Genetic Potential of Dissulfurimicrobium hydrothermale, an Obligate Sulfur-Disproportionating Thermophilic Microorganism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture, DNA Extraction and Genome Assembly
2.2. Genome Annotation
2.3. Data Availability
3. Results and Discussion
3.1. General Characteristics of Genome
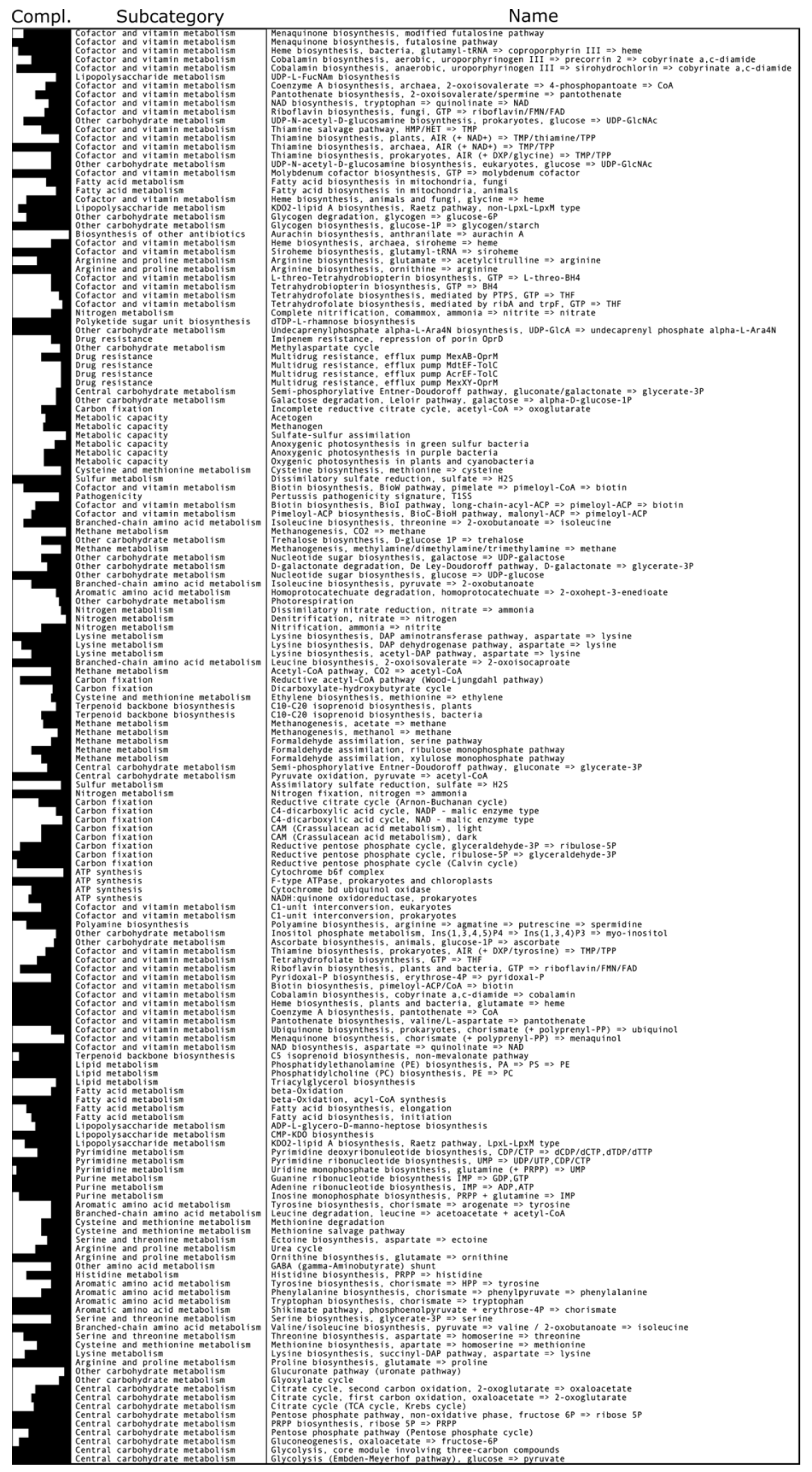
3.2. Carbon Metabolism
3.3. Nitrogen Metabolism
3.4. Hydrogen Metabolism
3.5. Sulfur Metabolism
3.6. Motility and Pili
3.7. Putative Secretion Systems
3.8. Probable Role of the Short Protein in ISC-Disproportionation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AQDS | 9,10-anthraquinone 2,6-disulfonate |
| APS | adenosine phosphosulfate |
| CDS | coding DNA sequences |
| COG | clusters of orthologous groups of proteins |
| DCT | DsrC-Trisulfide |
| ISC | inorganic sulfur compounds |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MIGS | minimum information about a genome sequence |
| NCBI | National Center for Biotechnology Information |
| PGAP | prokaryotic genome annotation pipeline |
| SDB | sulfur-disproportionating bacteria |
| SOR | sulfur oxygenase reductase |
| TCA | tricarboxylic acid cycle |
References
- Slobodkin, A.I.; Slobodkina, G.B. Diversity of Sulfur-Disproportionating Microorganisms. Microbiology 2019, 88, 509–522. [Google Scholar] [CrossRef]
- Umezawa, K.; Kojima, H.; Kato, Y.; Fukui, M. Disproportionation of inorganic sulfur compounds by a novel autotrophic bacterium belonging to Nitrospirota. Syst. Appl. Microbiol. 2020, 43, 126110. [Google Scholar] [CrossRef]
- Bak, F.; Cypionka, H. A novel type of energy metabolism involving fermentation of inorganic sulphur compounds. Nat. Cell Biol. 1987, 326, 891–892. [Google Scholar] [CrossRef]
- Finster, K. Microbiological disproportionation of inorganic sulfur compounds. J. Sulfur Chem. 2008, 29, 281–292. [Google Scholar] [CrossRef]
- Allioux, M.; Yvenou, S.; Slobodkina, G.; Slobodkin, A.; Shao, Z.; Jebbar, M.; Alain, K. Genomic Characterization and Environmental Distribution of a Thermophilic Anaerobe Dissulfurirhabdus thermomarina SH388T Involved in Disproportionation of Sulfur Compounds in Shallow Sea Hydrothermal Vents. Microorganisms 2020, 8, 1132. [Google Scholar] [CrossRef] [PubMed]
- Frederiksen, T.-M.; Finster, K. Sulfite-oxido-reductase is involved in the oxidation of sulfite in Desulfocapsa sulfoexigens during disproportionation of thiosulfate and elemental sulfur. Biodegradation 2003, 14, 189–198. [Google Scholar] [CrossRef]
