New Insights into the Genome Organization of Yeast Double-Stranded RNA LBC Viruses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Culture Media
2.2. Purification of V-LBC dsRNA from Killer Yeasts
2.3. cDNA Library Preparation and DNA Sequencing
2.4. Assembly of Virus Genome Sequences
2.5. Sequence Analysis Tools
2.6. Nucleotide Sequences
3. Results
3.1. Comparison of Nucleotide Canonical Sequences from TdV-LBC and ScV-LBC Genomes
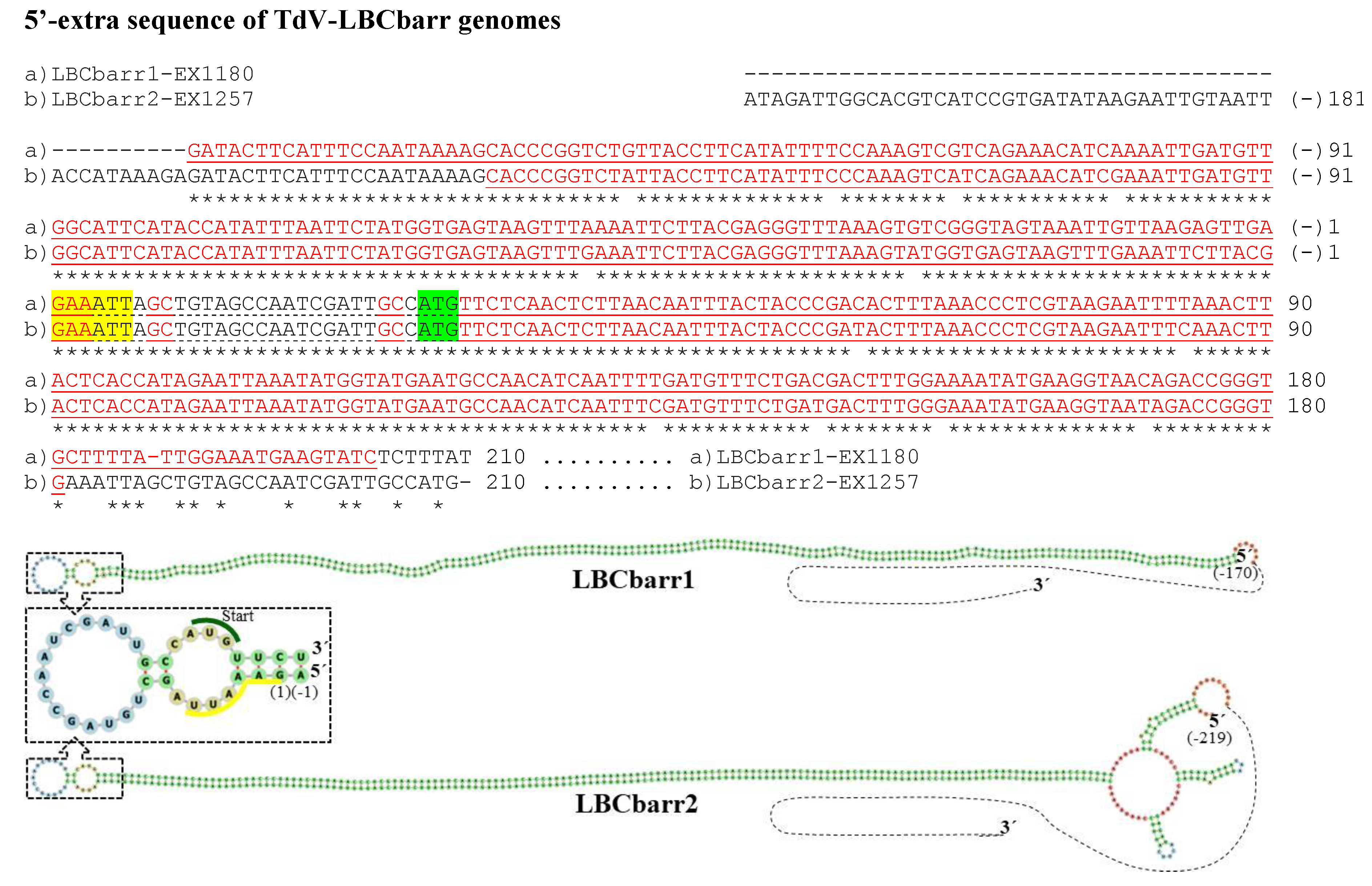
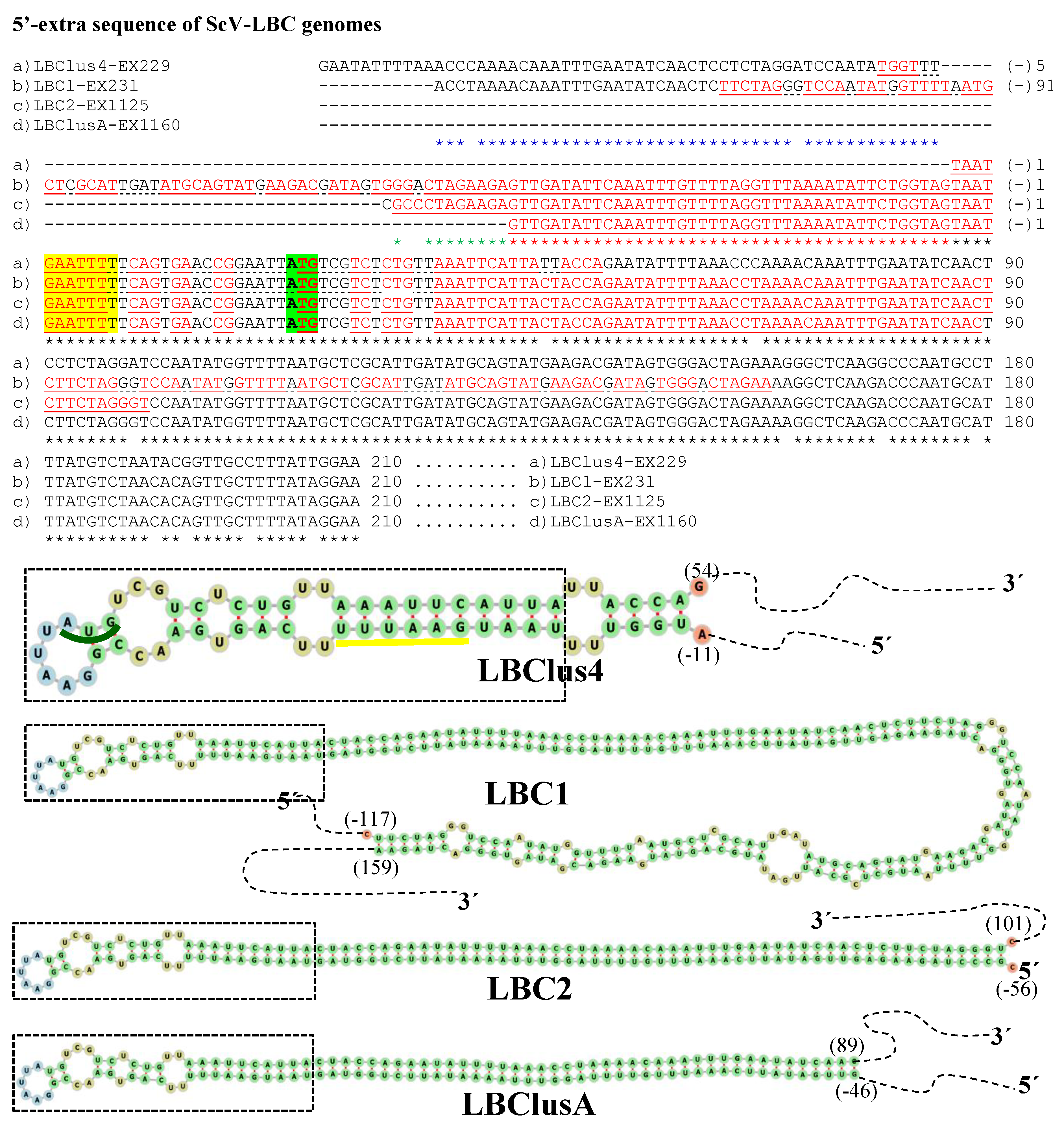
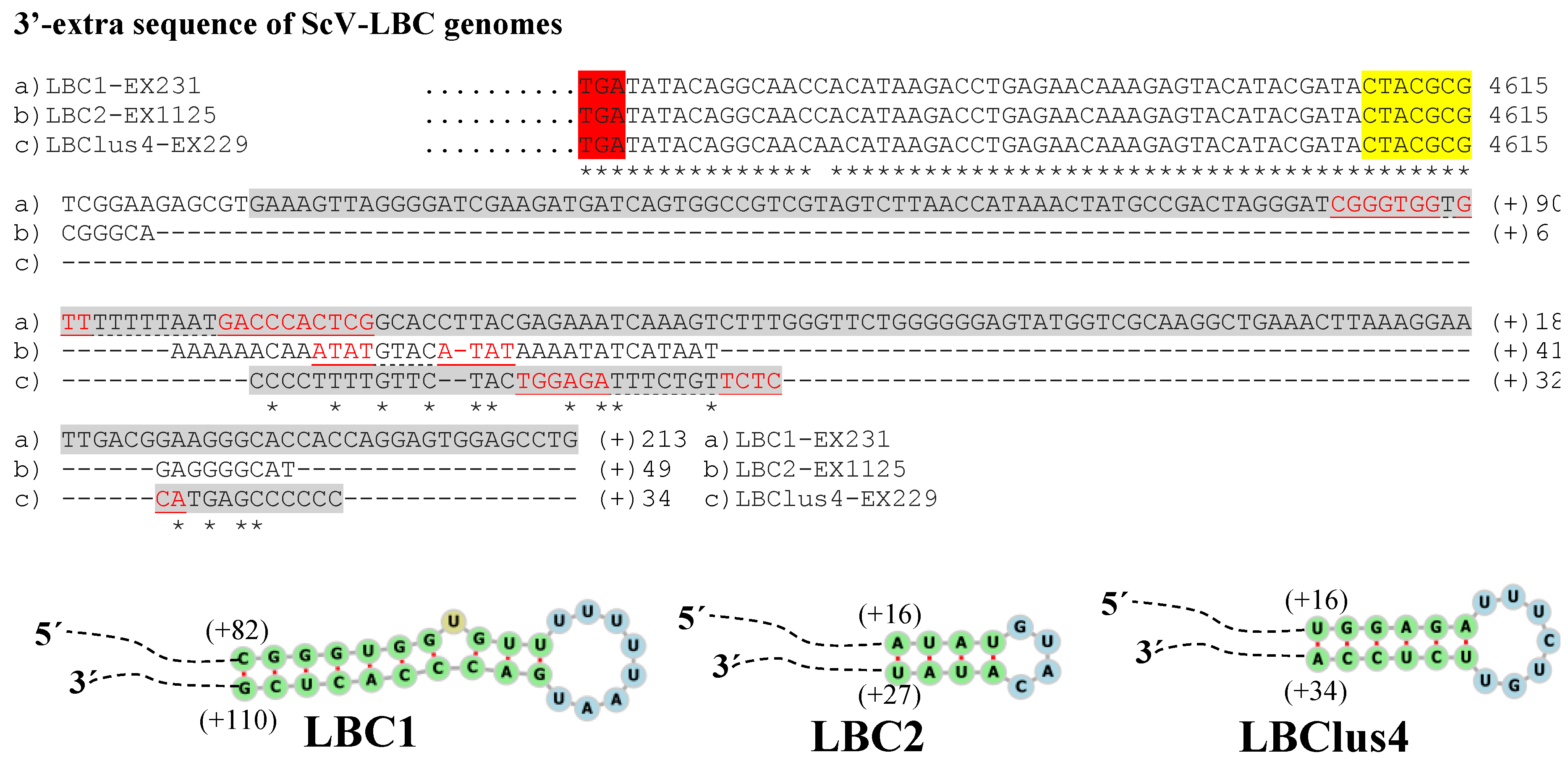
3.2. Analysis of 5′- and 3′-Extra Sequences of TdV-LBCbarr and ScV-LBC Genomes
4. Discussion
4.1. Canonical Nucleotide and Amino Acid Sequences of Td-LBC and Sc-LBC Viruses
4.2. Features Found in the 5′- and 3′-Extra Sequences from LBC Genomes
5. Conclusions
Supplementary Materials
 , ribosomal frameshift. Asterisks (*) indicates identical nucleotide positions. Sequence stretches of V-LBC genomes sharing high local identity with some mitochondrial or genomic sequences of several organisms are brown shaded, the sequences of other organisms are shown above each homologous stretch of LBC genome, and each percentage of identity is in parenthesis.
