Enzyme Profiling and Identification of Endophytic and Rhizospheric Bacteria Isolated from Arthrocnemum macrostachyum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Isolation of Rhizospheric and Endophytic Bacteria
2.3. Microscopic and Biochemical Characteristics
2.4. DNA Extraction and PCR Amplification of 16S rRNA
2.5. Identification by Phylogenetic Analysis
2.6. Determination of Growth Characteristics
2.7. Determination of Enzymatic Potential
2.8. Statistical Analysis
3. Results
3.1. Isolation and Microscopic Analysis
3.2. Molecular Identification
3.3. Analysis of Growth Parameters
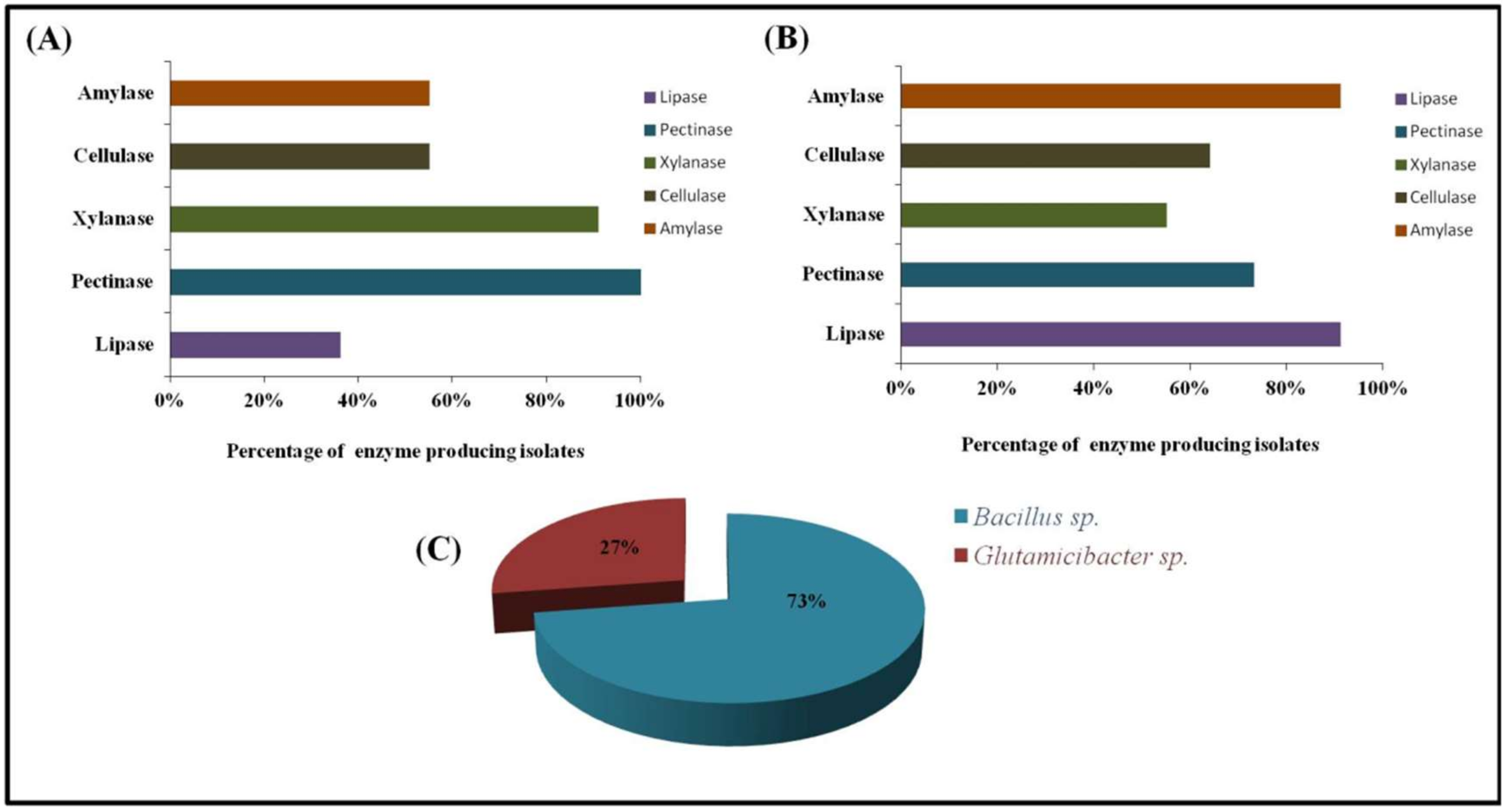
3.4. Enzyme Production
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Etesami, H.; Beattie, G.A. Mining Halophytes for Plant Growth-Promoting Halotolerant Bacteria to Enhance the Salinity Tolerance of Non-Halophytic Crops. Front. Microbiol. 2018, 9, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shurigin, V.; Egamberdieva, D.; Li, L.; Davranov, K.; Panosyan, H.; Birkeland, N.K.; Wirth, S.; Bellingrath-Kimura, S.D. Endophytic Bacteria Associated with Halophyte Seidlitzia rosmarinus Ehrenb. ex Boiss. from Saline Soil of Uzbekistan and Their Plant Beneficial Traits. J. Arid Land 2020, 12, 730–740. [Google Scholar] [CrossRef]
- Santoyo, G.; Moreno-Hagelsieb, G.; del Carmen Orozco-Mosqueda, M.; Glick, B.R. Plant Growth-Promoting Bacterial Endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Ruppel, S.; Franken, P.; Witzel, K. Properties of the Halophyte Microbiome and Their Implications for Plant Salt Tolerance. Funct. Plant Biol. 2013, 40, 940–951. [Google Scholar] [CrossRef] [Green Version]
- Hussain, A.; Arshad, M.; Zahir, Z.A.; Asghar, M. Prospects of Zinc Solubilizing Bacteria for Enhancing Growth of Maize. Pakistan J. Agric. Sci. 2015, 52, 915–922. [Google Scholar]
- Azevedo, J.L.; Araújo, W.L. Diversity and Applications of Endophytic Fungi Isolated from Tropical Plants. Fungi Multifaceted Microbes 2007, 9, 189–207. [Google Scholar]