- Frederiksen, T.-M.; Finster, K. The transformation of inorganic sulfur compounds and the assimilation of organic and inorganic carbon by the sulfur disproportionating bacterium Desulfocapsa sulfoexigens. Antonie Leeuwenhoek 2004, 85, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Finster, K.; Kjeldsen, K.U.; Kube, M.; Reinhardt, R.; Mussmann, M.; Amann, R.; Schreiber, L. Complete genome sequence of Desulfocapsa sulfexigens, a marine deltaproteobacterium specialized in disproportionating inorganic sulfur compounds. Stand. Genom. Sci. 2013, 8, 58–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mardanov, A.; Beletsky, A.; Kadnikov, V.V.; Slobodkin, A.I.; Ravin, N.V. Genome Analysis of Thermosulfurimonas dismutans, the First Thermophilic Sulfur-Disproportionating Bacterium of the Phylum Thermodesulfobacteria. Front. Microbiol. 2016, 7, 950. [Google Scholar] [CrossRef]
- Florentino, A.P.; Pereira, I.A.C.; Boeren, S.; Born, M.V.D.; Stams, A.J.M.; Sánchez-Andrea, I. Insight into the sulfur metabolism of Desulfurella amilsii by differential proteomics. Environ. Microbiol. 2019, 21, 209–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kojima, H.; Umezawa, K.; Fukui, M. Caldimicrobium thiodismutans sp. nov., a sulfur-disproportionating bacterium isolated from a hot spring, and emended description of the genus Caldimicrobium. Int. J. Syst. Evol. Microbiol. 2016, 66, 1828–1831. [Google Scholar] [CrossRef]
- Slobodkin, A.I.; Slobodkina, G.B.; Panteleeva, A.N.; Chernyh, N.A.; Novikov, A.A.; Bonch-Osmolovskaya, E.A. Dissulfurimicrobium hydrothermale gen. nov., sp. nov., a thermophilic, autotrophic, sulfur-disproportionating deltaproteobacterium isolated from a hydrothermal pond. Int. J. Syst. Evol. Microbiol. 2016, 66, 1022–1026. [Google Scholar] [CrossRef] [PubMed]
- Waite, D.W.; Chuvochina, M.; Pelikan, C.; Parks, D.H.; Yilmaz, P.; Wagner, M.; Loy, A.; Naganuma, T.; Nakai, R.; Whitman, W.B.; et al. Proposal to reclassify the proteobacterial classes Deltaproteobacteria and Oligoflexia, and the phylum Thermodesulfobacteria into four phyla reflecting major functional capabilities. Int. J. Syst. Evol. Microbiol. 2020, 70, 5972–6016. [Google Scholar] [CrossRef] [PubMed]
- Slobodkin, A.I.; Reysenbach, A.-L.; Slobodkina, G.B.; Baslerov, R.V.; Kostrikina, N.A.; Wagner, I.D.; Bonch-Osmolovskaya, E.A. Thermosulfurimonas dismutans gen. nov., sp. nov., an extremely thermophilic sulfur-disproportionating bacterium from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 2012, 62, 2565–2571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charbonnier, F.; Forterre, P.; Erauso, G.; Prieur, D. Purification of plasmids from thermophilic and hyperthermophilic archaea. Archaea Lab. Man. Thermophiles 1995, 87–90. [Google Scholar]
