of specific treatment by means of antivirals.
Supplementary Materials
Acknowledgment
Author Contributions
Funding
Conflicts of Interest
References
- Bishop, R.; Davidson, G.; Holmes, I.; Ruck, B. Virus particles in epithelial cells of duodenal mucosa from children with acute non-bacterial gastroenteritis. Lancet 1973, 302, 1281–1283. [Google Scholar] [CrossRef]
- Flewett, T.H.; Bryden, A.S.; Davies, H. Virus particles in gastroenteritis. Lancet 1973, 7844, 1497. [Google Scholar] [CrossRef]
- Adams, W.; Kraft, L. Epizootic diarrhea of infant mice: Identification of the etiologic agent. Science 1963, 141, 359–360. [Google Scholar] [CrossRef] [PubMed]
- Malherbe, H.; Harwin, R. The cytopathic effects of vervet monkey viruses. S. Afr. Med. J. 1963, 37, 407–411. [Google Scholar] [PubMed]
- Mebus, C.; Underdahl, N.; Rhodes, M.; Twiehaus, M. Further studies on neonatal calf diarrhea virus. In Proceedings of the Annual Meeting of the United States Animal Health Association, Louisville, KY, USA, 12–17 October 1969. [Google Scholar]
- Estes, M.; Greenberg, H. Rotaviruses. In Fields Virology, 6th ed.; David, M.K., Peter, M.H., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 1, pp. 1347–1401. [Google Scholar]
- Villarreal, L.; Uliana, G.; Valenzuela, C.; Chacón, J.L.V.; Saidenberg, A.B.S.; Sanches, A.A.; Brandão, P.E.; Jerez, J.A.; Ferreira, A.J.P. Rotavirus detection and isolation from chickens with or without symptoms. Rev. Bras. Ciênc. Avíc. 2006, 8, 187–191. [Google Scholar] [CrossRef]
- Dhama, K.; Saminathan, M.; Karthik, K.; Tiwari, R.; Shabbir, M.; Kumar, N.; Malik, Y.S.; Singh, R.K. Avian rotavirus enteritis—An updated review. Vet. Q. 2015, 35, 142–158. [Google Scholar] [CrossRef] [PubMed]
- Bergeland, M.; McAdaragh, J.; Stotz, I. Rotaviral enteritis in turkey poults. In Proceedings of the Western Poultry Disease Conference, Davis, CA, USA, 23–24 June 1977. [Google Scholar]
- McNulty, M.; Allan, G.; Todd, D.; McFerran, J. Isolation and cell culture propagation of rotaviruses from turkeys and chickens. Arch. Virol. 1979, 61, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Takase, K.; Nonaka, F.; Sakaguchi, M.; Yamada, S. Cytopathic avian rotavirus isolated from duck faeces in chicken kidney cell cultures. Avian Pathol. 1986, 15, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, D.; Theil, K.; Saif, Y.M. Demonstration of rotavirus and rotavirus-like virus in the intestinal contents of diarrheic pheasant chicks. Avian Dis. 1987, 31, 376–379. [Google Scholar] [CrossRef] [PubMed]
- Minamoto, N.; Oki, K.; Tomita, M.; Kinjo, T.; Suzuki, Y. Isolation and characterization of rotavirus from feral pigeon in mammalian cell cultures. Epidemiol. Infect. 1988, 100, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Takehara, K.; Kiuchi, H.; Kuwahara, M.; Yanagisawa, F.; Mizukami, M.; Matsuda, H.; Yoshimura, M. Identification and characterization of a plaque forming avian rotavirus isolated from a wild bird in Japan. J. Vet. Med. Sci. 1991, 53, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Pantin-Jackwood, M.J.; Spackman, E.; Michael-Day, J.; Rives, D. Periodic monitoring of commercial turkeys for enteric viruses indicates continuous presence of astrovirus and rotavirus on the farms. Avian Dis. 2007, 51, 674–680. [Google Scholar] [CrossRef]
- McNulty, M.; Allan, G.; Stuart, J. Rotavirus infection in avian species. Vet. Rec. 1978, 103, 319–320. [Google Scholar] [CrossRef] [PubMed]
- Theil, K.W.; Reynolds, D.L.; Saif, Y.M. Genomic variation among avian rotavirus-like viruses detected by polyacrylamide gel electrophoresis. Avian Dis. 1986, 30, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Hemida, M.G. Detection and molecular characterization of group D avian rota virus in Kafrelsheikh and Gharbia Governorates. J. Vet. Sci. 2013, 39, 145–154. [Google Scholar]