- Soltani, J.; Zaheri-Shoja, M.; Hamzei, J.; Hosseyni-Moghaddam, M.S.; Pakvaz, S. Diversity and Bioactivity of Bacterial Endophyte Community of Cupressaceae. For. Pathol. 2016, 46, 353–361. [Google Scholar] [CrossRef]
- Clay, K.; Holah, J. Fungal Endophyte Symbiosis and Plant Diversity in Successional Fields. Science 1999, 285, 1742–1744. [Google Scholar] [CrossRef]
- Sturz, A.V.; Matheson, B.G. Populations of Endophytic Bacteria Which Influence Host-Resistance to Erwinia-Induced Bacterial Soft Rot in Potato Tubers. Plant Soil 1996, 184, 265–271. [Google Scholar] [CrossRef]
- Schulz, B.; Boyle, C.; Draeger, S.; Römmert, A.-K.; Krohn, K. Endophytic Fungi: A Source of Novel Biologically Active Secondary Metabolites. Mycol. Res. 2002, 106, 996–1004. [Google Scholar] [CrossRef]
- Khan, A.L.; Hussain, J.; Al-Harrasi, A.; Al-Rawahi, A.; Lee, I.-J. Endophytic Fungi: Resource for Gibberellins and Crop Abiotic Stress Resistance. Crit. Rev. Biotechnol. 2015, 35, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.M.; Hong, S.Y.; Lee, S.M.; Kim, Y.H.; Kahng, G.G.; Lim, Y.P.; Kim, H.; Yun, H.D. Endophytic Bacterial Communities in Ginseng and Their Antifungal Activity against Pathogens. Microb. Ecol. 2007, 54, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial Endophyte Colonization and Distribution within Plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.; Kumar, M.; Mittal, A.; Mehta, P.K. Microbial Enzymes: Industrial Progress in 21st Century. 3 Biotech 2016, 6, 174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Xu, Y.; Lai, X.H.; Shan, C.; Deng, Z.; Ji, Y. Screening and Characterization of Endophytic Bacillus and Paenibacillus Strains from Medicinal Plant Lonicera Japonica for Use as Potential Plant Growth Promoters. Brazilian J. Microbiol. 2015, 46, 977–989. [Google Scholar] [CrossRef]
- Dastgheib, S.M.M.; Amoozegar, M.A.; Khajeh, K.; Ventosa, A. A Halotolerant Alcanivorax sp. Strain with Potential Application in Saline Soil Remediation. Appl. Microbiol. Biotechnol. 2011, 90, 305–312. [Google Scholar] [CrossRef]