, ribosomal frameshift. Asterisks (*) indicates identical nucleotide positions. Sequence stretches of V-LBC genomes sharing high local identity with some mitochondrial or genomic sequences of several organisms are brown shaded, the sequences of other organisms are shown above each homologous stretch of LBC genome, and each percentage of identity is in parenthesis.Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aitmanaitė, L.; Konovalovas, A.; Medvedevas, P.; Servienė, E.; Serva, S. Specificity determination in Saccharomyces cerevisiae killer virus systems. Microorganisms 2021, 9, 236. [Google Scholar] [CrossRef]
- Schmitt, M.J.; Breinig, F. Yeast viral killer toxins: Lethality and self-protection. Nat. Rev. Microbiol. 2006, 4, 212–221. [Google Scholar] [CrossRef]
- Wickner, R.B.; Fujimura, T.; Esteban, R. Viruses and Prions of Saccharomyces cerevisiae. Adv. Appl. Microbiol. 2013, 86, 1–36. [Google Scholar] [CrossRef] [Green Version]
- Ramírez, M.; Velázquez, R.; López-Piñeiro, A.; Naranjo, B.; Roig, F.; Llorens, C. New Insights into the Genome Organization of Yeast Killer Viruses Based on “Atypical” Killer Strains Characterized by High-Throughput Sequencing. Toxins 2017, 9, 292. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Cousiño, N.; Esteban, R. Relationships and Evolution of Double-Stranded RNA Totiviruses of Yeasts Inferred from Analysis of L-A-2 and L-BC Variants in Wine Yeast Strain Populations. Appl. Environ. Microbiol. 2017, 83, e02991-16. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Cousiño, N.; Maqueda, M.; Ambrona, J.; Zamora, E.; Esteban, E.; Ramírez, M. A new wine Saccharomyces cerevisiae double-stranded RNA virus encoded killer toxin (Klus) with broad antifungal activity is evolutionarily related to a chromosomal host gene. Appl. Environ. Microbiol. 2011, 77, 1822–1832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vepštaitė-Monstavičė, I.; Lukša, J.; Konovalovas, A.; Ežerskytė, D.; Stanevičienė, R.; Strazdaitė-Žielienė, Ž.; Serva, S.; Servienė, E. Saccharomyces paradoxus K66 Killer System Evidences Expanded Assortment of Helper and Satellite Viruses. Viruses 2018, 10, 564. [Google Scholar] [CrossRef] [Green Version]
- Park, C.-M.; Lopinski, J.D.; Masuda, J.; Tzeng, T.-H.; Bruenn, J. A Second Double-Stranded RNA Virus from Yeast. Virology 1996, 216, 451–454. [Google Scholar] [CrossRef]
- Ramírez, M.; Velázquez, R.; López-Piñeiro, A.; Martínez, A. Genome Features of a New Double-Stranded RNA Helper Virus (LBCbarr) from Wine Torulaspora delbrueckii Killer Strains. Int. J. Mol. Sci. 2021, 22, 13492. [Google Scholar] [CrossRef]
- Wickner, R.B. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol. Rev. 1996, 60, 250–265. [Google Scholar] [CrossRef]
- Dinman, J.D.; Wickner, R.B. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 1992, 66, 3669–3676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujimura, T.; Ribas, J.; Makhov, A.M.; Wickner, R.B. Pol of gag–pol fusion protein required for encapsidation of viral RNA of yeast L-A virus. Nature 1992, 359, 746–749. [Google Scholar] [CrossRef] [PubMed]