- Beserra, L.A.; Bernardes, N.T.; Brandão, P.E.; Gregori, F. Monitoring and molecular characterization of group Drotavirus in Brazilian poultry farms. Pesqui. Vet. Bras. 2015, 35, 536–540. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Otto, P.H.; Ciarlet, M.; Desselberger, U.; Van, R.M.; Johne, R. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch. Virol. 2012, 157, 1177–1182. [Google Scholar] [CrossRef] [PubMed]
- Mihalov-Kovács, E.; Gellért, Á.; Marton, S.; Szilvia, L.; Farkas, E.F.; Oldal, M.; Jakab, F.; Martella, V.; Bányai, K. Candidate new rotavirus species in sheltered dogs, Hungary. Emerg. Infect. Dis. 2015, 21, 660. [Google Scholar]
- Phan, T.G.; Leutenegger, C.M.; Chan, R.; Delwart, E. Rotavirus I in feces of a cat with diarrhea. Virus Genes 2017, 53, 487–490. [Google Scholar] [CrossRef] [PubMed]
- Bányai, K.; Kemenesi, G.; Budinski, I.; Földes, F.; Zana, B.; Marton, S.; Varga-Kugler, R.; Oldal, M.; Kurucz, K.; Jakab, F. Candidate new rotavirus species in Schreiber’s bats, Serbia. Infect. Genet. Evol. 2017, 48, 19–26. [Google Scholar]
- Otto, P.; Liebler-Tenorio, E.M.; Elschner, M.; Reetz, J.; Löhren, U.; Diller, R. Detection of rotaviruses and intestinal lesions in broiler chicks from flocks with runting and stunting syndrome (RSS). Avian Dis. 2006, 50, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Johne, R.; Otto, P.; Roth, B.; Löhren, U.; Belnap, D.; Reetz, J.; Trojnar, E. Sequence analysis of the VP6-encoding genome segment of avian group F and G rotaviruses. Virology 2011, 412, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Nuñez, L.F.N.; Parra, S.H.S.; Astolfi-Ferreira, C.S.; Claudia, C.; De La Torre, D.I.D.; Pedroso, A.C.; Ferreira, A.J.P. Detection of enteric viruses in pancreas and spleen of broilers with runting-stunting syndrome (RSS). Pesqui. Vet. 2016, 36, 595–599. [Google Scholar]
- Rosenberger, J.; Direction, C.S. Update on the Runting-Stunting Syndrome. Ceva Eggs Program Online. Available online: http://fs-1.5mpublishing.com/images/ceva/EPO_No3-May2012.pdf (accessed on 8 August 2011).
- Haynes, J.; Reynolds, D.; Fagerland, J.; Fix, A. Morphogenesis of enteric lesions induced by group D rotavirus in ringneck pheasant chicks (Phasianus colchicus). Vet. Pathol. 1994, 31, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Otto, P.H.; Ahmed, M.U.; Hotzel, H.; Machnowska, P.; Reetz, J.; Roth, B.; Trojnar, E.; Johne, R. Detection of avian rotaviruses of groups A, D, F and G in diseased chickens and turkeys from Europe and Bangladesh. Vet. Microbiol. 2012, 156, 8–15. [Google Scholar] [CrossRef] [PubMed]
- McNulty, M.; Allan, G.; Todd, D.; McFerran, J.; McCracken, R. Isolation from chickens of a rotavirus lacking the rotavirus group antigen. J. Gen. Virol. 1981, 55, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Pedley, S.; Bridger, J.; Chasey, D.; McCrae, M. Definition of two new groups of atypical rotaviruses. J. Gen. Virol. 1986, 67, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Pauly, M.; Oni, O.O.; Sausy, A.; Owoade, A.A.; Adeyefa, C.A.O.; Muller, C.P.; Hübschen, J.M.; Snoeck, C.J. Molecular epidemiology of avian rotaviruses group A and D shed by different bird species in Nigeria. Virol. J. 2017, 14, 111. [Google Scholar]
- Flewett, T.; Bryden, A.; Davies, H. Virus diarrhoea in foals and other animals. Vet. Rec. 1975, 96, 477. [Google Scholar]
- Estes, M.K.; Graham, D.Y.; Mason, B.B. Proteolytic enhancement of rotavirus infectivity: Molecular mechanisms. J. Virol. 1981, 3, 879–888. [Google Scholar]