- Thamdrup, B.; Finster, K.; Hansen, J.W.; Bak, F. Bacterial Disproportionation of Elemental Sulfur Coupled to Chemical Reduction of Iron or Manganese. Appl. Environ. Microbiol. 1993, 59, 101–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, S. FastQC: A quality control tool for high throughput sequence data. 2010. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Vallenet, D.; Calteau, A.; Dubois, M.; Amours, P.; Bazin, A.; Beuvin, M.; Burlot, L.; Bussell, X.; Fouteau, S.; Gautreau, G.; et al. MicroScope: An integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res. 2019, 48, D579–D589. [Google Scholar] [CrossRef] [Green Version]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [Green Version]
- Eren, A.M.; Kiefl, E.; Shaiber, A.; Veseli, I.; Miller, S.E.; Schechter, M.S.; Fink, I.; Pan, J.N.; Yousef, M.; Fogarty, E.C.; et al. Community-led, integrated, reproducible multi-omics with anvi’o. Nat. Microbiol. 2021, 6, 3–6. [Google Scholar] [CrossRef]
- Slobodkin, A.I.; Reysenbach, A.-L.; Slobodkina, G.B.; Kolganova, T.V.; Kostrikina, N.A.; Bonch-Osmolovskaya, E.A. Dissulfuribacter thermophilus gen. nov., sp. nov., a thermophilic, autotrophic, sulfur-disproportionating, deeply branching deltaproteobacterium from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 2013, 63, 1967–1971. [Google Scholar] [CrossRef]
- Frolova, A.A.; Slobodkina, G.B.; Baslerov, R.V.; Novikov, A.A.; Bonch-Osmolovskaya, E.A.; Slobodkin, A.I. Thermosulfurimonas marina sp. nov., an Autotrophic Sulfur-Disproportionating and Nitrate-Reducing Bacterium Isolated from a Shallow-Sea Hydrothermal Vent. Microbiology 2018, 87, 502–507. [Google Scholar] [CrossRef]
- Slobodkina, G.B.; Kolganova, T.V.; Kopitsyn, D.S.; Viryasov, M.B.; Bonch-Osmolovskaya, E.A.; Slobodkin, A.I. Dissulfurirhabdus thermomarina gen. nov., sp. nov., a thermophilic, autotrophic, sulfite-reducing and disproportionating deltaproteobacterium isolated from a shallow-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 2016, 66, 2515–2519. [Google Scholar] [CrossRef]
- Widdel, F.; Pfennig, N. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids II. Incomplete oxidation of propionate by Desulfobulbus propionicus gen. nov., sp. nov. Arch. Microbiol. 1982, 129, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, M.; Springer, N.; Ludwig, W.; Schink, B. Phylogenetic Positions of Desulfofustis glycolicus gen. nov., sp. nov. and Syntrophobotulus glycolicus gen. nov., sp. nov., Two New Strict Anaerobes Growing with Glycolic Acid. Int. J. Syst. Evol. Microbiol. 1996, 46, 1065–1069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorokin, D.Y.; Tourova, T.P.; Henstra, A.M.; Stams, A.; Galinski, E.A.; Muyzer, G. Sulfidogenesis under extremely haloalkaline conditions by Desulfonatronospira thiodismutans gen. nov., sp. nov., and Desulfonatronospira delicata sp. nov.—A novel lineage of Deltaproteobacteria from hypersaline soda lakes. Microbiology 2008, 154, 1444–1453. [Google Scholar] [CrossRef] [Green Version]
- Pikuta, E.V.; Zhilina, T.N.; Zavarzin, G.A.; Kostrikina, N.A.; Osipov, G.A. Desulfonatronum lacustre gen. nov., sp. nov.: A new alkaliphilic sulfate-reducing bacterium utilizing ethanol. Microbiology 1998, 67, 105–113. [Google Scholar]