- Kaleem Sarwar, M.; Azam, I.; Iqbal, T. Biology and Applications of Halophilic Bacteria and Archaea: A Review. Electron. J. Biol. 2015, 11, 98–103. [Google Scholar]
- Marhuenda-Egea, F.C.; Bonete, M.J. Extreme Halophilic Enzymes in Organic Solvents. Curr. Opin. Biotechnol. 2002, 13, 385–389. [Google Scholar] [CrossRef]
- Redondo-Gómez, S.; Mateos-Naranjo, E.; Figueroa, M.E.; Davy, A.J. Salt Stimulation of Growth and Photosynthesis in an Extreme Halophyte, Arthrocnemum macrostachyum. Plant Biol. 2010, 12, 79–87. [Google Scholar] [CrossRef]
- Mora-Ruiz, M.d.R.; Font-Verdera, F.; Orfila, A.; Rita, J.; Rosselló-Móra, R. Endophytic Microbial Diversity of the Halophyte Arthrocnemum macrostachyum across Plant Compartments. FEMS Microbiol. Ecol. 2016, 92, fiw145. [Google Scholar] [CrossRef] [Green Version]
- Mora-Ruiz, M.d.R.; Alejandre-Colomo, C.; Ledger, T.; González, B.; Orfila, A.; Rosselló-Móra, R. Non-Halophilic Endophytes Associated with the Euhalophyte Arthrocnemum macrostachyum and Their Plant Growth Promoting Activity Potential. FEMS Microbiol. Lett. 2018, 365, fny208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarro-Torre, S.; Mateos-Naranjo, E.; Caviedes, M.A.; Pajuelo, E.; Rodríguez-Llorente, I.D. Isolation of Plant-Growth-Promoting and Metal-Resistant Cultivable Bacteria from Arthrocnemum macrostachyum in the Odiel Marshes with Potential Use in Phytoremediation. Mar. Pollut. Bull. 2016, 110, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Torre, S.; Barcia-Piedras, J.M.; Mateos-Naranjo, E.; Redondo-Gómez, S.; Camacho, M.; Caviedes, M.A.; Pajuelo, E.; Rodríguez-Llorente, I.D. Assessing the Role of Endophytic Bacteria in the Halophyte Arthrocnemum macrostachyum Salt Tolerance. Plant Biol. 2017, 19, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Fouda, A.H.; Hassan, S.E.-D.; Hany, A.M.; Mohamed, A. Plant growth promoting activities of endophytic bacteria associated with the halophyte Arthrocnemum macrostachyum (moric) k. koch. plant. J. Microbiol. 2018, 49, 123–141. [Google Scholar]