- Estes, M.K.; Cohen, J. Rotavirus gene structure and function. Microbiol. Rev. 1989, 53, 410–449. [Google Scholar] [PubMed]
- Guy, J.S. Virus infections of the gastrointestinal tract of poultry. Poult. Sci. 1998, 77, 1166–1175. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Borgan, M.A.; Ito, N.; Sugiyama, M.; Minamoto, N. Diarrhea-inducing activity of avian rotavirus NSP4 glycoproteins, which differ greatly from mammalian rotavirus NSP4 glycoproteins in deduced amino acid sequence, in suckling mice. J. Virol. 2002, 76, 5829–5834. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Crawford, S.E.; Czako, R.; Cortes-Penfield, N.W.; Smith, D.F.; Le Pendu, J.; Estes, M.K.; Prasad, B.V. Cell attachment protein VP8 * of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature 2012, 485, 256. [Google Scholar] [CrossRef] [PubMed]
- Trojnar, E.; Otto, P.; Roth, B.; Reetz, J.; Johne, R. The genome segments of a group D rotavirus possess group A-like conserved termini but encode group-specific proteins. J. Virol. 2010, 84, 10254–10265. [Google Scholar] [CrossRef] [PubMed]
- Matthijnssens, J.; Ciarlet, M.; Heiman, E.; Arijs, I.; Delbeke, T.; McDonald, S.M.; Palombo, E.A.; Iturriza-Gómara, M.; Maes, P.; Patton, J.T.; et al. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J. Virol. 2008, 7, 3204–3219. [Google Scholar] [CrossRef] [PubMed]
- Malik, Y.S.; Sharma, K.; Vaid, N.; Chandrashekar, K.M.; Basera, S.S.; Singh, R.; Prasad, G.; Gulati, B.R.; Bhilegaonkar, K.N.; Pandey, A.B. Frequency of group A rotavirus with mixed G and P genotypes in bovines: Predominance of G3 genotype and its emergence in combination with G8/G10 types. J. Vet. Sci. 2012, 13, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Marthaler, D.; Suzuki, T.; Rossow, K.; Culhane, M.; Collins, J.; Goyal, S.; Tsunemitsu, H.; Ciarlet, M.; Matthijnssens, J. VP6 genetic diversity, reassortment, intragenic recombination and classification of rotavirus B in American and Japanese pigs. Vet. Microbiol. 2014, 3–4, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Marthaler, D.; Rossow, K.; Gramer, M.; Collins, J.; Goyal, S.; Tsunemitsu, H.; Kuga, K.; Suzuki, T.; Ciarlet, M.; Matthijnssens, J. Detection of substantial porcine group B rotavirus genetic diversity in the United States, resulting in a modified classification proposal for G genotypes. Virology 2012, 1, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Moutelíková, R.; Prodělalová, J.; Dufková, L. Diversity of VP7, VP4, VP6, NSP2, NSP4 and NSP5 genes of porcine rotavirus C: Phylogenetic analysis and description of potential new VP7, VP4, VP6, and NSP4 genotypes. Arch. Virol. 2015, 7, 1715–1727. [Google Scholar] [CrossRef] [PubMed]
- Niira, K.; Ito, M.; Masuda, T.; Saitou, T.; Abe, T.; Komoto, S.; Sato, M.; Yamasato, H.; Kishimoto, M.; Naoi, Y.; et al. Whole genome sequences of Japanese porcine species C rotaviruses reveal a high diversity of genotypes of individual genes and will contribute to a comprehensive, generally accepted classification system. Infect. Genet. Evol. 2016, 44, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Kusumakar, A.; Malik, Y.; Prasad, G. Detection and characterization of group A and D avian rotaviruses in India. Indian J. Biotechnol. 2008, 7, 554–556. [Google Scholar]
- Beserra, L.; Gregori, F. Description of rotavirus F in broilers from Brazilian poultry farms. Avian Dis. 2014, 58, 458–461. [Google Scholar] [CrossRef] [PubMed]