- Sorokin, D.Y.; Tourova, T.P.; Kolganova, T.V.; Detkova, E.N.; Galinski, E.A.; Muyzer, G. Culturable diversity of lithotrophic haloalkaliphilic sulfate-reducing bacteria in soda lakes and the description of Desulfonatronum thioautotrophicum sp. nov., Desulfonatronum thiosulfatophilum sp. nov., Desulfonatronovibrio thiodismutans sp. nov., and Desulfonatronovibrio magnus sp. nov. Extremophiles 2011, 15, 391–401. [Google Scholar] [CrossRef] [Green Version]
- Pikuta, E.V.; Hoover, R.B.; Bej, A.K.; Marsic, D.; Whitman, W.; Cleland, D.; Krader, P. Desulfonatronum thiodismutans sp. nov., a novel alkaliphilic, sulfate-reducing bacterium capable of lithoautotrophic growth. Int. J. Syst. Evol. Microbiol. 2003, 53, 1327–1332. [Google Scholar] [CrossRef] [Green Version]
- Bak, F.; Pfennig, N. Chemolithotrophic growth of Desulfovibrio sulfodismutans sp. nov. by disproportionation of inorganic sulfur compounds. Arch. Microbiol. 1987, 147, 184–189. [Google Scholar] [CrossRef]
- Finster, K.; Liesack, W.; Thamdrup, B. Elemental Sulfur and Thiosulfate Disproportionation by Desulfocapsa sulfoexigens sp. nov., a New Anaerobic Bacterium Isolated from Marine Surface Sediment. Appl. Environ. Microbiol. 1998, 64, 119–125. [Google Scholar] [CrossRef] [Green Version]
- Florentino, A.P.; Brienza, C.; Stams, A.J.M.; Sánchez-Andrea, I. Desulfurella amilsii sp. nov., a novel acidotolerant sulfur-respiring bacterium isolated from acidic river sediments. Int. J. Syst. Evol. Microbiol. 2016, 66, 1249–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorokin, D.Y.; Tourova, T.P.; Mussmann, M.; Muyzer, G. Dethiobacter alkaliphilus gen. nov. sp. nov., and Desulfurivibrio alkaliphilus gen. nov. sp. nov.: Two novel representatives of reductive sulfur cycle from soda lakes. Extremophiles 2008, 12, 431–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slobodkina, G.B.; Reysenbach, A.-L.; Kolganova, T.V.; Novikov, A.A.; Bonch-Osmolovskaya, E.A.; Slobodkin, A.I. Thermosulfuriphilus ammonigenes gen. nov., sp. nov., a thermophilic, chemolithoautotrophic bacterium capable of respiratory ammonification of nitrate with elemental sulfur. Int. J. Syst. Evol. Microbiol. 2017, 67, 3474–3479. [Google Scholar] [CrossRef]
- Alain, K.; Postec, A.; Grinsard, E.; Lesongeur, F.; Prieur, D.; Godfroy, A. Thermodesulfatator atlanticus sp. nov., a thermophilic, chemolithoautotrophic, sulfate-reducing bacterium isolated from a Mid-Atlantic Ridge hydrothermal vent. Int. J. Syst. Evol. Microbiol. 2010, 60, 33–38. [Google Scholar] [CrossRef]
- Isaksen, M.F.; Teske, A. Desulforhopalus vacuolatus gen. nov., sp. nov., a new moderately psychrophilic sulfate-reducing bacterium with gas vacuoles isolated from a temperate estuary. Arch. Microbiol. 1996, 166, 160–168. [Google Scholar] [CrossRef]