- Icho, T.; Wickner, R.B. The Double-stranded RNA Genome of Yeast Virus L-A Encodes Its Own Putative RNA Polymerase by Fusing Two Open Reading Frames. J. Biol. Chem. 1989, 264, 6716–6723. [Google Scholar] [CrossRef]
- Ramírez, M.; Velázquez, R.; Maqueda, M.; Martínez, A. Genome Organization of a New Double-Stranded RNA LA Helper Virus from Wine Torulaspora delbrueckii Killer Yeast as Compared with Its Saccharomyces Counterparts. Front. Microbiol. 2020, 11, 2977. [Google Scholar] [CrossRef]
- Rodríguez-Cousiño, N.; Gómez, P.; Esteban, R. L-A-lus, a New Variant of the L-A Totivirus Found in Wine Yeasts with Klus Killer Toxin-Encoding Mlus Double-Stranded RNA: Possible Role of Killer Toxin-Encoding Satellite RNAs in the Evolution of Their Helper Viruses. Appl. Environ. Microbiol. 2013, 79, 4661–4674. [Google Scholar] [CrossRef] [Green Version]
- Fujimura, T.; Esteban, R.; Esteban, L.M.; Wickner, R.B. Portable encapsidation signal of the L-A double-stranded-RNA virus of Saccharomyces cerevisiae. Cell 1990, 62, 819–828. [Google Scholar] [CrossRef]
- Ramírez, M.; Velázquez, R.; Maqueda, M.; López-Piñeiro, A.; Ribas, J.C. A new wine Torulaspora delbrueckii killer strain with broad antifungal activity and its toxin-encoding double-stranded RNA virus. Front. Microbiol. 2015, 6, 983. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Cousiño, N.; Gómez, P.; Esteban, R. Variation and Distribution of L-A Helper Totiviruses in Saccharomyces sensu stricto Yeasts Producing Different Killer Toxins. Toxins 2017, 9, 313. [Google Scholar] [CrossRef]
- Thiele, D.J.; Leibowitz, M.J. Structural and functional analysis of separated strands of killer double-stranded RNA of yeast. Nucleic Acids Res. 1982, 10, 6903–6918. [Google Scholar] [CrossRef] [Green Version]
- Thiele, D.J.; Wang, R.W.; Leibowitz, M.J. Separation and sequence of the 3′ termini of M double-stranded RNA from killer yeast. Nucleic Acids Res. 1982, 10, 1661–1678. [Google Scholar] [CrossRef] [Green Version]
- Guthrie, C.; Fink, G.R. Guide to Yeast Genetics and Molecular Biology. Methods Enzymol. 1991, 194, 3–57. [Google Scholar]
- Maqueda, M.; Zamora, E.; Rodríguez-Cousiño, N.; Ramírez, M. Wine yeast molecular typing using a simplified method for simultaneously extracting mtDNA, nuclear DNA and virus dsRNA. Food Microbiol. 2010, 27, 205–209. [Google Scholar] [CrossRef]
- Toh-E, A.; Guerry, P.; Wickner, R.B. Chromosomal superkiller mutants of Saccharomyces cerevisiae. J. Bacteriol. 1978, 136, 1002–1007. [Google Scholar] [CrossRef] [Green Version]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience 2012, 1. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Futami, R.; Muñoz-Pomer, L.; Dominguez-Escriba, L.; Covelli, L.; Bernet, G.P.; Sempere, J.M. GPRO The professional tool for annotation, management and functional analysis of omic databases. Biotechvana Bioinform. 2011, 2011, SOFT3. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- Zuker, M.; Mathews, D.H.; Turner, D.H.; Barciszewski, J.; Clark, B.F.C. Algorithms and thermodynamics for RNA secondary structure prediction: A practical guide. In RNA Biochemistry and Biotechnology; Springer: Dordrecht, The Netherlands, 1999. [Google Scholar]