- Kindler, E.; Trojnar, E.; Heckel, G.; Otto, P.H.; Johne, R. Analysis of rotavirus species diversity and evolution including the newly determined full-length genome sequences of rotavirus F and G. Infect. Genet. Evol. 2013, 14, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Kattoor, J.J.; Malik, Y.S.; Sasidharan, A.; Rajan, V.M.; Dhama, K.; Ghosh, S.; Bányai, K.; Kobayashi, N.; Singh, R.K. Analysis of codon usage pattern evolution in avian rotaviruses and their preferred host. Infect. Genet. Evol. 2015, 34, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Kattoor, J.J.; Malik, Y.S.; Sharma, K.; Kumar, N.; Batra, M.; Jindal, N.; Yadav, A.S. Molecular evidence of group D rotavirus in commercial broiler chicks in India. Avian Biol. Res. 2013, 6, 313–316. [Google Scholar] [CrossRef]
- Bezerra, D.A.M.; da Silva, R.R.; Kaiano, J.H.L.; Oliveira, D.D.S.; Gabbay, Y.B.; Linhares, A.C.; Mascarenhas, J.D.P. Detection, epidemiology and characterization of VP6 and VP7 genes of group D rotavirus in broiler chickens. Avian Pathol. 2014, 43, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Ahmed, M. Detection of avian rotavirus-like virus in broiler chickens in Bangladesh. Bangladesh J. Vet. Med. 2006, 4, 73–77. [Google Scholar] [CrossRef]
- Saif, Y.; Saif, L.; Hofacre, C.; Hayhow, C.; Swayne, D.; Dearth, R. A small round virus associated with enteritis in turkey poults. Avian Dis. 1990, 34, 762–764. [Google Scholar] [CrossRef] [PubMed]
- Roth, B. Avian Group D Rotavirus. U.S. Patent 9243299 B2, 26 January 2016. [Google Scholar]
- Crawford, S.E.; Patel, D.G.; Cheng, E.; Berkova, Z.; Hyser, J.M.; Ciarlet, M.; Finegold, M.J.; Conner, M.E.; Estes, M.K. Rotavirus viremia and extraintestinal viral infection in the neonatal rat model. J. Virol. 2006, 80, 4820–4832. [Google Scholar] [CrossRef] [PubMed]
- Niture, G.; Karpe, A.; Prasad, M.; Zade, N. Electrophoretic pattern analysis of rotaviruses of buffalo, poultry and human. J. Vet. Public Health 2010, 8, 7–10. [Google Scholar]
- Kattoor, J.J.; Malik, Y.S.; Kumar, N.; Sharma, K.; Sircar, S. Development of VP6 gene specific reverse transcription (RT)-PCR assay for detection of avian group D rotavirus in diarrheic chickens. J. Vet. Sci. Med. Diagn. 2014, 3, 2. [Google Scholar]
- Kattoor, J.J.; Malik, Y.S.; Sharma, K.; Sircar, S.; Batra, M.; Jindal, N.; Dhama, K. Frequency distribution of avian group-D rotavirus in Southern and Northern parts of India. J. Pur. Appl. Microbiol. 2015, 9, 449–452. [Google Scholar]
- Collett, S. Principles of disease prevention, diagnosis, and control introduction. In Diseases of Poultry, 13th ed.; Wiley-Blackwell: Oxford, UK, 2013; pp. 4–60. ISBN 978-0-470-95899-5. [Google Scholar]
- Nuñez, L.F.N.; Parra, S.H.S.; Mettifogo, E.; Catroxo, M.H.B.; Ferreira, C.S.; Ferreira, A.J. Isolation of chicken astrovirus from specific pathogen-free chicken embryonated eggs. Poult. Sci. 2015, 94, 947–954. [Google Scholar] [CrossRef] [PubMed]
- Devitt, C.; Reynolds, D. Characterization of a group D rotavirus. Avian Dis. 1993, 37, 749–755. [Google Scholar] [CrossRef] [PubMed]
- Espejo, R.T.; Calderón, E.; González, N.; Salomon, A.; Martuscelli, A.; Romero, P. Presence of two distinct types of rotavirus in infants and young children hospitalized with acute gastroenteritis in Mexico City, 1977. J. Infect. Dis. 1979, 139, 474–477. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, D.A.M.; da Silva, R.R.; Kaiano, J.H.L.; Silvestre, R.V.D.; de Souza Oliveira, D.; Linhares, A.C.; Gabbay, Y.B.; Mascarenhas, J.D.A.P. Detection of avian group D rotavirus using the polymerase chain reaction for the VP6 gene. J. Virol. Methods 2012, 2, 189–192. [Google Scholar] [CrossRef] [PubMed]