- Golinska, P.; Wypij, M.; Agarkar, G.; Rathod, D.; Dahm, H.; Rai, M. Endophytic Actinobacteria of Medicinal Plants: Diversity and Bioactivity. Antonie Leeuwenhoek 2015, 108, 267–289. [Google Scholar] [CrossRef] [Green Version]
- Corrêa, R.C.G.; Rhoden, S.A.; Mota, T.R.; Azevedo, J.L.; Pamphile, J.A.; de Souza, C.G.M.; Polizeli, M.d.L.T.d.M.; Bracht, A.; Peralta, R.M. Endophytic Fungi: Expanding the Arsenal of Industrial Enzyme Producers. J. Ind. Microbiol. Biotechnol. 2014, 41, 1467–1478. [Google Scholar] [CrossRef]
- Traving, S.J.; Thygesen, U.H.; Riemann, L.; Stedmon, C.A. A Model of Extracellular Enzymes in Free-Living Microbes: Which Strategy Pays Off? Appl. Environ. Microbiol. 2015, 81, 7385–7393. [Google Scholar] [CrossRef] [Green Version]
- Ferreira Filho, A.S.; Quecine, M.C.; Bogas, A.C.; Rossetto, P.d.B.; Lima, A.O.d.S.; Lacava, P.T.; Azevedo, J.L.; Araújo, W.L. Endophytic Methylobacterium Extorquens Expresses a Heterologous β-1,4-Endoglucanase A (EglA) in Catharanthus Roseus Seedlings, a Model Host Plant for Xylella Fastidiosa. World J. Microbiol. Biotechnol. 2012, 28, 1475–1481. [Google Scholar] [CrossRef]
- Bai, Y.; Müller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Münch, P.C.; Spaepen, S.; Remus-Emsermann, M. Functional Overlap of the Arabidopsis Leaf and Root Microbiota. Nature 2015, 528, 364–369. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Nikolaidis, M.; Hesketh, A.; Mossialos, D.; Iliopoulos, I.; Oliver, S.G.; Amoutzias, G.D. A Comparative Analysis of the Core Proteomes within and among the Bacillus Subtilis and Bacillus Cereus Evolutionary Groups Reveals the Patterns of Lineage- and Species-Specific Adaptations. Microorganisms 2022, 10, 1720. [Google Scholar] [CrossRef] [PubMed]
- Ghani, M.; Ansari, A.; Aman, A.; Zohra, R.R.; Siddiqui, N.; Qader, S.A.U. Isolation and Characterization of Different Strains of Bacillus Licheniformis for the Production of Commercially Significant Enzymes. Pak. J. Pharm. Sci. 2013, 26, 691–697. [Google Scholar] [PubMed]