- Umezawa, K.; Kojima, H.; Kato, Y.; Fukui, M. Dissulfurispira thermophila gen. nov., sp. nov., a thermophilic chemolithoautotroph growing by sulfur disproportionation, and proposal of novel taxa in the phylum Nitrospirota to reclassify the genus Thermodesulfovibrio. Syst. Appl. Microbiol. 2021, 44, 126184. [Google Scholar] [CrossRef]
- Moussard, H.; Haridon, S.L.; Tindall, B.J.; Banta, A.; Schumann, P.; Stackebrandt, E.; Reysenbach, A.-L.; Jeanthon, C. Thermodesulfatator indicus gen. nov., sp. nov., a novel thermophilic chemolithoautotrophic sulfate-reducing bacterium isolated from the Central Indian Ridge. Int. J. Syst. Evol. Microbiol. 2004, 54, 227–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slobodkina, G.; Allioux, M.; Merkel, A.; Alain, K.; Jebbar, M.; Slobodkin, A. Genome analysis of Thermosulfuriphilus ammonigenes ST65T, an anaerobic thermophilic chemolithoautotrophic bacterium isolated from a deep-sea hydrothermal vent. Mar. Genom. 2020, 54, 100786. [Google Scholar] [CrossRef] [PubMed]
- Berg, I.A. Ecological Aspects of the Distribution of Different Autotrophic CO2 Fixation Pathways. Appl. Environ. Microbiol. 2011, 77, 1925–1936. [Google Scholar] [CrossRef] [Green Version]
- Hügler, M.; Sievert, S.M. Beyond the Calvin Cycle: Autotrophic Carbon Fixation in the Ocean. Annu. Rev. Mar. Sci. 2011, 3, 261–289. [Google Scholar] [CrossRef] [Green Version]
- Gaby, J.; Buckley, D.H. A comprehensive aligned nifH gene database: A multipurpose tool for studies of nitrogen-fixing bacteria. Database 2014, 2014, bau001. [Google Scholar] [CrossRef] [Green Version]
- Halbleib, C.M.; Ludden, P.W. Regulation of Biological Nitrogen Fixation. J. Nutr. 2000, 130, 1081–1084. [Google Scholar] [CrossRef] [Green Version]
- Wasmund, K.; Mußmann, M.; Loy, A. The life sulfuric: Microbial ecology of sulfur cycling in marine sediments. Environ. Microbiol. Rep. 2017, 9, 323–344. [Google Scholar] [CrossRef]
- Krämer, M.; Cypionka, H. Sulfate formation via ATP sulfurylase in thiosulfate- and sulfite-disproportionating bacteria. Arch. Microbiology 1989, 151, 232–237. [Google Scholar] [CrossRef]
- Thorup, C.; Schramm, A.; Findlay, A.J.; Finster, K.W.; Schreiber, L. Disguised as a Sulfate Reducer: Growth of the Deltaproteobacterium Desulfurivibrio alkaliphilus by Sulfide Oxidation with Nitrate. mBio 2017, 8, e00671-17. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Ochman, H. Stepwise formation of the bacterial flagellar system. Proc. Natl. Acad. Sci. USA 2007, 104, 7116–7121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diepold, A.; Wagner, S. Assembly of the bacterial type III secretion machinery. FEMS Microbiol. Rev. 2014, 38, 802–822. [Google Scholar] [CrossRef] [Green Version]