- Konovalovas, A.; Serviené, E.; Serva, S. Genome Sequence of Saccharomyces cerevisiae Double-Stranded RNA Virus L-A-28. Genome Announc. 2016, 4, e00549-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribas, J.C.; Fujimura, T.; Wickner, R.B. Essential RNA binding and packaging domains of the Gag-Pol fusion protein of the L-A double-stranded RNA virus of Saccharomyces cerevisiae. J. Biol. Chem. 1994, 269, 28420–28428. [Google Scholar] [CrossRef]
- Park, D.; Hahn, Y. Rapid protein sequence evolution via compensatory frameshift is widespread in RNA virus genomes. BMC Bioinform. 2021, 22, 251. [Google Scholar] [CrossRef]
- Lim, C.S.; Brown, C.M. Know Your Enemy: Successful Bioinformatic Approaches to Predict Functional RNA Structures in Viral RNAs. Front. Microbiol. 2018, 8, 2582. [Google Scholar] [CrossRef]
- Gmyl, A.P.; Korshenko, S.A.; Belousov, E.V.; Khitrina, E.V.; Agol, V.I. Nonreplicative homologous RNA recombination: Promiscuous joining of RNA pieces? RNA 2003, 9, 1221–1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sztuba-Solińska, J.; Urbanowicz, A.; Figlerowicz, M.; Bujarski, J.J. RNA-RNA Recombination in Plant Virus Replication and Evolution. Annu. Rev. Phytopathol. 2011, 49, 415–443. [Google Scholar] [CrossRef] [PubMed]
- Flynn, P.J.; Moreau, C.S. Assessing the Diversity of Endogenous Viruses Throughout Ant Genomes. Front. Microbiol. 2019, 10, 1139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, Q.; Stockinger, M.P.; Chang, Y.; Tashiro, H.; Lin, C.-L.G. The presence of rRNA sequences in polyadenylated RNA and its potential functions. Biotechnol. J. 2008, 3, 1041–1046. [Google Scholar] [CrossRef]
- Mauro, V.P.; Edelman, G.M. rRNA-like sequences occur in diverse primary transcripts: Implications for the control of gene expression. Proc. Natl. Acad. Sci. USA 1997, 94, 422–427. [Google Scholar] [CrossRef] [Green Version]
- Pánek, J.; Kolář, M.; Vohradský, J.; Valášek, L.S. An evolutionary conserved pattern of 18S rRNA sequence complementarity to mRNA 5′ UTRs and its implications for eukaryotic gene translation regulation. Nucleic Acids Res. 2013, 41, 7625–7634. [Google Scholar] [CrossRef] [Green Version]
- Fujimura, T.; Esteban, R. Cap-snatching mechanism in yeast L-A double-stranded RNA virus. Proc. Natl. Acad. Sci. USA 2011, 108, 17667–17671. [Google Scholar] [CrossRef] [Green Version]


| Yeast Strain | Killer Phenotype/LBC dsRNA Type | Sequenced Length (bp) (Canonical) | 5′-Extra Sequence (bp)/% Identity to, Size [Position] | 3′-Extra Sequence (bp)/% Identity to, Size [Position] | Stem-Loop Involving 5′-Extra Sequence [Position] (ΔG) | Stem-Loop Involving 3′-Extra Sequence [Position] (ΔG) |
|---|---|---|---|---|---|---|