- Sadeghi, H.; Arjmand, S.; Ranaei Siadat, S.O.; Fooladi, J.; Ebrahimipour, G. A Novel Thermostable Alkaline Histamine Oxidase from Glutamicibacter sp. N1A3101, Induced by Histamine and Its Analogue Betahistine. AMB Express 2020, 10, 176. [Google Scholar] [CrossRef] [PubMed]
- Busse, H.J.; Schumann, P. Reclassification of Arthrobacter Enclensis as Pseudarthrobacter Enclensis Comb. Nov., and Emended Descriptions of the Genus Pseudarthrobacter, and the Species Pseudarthrobacter Phenanthrenivorans and Pseudarthrobacter Scleromae. Int. J. Syst. Evol. Microbiol. 2019, 69, 3508–3511. [Google Scholar] [CrossRef]
- Szymańska, S.; Płociniczak, T.; Piotrowska-Seget, Z.; Hrynkiewicz, K. Endophytic and Rhizosphere Bacteria Associated with the Roots of the Halophyte Salicornia europaea L.—Community Structure and Metabolic Potential. Microbiol. Res. 2016, 192, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; DeBolt, S.; Dreyer, J.; Scott, D.; Williams, M.A. Characterization of Culturable Bacterial Endophytes and Their Capacity to Promote Plant Growth from Plants Grown Using Organic or Conventional Practices. Front. Plant Sci. 2015, 6, 490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopes, R.; Tsui, S.; Gonçalves, P.J.R.O.; de Queiroz, M.V. A Look into a Multifunctional Toolbox: Endophytic Bacillus Species Provide Broad and Underexploited Benefits for Plants. World J. Microbiol. Biotechnol. 2018, 34, 94. [Google Scholar] [CrossRef] [PubMed]
- Bunk, B.; Schulz, A.; Stammen, S.; Münch, R.; Warren, M.J.; Jahn, D.; Biedendieck, R. A Short Story about a Big Magic Bug. Bioeng. Bugs 2010, 1, 85–91. [Google Scholar] [CrossRef] [Green Version]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant Beneficial Endophytic Bacteria: Mechanisms, Diversity, Host Range and Genetic Determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar] [CrossRef]
- Wang, H.-F.; Li, L.; Zhang, Y.-G.; Hozzein, W.N.; Zhou, X.-K.; Liu, W.-H.; Duan, Y.-Q.; Li, W.-J. Arthrobacter Endophyticus sp. Nov., an Endophytic Actinobacterium Isolated from Root of Salsola Affinis CA Mey. Int. J. Syst. Evol. Microbiol. 2015, 65, 2154–2160. [Google Scholar] [CrossRef]