- Van Gijsegem, F.; Gough, C.; Zischek, C.; Niqueux, E.; Arlat, M.; Genin, S.; Barberis, P.; German, S.; Castello, P.; Boucher, C. The hrp gene locus of Pseudomonas solanacearum, which controls the production of a type III secretion system, encodes eight proteins related to components of the bacterial flagellar biogenesis complex. Mol. Microbiol. 1995, 15, 1095–1114. [Google Scholar] [CrossRef] [PubMed]
- Tseng, T.-T.; Tyler, B.M.; Setubal, J.C. Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 2009, 9, S2. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Qiu, Y.-Y.; Zhou, Y.; Chen, G.-H.; van Loosdrecht, M.C.; Jiang, F. Elemental sulfur as electron donor and/or acceptor: Mechanisms, applications and perspectives for biological water and wastewater treatment. Water Res. 2021, 202, 117373. [Google Scholar] [CrossRef] [PubMed]
- Boyd, E.S.; Druschel, G.K. Involvement of Intermediate Sulfur Species in Biological Reduction of Elemental Sulfur under Acidic, Hydrothermal Conditions. Appl. Environ. Microbiol. 2013, 79, 2061–2068. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Item | Description |
|---|---|
| Investigation | |
| Strain | Dissulfurimicrobium hydrothermale Sh68T |
| Submitted to INSDC | GenBank |
| Investigation type | Bacteria |
| Project name | PRJNA769390 |
| Geographic location (latitude and longitude) | 54°49.4′ N, 160°01.0′ E |
| Geographic location (country and/or sea, region) | Uzon Caldera, Kamchatka, Russia |
| Collection date | September 20009 |
| Environment (biome) | Hot spring ENVO:00000051 |
| Environment (feature) | Hot spring ENVO:00000051 |
| Environment (material) | Hydrothermally influenced sediment ENVO:01001821 |
| Depth | 30 cm |
| General features | |
| Classification | Domain: Bacteria |
| Phylum: Desulfobacterota | |
| Class: Dissulfuribacteria | |
| Order: Dissulfuribacterales | |
| Family: Dissulfuribacteraceae | |
| Genus: Dissulfurimicrobium | |
| Species: Dissulfurimicrobium hydrothermale | |
| Sh68T | |
| Gram stain | Negative |
| Cell shape | short rod with rounded ends |
| Motility | Motile |
| Growth temperature | 30–65 °C |
| Relationship to oxygen | Anaerobic |
| Trophic level | Chemolithoautotrophic |
| Biotic relationship | free-living |
| Isolation and growth conditions | DOI 10.1099/ijsem.0.000828 |
| Sequencing | |
| Sequencing technology | Illumina Miseq Nano 2 × 150 bp and Oxford MinION (R9 flow cell and Rapid Sequencing kit) |
| Sequencing platform | Fasteris and in house |
| Assembler | Unicycler (v0.4.9) |
| Contig number | 1 |
| N50 | 2,025,450 |
| Genome coverage | 50.6 × (based only on short reads) |
| 131.1 × (based on short and long reads) | |
| Genome Accession NCBI | CP085041 |
| Assembly level | Complete |
| Genomic features | |
| Genome size (bp) | 2,025,450 |
| GC content (%) | 49.66 |