| Sc EX231 | K1/LBCM1-1 | 4971 (4615) | 143/100% identity to (+) strand, 95 nt [A(−)143–A(−)49] to 95 nt [A65–A159] | 213/99% to (+) strand Sc 18S rRNA, 201 nt [G(+)13–G(+)213] | [T(−)116–A159] (−604) | [C(+)82–G(+)110] (−49) |
| Sc EX1125 | K2/LBCM2-4 | 4722 (4615) | 58/100% identity to (−) strand, 62 nt [C(−)56–T6] to 62 nt [A38–G99] | 49/no identity found | [G(−)55–T100] (−429) | [A(+)16–T(+)27] (−6) |
| Sc EX229 | Klus/LBCMlus4 | 4839 (4615) | 63/100% identity to (+) strand, 64 nt [G(−)63–G1] to 64 nt [G54–G116] | 43/100% to (−) strand Sc 25S rRNA, 43 nt [C(+)2–C(+)44] | [T(−)10–A53] (−86) | [T(+)15–A(+)34] (−30) |
| Sc EX1160 | Klus/LBCMlusA | 4661 (4615) | 46/100% identity to (−) strand, 52 nt [G(−)46–T5] to 52 nt [A38–C89] | None | [G(−)46–C89] (−327) | None |
| Td EX1180 | Kbarr1/LBCbarr1 | 4763(4565) | 170/100% identity to (−) strand, 173 nt [G(−)170–A3] to 173 nt [T31–C203] | 28/100% identity to (+) strand, 28 nt [C(+)1–C(+)28] to 28 nt [C4466–C4493] | [G(−)170–C203] (−1269) | [G(+)8–C(+)21] (−15) |
| Td EX1257 | Kbarr2/LBCbarr2 | 5115 (4565) | 219/100% identity to (−) strand, 151 nt [C(−)148–A3] to 151 nt [T31–G181] | 331/100% identity to (+) strand, 22 nt [A(+)7–A(+)28] to 22 nt [A4471–A4492]; and 307 nt [T(+)25–A(+)331] to 307 nt [T3175–A3481] | [C(−)148–G181] (−1121) | [A(+)33–T(+)53] (−25)[C(+)158–G(+)196] (−39) |
| Virus-Yeast Strain | Accession Number | Reference/Comment |
|---|---|---|
| TdV-LBCbarr1-EX1180 | OL469171 | [9] |
| TdV-LBCbarr2-EX1257 | OL469172 | [9] |
| ScV-LBC1-original | U01060.1 | [8]. Previously known as ScV-La or L-B-C. Here renamed to distinguish it from other LBC viruses from different K1 killer strains |
| ScV-LBClus4-EX229 | KT784813.1 | [5] |
| ScV-LBC2-S3920 | KX906605.1 | [5] |
| ScV-LBC1-EX231 | OL469175 | [9] |
| ScV-LBC2-EX1125 | OL469176 | [9] |
| ScV-LBClusA-EX1160 | OL469174 | [9] |
| ScV-LBClus4-EX229 | OL469173 | [9] |
| TdV-LAbarr1-EX1180 | MW174763 | [14] |
| ScV-LA1-original | J04692.1 | [13]. Previously known as ScV-LA. Here renamed to distinguish it from other LA viruses from different K1 killer strains |
| ScV-LA1-EX231 | MW174760 | [14] |
| ScV-LAlus4-EX229 | MW174758 | [14] |
| ScV-LAlus1-EX436 | MW174761 | [14] |
| ScV-LAlusA-EX1160 | MW174762 | [14] |
| ScV-LA2-S3920 | KC677754.1 | [5] |
| ScV-LA2-EX1125 | MW174759 | [14] |
| SpV-LA28 | KU845301.2 | [30]. Previously assigned to S. cerevisiae but thereafter re-assigned to S. paradoxus [7] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramírez, M.; Martínez, A.; Molina, F. New Insights into the Genome Organization of Yeast Double-Stranded RNA LBC Viruses. Microorganisms 2022, 10, 173. https://doi.org/10.3390/microorganisms10010173
Ramírez M, Martínez A, Molina F. New Insights into the Genome Organization of Yeast Double-Stranded RNA LBC Viruses. Microorganisms. 2022; 10(1):173. https://doi.org/10.3390/microorganisms10010173
Chicago/Turabian StyleRamírez, Manuel, Alberto Martínez, and Felipe Molina. 2022. "New Insights into the Genome Organization of Yeast Double-Stranded RNA LBC Viruses" Microorganisms 10, no. 1: 173. https://doi.org/10.3390/microorganisms10010173