- Christakis, C.A.; Daskalogiannis, G.; Chatzaki, A.; Markakis, E.A.; Mermigka, G.; Sagia, A.; Rizzo, G.F.; Catara, V.; Lagkouvardos, I.; Studholme, D.J. Endophytic Bacterial Isolates from Halophytes Demonstrate Phytopathogen Biocontrol and Plant Growth Promotion under High Salinity. Front. Microbiol. 2021, 12, 1001. [Google Scholar] [CrossRef]
- Presta, L.; Fondi, M.; Perrin, E.; Maida, I.; Miceli, E.; Chiellini, C.; Maggini, V.; Bogani, P.; Di Pilato, V.; Rossolini, G.M. Arthrobacter sp. EpRS66 and Arthrobacter sp. EpRS71: Draft Genome Sequences from Two Bacteria Isolated from Echinacea Purpurea Rhizospheric Soil. Front. Microbiol. 2016, 7, 1417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-González, A.J.; Martínez-Hidalgo, P.; Cobo-Díaz, J.F.; Villadas, P.J.; Martínez-Molina, E.; Toro, N.; Tringe, S.G.; Fernández-López, M. The Rhizosphere Microbiome of Burned Holm-Oak: Potential Role of the Genus Arthrobacter in the Recovery of Burned Soils. Sci. Rep. 2017, 7, 6008. [Google Scholar]
- Bazhanov, D.P.; Yang, K.; Li, H.; Li, C.; Li, J.; Chen, X.; Yang, H. Colonization of Plant Roots and Enhanced Atrazine Degradation by a Strain of Arthrobacter Ureafaciens. Appl. Microbiol. Biotechnol. 2017, 101, 6809–6820. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.P.; Rekha, P.D.; Arun, A.B.; Shen, F.T.; Lai, W.-A.; Young, C.C. Phosphate Solubilizing Bacteria from Subtropical Soil and Their Tricalcium Phosphate Solubilizing Abilities. Appl. Soil Ecol. 2006, 34, 33–41. [Google Scholar] [CrossRef]
- Camargo, F.A.O.; Bento, F.M.; Okeke, B.C.; Frankenberger, W.T. Hexavalent Chromium Reduction by an Actinomycete, Arthrobacter Crystallopoietes ES 32. Biol. Trace Elem. Res. 2004, 97, 183–194. [Google Scholar] [CrossRef]