| Protein coding genes | 1925 |
| Number of RNAs | 54 |
| tRNAs | 47 |
| 16S-23S-5S rRNAs | 1-1-1 |
| Strain | S0 Disproportionation Ability | Short Protein of Unknown Function (Automatically Annotated as an “EscU/YscU/HrcU Family Type III Secretion System Export Apparatus Switch Protein”) | Locus Tag of the Short Protein | Motility |
|---|---|---|---|---|
| Dissulfurimicrobium hydrothermale Sh68T | + | + | LGS26_00065 | + |
| Dissulfuribacter thermophilus S69T | + | + | DBT_RS04205 | + |
| Thermosulfurimonas marina SU872T | + | + | FVE67_RS02390 | + |
| Thermosulfurimonas dismutans S95T | + | + | TDIS_RS03420 | + |
| Dissulfurirhabdus thermomarina SH388T | + | + | HCU62_RS02240 | + |
| Caldimicrobium thiodismutans TF1T | + | + | THC_RS00840 | + |
| Desulfobulbus propionicus DSM 2032T | + | + | DESPR_RS10825 | − |
| Desulfofustis glycolicus DSM 9705T | + | + | BUC26_RS20670 | + |
| Desulfocapsa sulfoexigens DSM 10523T | + | + | UWK_RS15510 | + |
| Desulfurella amilsii TR1T | + | + | DESAMIL20_RS08330 | + |
| Desulfurivibrio alkaliphilus AHT 2T | + | + | DAAHT2_RS11940 | − |
| Dethiobacter alkaliphilus AHT 1T | + | + | DEALDRAFT_RS03220 | + |
| Thermosulfuriphilus ammonigenes ST65T | + | + | G4V39_RS06430 | − |
| Thermodesulfatator atlanticus DSM 21156T | + | + | H528_RS0110240 | + |
| Dissulfurispira thermophila T55JT | + | + | JTV28_RS00420 | + |
| Thermodesulfatator indicus DSM 15286T | − | + | THEIN_RS08670 | + |
| Desulfonatronospira thiodismutans ASO3-1T | − | − | − | + |
| Desulfonatronum lacustre DSM 10312T | − | − | − | + |
| Desulfonatronum thioautotrophicum ASO4-1T | − | − | − | + |
| Desulfonatronum thiodismutans MLF1T | − | − | − | + |
| Desulfonatronum thiosulfatophilum ASO4-2T | − | − | − | + |
| Desulfolutivibrio sulfodismutans DSM 3696T | − | − | − | + |
| Desulforhopalus vacuolatus strain DSM 9700T | − | − | − | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yvenou, S.; Allioux, M.; Slobodkin, A.; Slobodkina, G.; Jebbar, M.; Alain, K. Genetic Potential of Dissulfurimicrobium hydrothermale, an Obligate Sulfur-Disproportionating Thermophilic Microorganism. Microorganisms 2022, 10, 60. https://doi.org/10.3390/microorganisms10010060
Yvenou S, Allioux M, Slobodkin A, Slobodkina G, Jebbar M, Alain K. Genetic Potential of Dissulfurimicrobium hydrothermale, an Obligate Sulfur-Disproportionating Thermophilic Microorganism. Microorganisms. 2022; 10(1):60. https://doi.org/10.3390/microorganisms10010060
Chicago/Turabian StyleYvenou, Stéven, Maxime Allioux, Alexander Slobodkin, Galina Slobodkina, Mohamed Jebbar, and Karine Alain. 2022. "Genetic Potential of Dissulfurimicrobium hydrothermale, an Obligate Sulfur-Disproportionating Thermophilic Microorganism" Microorganisms 10, no. 1: 60. https://doi.org/10.3390/microorganisms10010060