- Middleton, P.; Petric, M.; Hewitt, C.; Szymanski, M.; Tam, J. Counter-immunoelectro-osmophoresis for the detection of infantile gastroenteritis virus (orbi-group) antigen and antibody. J. Clin. Pathol. 1976, 29, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Middleton, P.; Holdaway, M.; Petric, M.; Szymanski, M.; Tam, J. Solid-phase radioimmunoassay for the detection of rotavirus. Infect. Immun. 1977, 16, 439–444. [Google Scholar] [PubMed]
- Sanekata, T.; Yoshida, Y.; Okada, H. Detection of rotavirus in faeces by latex agglutination. J. Immunol. Methods 1981, 41, 377–385. [Google Scholar] [CrossRef]
- Yolken, R.; Kim, H.; Clem, T.; Wyatt, R.; Kalica, A.; Chanock, R.; Kapikian, A. Enzyme-linked immunosorbent assay (ELISA) for detection of human reovirus-like agent of infantile gastroenteritis. Lancet 1977, 310, 263–267. [Google Scholar] [CrossRef]
- Hafez, H.M. Enteric diseases of poultry with special attention to Clostridium perfringens. Pak. Vet. J. 2011, 31, 175–184. [Google Scholar]
- Koo, B.S.; Lee, H.R.; Jeon, E.O.; Jang, H.S.; Han, M.S.; Mo, I.P. An unusual case of concomitant infection with chicken astrovirus and group A avian rotavirus in broilers with a history of severe clinical signs. J. Vet. Sci. 2013, 14, 231–233. [Google Scholar] [CrossRef] [PubMed]
- Jindal, N.; Patnayak, D.; Chander, Y.; Ziegler, A.; Goyal, S. Detection and molecular characterization of enteric viruses from poult enteritis syndrome in turkeys. Poult. Sci. 2010, 89, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Jindal, N.; Patnayak, D.P.; Chander, Y.; Ziegler, A.F.; Goyal, S.M. Detection and molecular characterization of enteric viruses in breeder turkeys. Avian Pathol. 2010, 39, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Nagaraja, K.; Newman, J. Physical, chemical, and serological characterization of avian rotaviruses. Avian Dis. 1988, 32, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Patnayak, D.P.; Prasad, M.; Malik, Y.S.; Ramakrishnan, M.A.; Goyal, S.M. Efficacy of disinfectants and hand sanitizers against avian respiratory viruses. Avian Dis. 2008, 52, 199–202. [Google Scholar] [CrossRef] [PubMed]

| Gene Segment | RNA Segment Number Coding for the Gene | Size of Coding Sequence (in bp #) | No. of Complete Sequences * (Accession No.) | No. of Partial Sequences | Total Number of Nucleotide Sequences |
|---|---|---|---|---|---|
| VP1 | 1 | 3237 | 1 (NC_014511) | 4 | 5 |
| VP2 | 2 | 2739 | 1 (NC_014512) | 5 | 6 |
| VP3 | 4 | 2055 | 2 (NC_014514, KF142491) | 6 | 8 |
| VP4 | 3 | 2331 | 1 (NC_014513) | 6 | 7 |
| VP6 | 6 | 1194 | 3 (NC_014516, KX374470, JX187435) | 36 | 39 |
| VP7 | 9 | 948 | 3 (KM254196, NC_014519, KF142489) | 21 | 24 |
| NSP1 | 5 | 1722 | 1 (NC_014515) | 5 | 6 |
| NSP2 | 8 | 930 | 1 (NC_014518) | 3 | 4 |
| NSP3 | 7 | 1110 | 1(NC_014517) | 3 | 4 |
| NSP4 | 10 | ORF1: 381 ORF2: 279 | 4 (NC_014520,KF142490, KX374472, KX374471 ) | 3 | 7 |
| NSP5 | 11 | 585 | 1 (NC_014521) | 1 | 2 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deol, P.; Kattoor, J.J.; Sircar, S.; Ghosh, S.; Bányai, K.; Dhama, K.; Malik, Y.S. Avian Group D Rotaviruses: Structure, Epidemiology, Diagnosis, and Perspectives on Future Research Challenges. Pathogens 2017, 6, 53. https://doi.org/10.3390/pathogens6040053
Deol P, Kattoor JJ, Sircar S, Ghosh S, Bányai K, Dhama K, Malik YS. Avian Group D Rotaviruses: Structure, Epidemiology, Diagnosis, and Perspectives on Future Research Challenges. Pathogens. 2017; 6(4):53. https://doi.org/10.3390/pathogens6040053
Chicago/Turabian StyleDeol, Pallavi, Jobin Jose Kattoor, Shubhankar Sircar, Souvik Ghosh, Krisztián Bányai, Kuldeep Dhama, and Yashpal Singh Malik. 2017. "Avian Group D Rotaviruses: Structure, Epidemiology, Diagnosis, and Perspectives on Future Research Challenges" Pathogens 6, no. 4: 53. https://doi.org/10.3390/pathogens6040053