- Kearl, J.; McNary, C.; Lowman, J.S.; Mei, C.; Aanderud, Z.T.; Smith, S.T.; West, J.; Colton, E.; Hamson, M.; Nielsen, B.L. Salt-Tolerant Halophyte Rhizosphere Bacteria Stimulate Growth of Alfalfa in Salty Soil. Front. Microbiol. 2019, 10, 1849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Z.-Y.; Rao, M.P.N.; Wang, H.-F.; Fang, B.-Z.; Liu, Y.-H.; Li, L.; Xiao, M.; Li, W.-J. Transcriptomic Analysis of Two Endophytes Involved in Enhancing Salt Stress Ability of Arabidopsis Thaliana. Sci. Total Environ. 2019, 686, 107–117. [Google Scholar] [CrossRef]
- Margesin, R.; Schinner, F. Potential of Halotolerant and Halophilic Microorganisms for Biotechnology. Extremophiles 2001, 5, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Carroll, G.; Petrini, O. Patterns of Substrate Utilization by Some Fungal Endophytes from Coniferous Foliage. Mycologia 1983, 75, 53–63. [Google Scholar] [CrossRef]

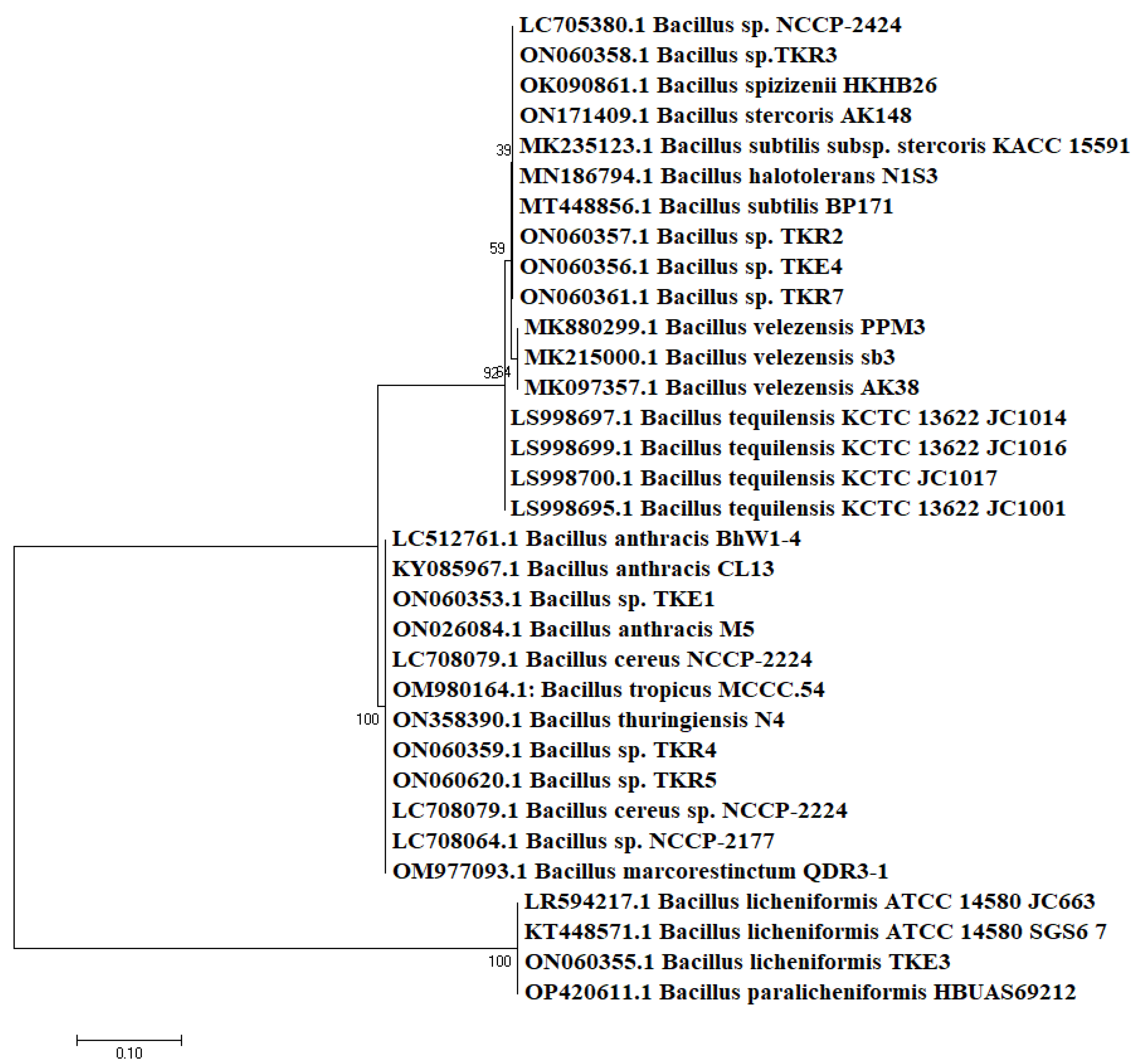
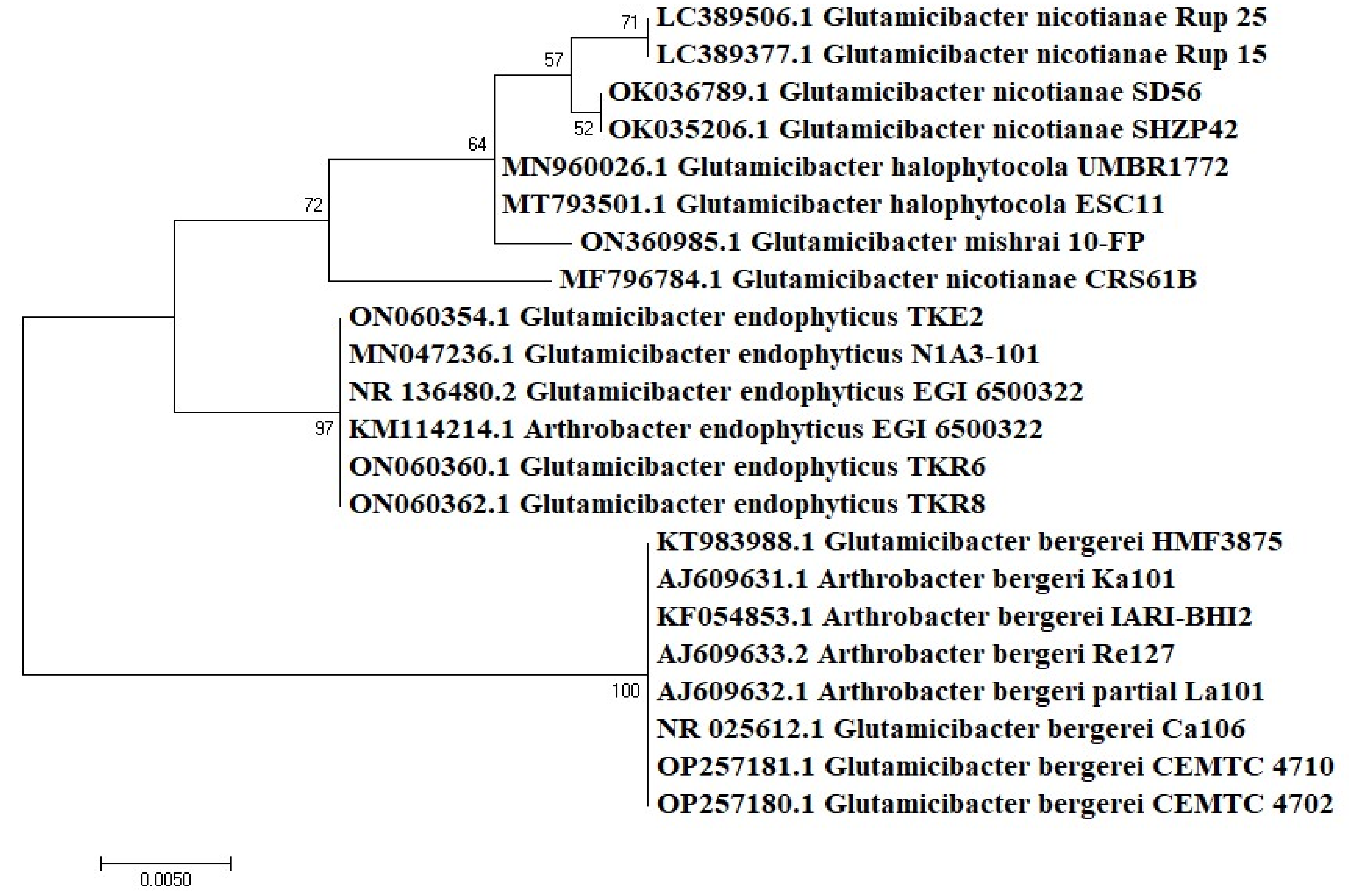
| Isolates | Source | Phylogenetic Identification | Accession Number (GenBank) | Length (bp) | Identity % | Temperature Range (°C) | pH Range | Salt Range (%) |
|---|---|---|---|---|---|---|---|---|
| TKE1 | Roots | Bacillus sp. | ON060353 | 448 | 100 | 15–55, opt. 25 | 5–11, opt. 7 | 2–10, opt. 4 |
| TKE2 | Roots | Glutamicibacter endophyticus | ON060354 | 436 | 99.5 | 15–45, opt. 45 | 5–11, opt. 11 | 2–12, opt. 4 |
| TKE3 | Roots | Bacillus licheniformis | ON060355 | 596 | 100 | 15–45, opt. 25 | 3–11, opt. 7 | 2–8, opt. 4 |
| TKE4 | Roots | Bacillus sp. | ON060356 | 310 | 100 | 15–45, opt. 45 | 3–11, opt. 7 | 2–12, opt. 6 |
| TKR2 | Soil | Bacillus sp. | ON060357 | 455 | 100 | 15–55, opt. 37 | 1–9, opt. 5 | 2–12, opt. 4 |
| TKR3 | Soil | Bacillus sp. | ON060358 | 457 | 100 | 15–55, opt. 37 | 1–9, opt. 7 | 2–8, opt. 2 |
| TKR4 | Soil | Bacillus sp. | ON060359 | 443 | 100 | 15–45, opt. 37 | 3–11, opt. 9 | 2–6, opt. 4 |
| TKR5 | Soil | Bacillus sp. | ON060620 | 189 | 100 | 15–45, opt. 37 | 3–11, opt. 7 | 2–8, opt. 4 |
| TKR6 | Soil | Glutamicibacter endophyticus | ON060360 | 392 | 100 | 15–55, opt. 37 | 1–9, opt. 5 | 2–8, opt. 4 |
| TKR7 | Soil | Bacillus sp. | ON060361 | 450 | 100 | 15–45, opt. 37 | 5–11, opt. 7 | 2–6, opt. 2 |
| TKR8 | Soil | Glutamicibacter endophyticus | ON060362 | 422 | 98.2 | 15–55, opt. 45 | 3–11, opt. 11 | 2–12, opt. 2 |
| Bacillus licheniformis KIBGE-IB4 * [32] | Bacillus licheniformis TKE3 | |
|---|---|---|
| Lactose | − | + |
| Fructose | ND | + |
| Maltose | + | + |
| Glucose | + | + |
| Mannose | + | − |
| Sucrose | + | + |
| Catalase | + | + |
| Oxidase | ND | − |
| MR | ND | + |
| VP | ND | + |
| Citrate | + | + |
| Starch | + | + |
| NaCl range | 7–12% | 2–8% |
| Temperature | 30–55 °C | 15–45 °C |
| pH | 4–11 | 3–11 |
| G. endophyticus [33,34] | G. endophyticus TKE2 | G. endophyticus TKR6 | G. endophyticus TKR8 | |
|---|---|---|---|---|
| Lactose | − | − | − | − |
| Fructose | + | + | + | + |
| Maltose | ND | + | − | − |
| Glucose | + | + | + | + |
| Mannose | ND | − | − | − |
| Sucrose | ND | − | − | + |
| Catalase | + | + | + | + |
| Oxidase | − | − | − | − |
| MR | ND | − | − | − |
| VP | ND | + | + | + |
| Citrate | + | + | + | + |
| Starch | + | + | + | + |
| NaCl range | 0–13% | 2–12% | 2–8% | 2–12% |
| Temperature | 5–35 °C | 15–45 °C | 15–55 °C | 15–55 °C |
| Isolates | Enzyme Activity (IU mL−1) | ||||
|---|---|---|---|---|---|
| Xylanase | Amylase | Cellulase | Pectinase | Lipase | |
| TKE1 | N/D | 0.76 ± 0.03 | 0.67 ± 0.03 | 3.12 ± 0.04 | 5.77 ± 5.7 |
| TKE2 | N/D | 4.39 ± 0.09 | 0.37 ± 0.03 | 3.49 ± 0.06 | 3.22 ± 4.7 |
| TKE3 | N/D | N/D | N/D | N/D | 4.11 ± 3.4 |
| TKE4 | 9.35 ± 0.11 | 2.04 ± 0.1 | N/D | 3.77 ± 0.04 | 2.8 ± 0.23 |
| TKR2 | 2.31 ± 0.07 | 2.63 ± 0.06 | 1.18 ± 0.02 | N/D | 0.35 ± 0.52 |
| TKR3 | N/D | 1.24 ± 0.02 | N/D | 2.82 ± 0.05 | N/D |
| TKR4 | 3.08 ± 0.03 | N/D | 0.76 ± 0.01 | 2.81 ± 0.1 | 17.32 ± 2.8 |
| TKR5 | 6.61 ± 0.04 | 2.75 ± 0.01 | 0.76 ± 0.09 | N/D | 5.07 ± 2.06 |
| TKR6 | 5.06 ± 0.01 | 2.45 ± 0.07 | N/D | 1.29 ± 0.075 | 7.79 ± 1.56 |
| TKR7 | 2.07 ± 0.003 | 0.89 ± 0.02 | 1.18 ± 0.01 | 1.99 ± 0.032 | 12.79 ± 1.98 |
| TKR8 | N/D | 3.55 ± 0.05 | 1.68 ± 0.03 | 1.15 ± 0.028 | 15.81 ± 1.48 |
| Isolates | Enzyme Activity (IU mL−1) | ||||
|---|---|---|---|---|---|
| Xylanase | Amylase | Cellulase | Pectinase | Lipase | |
| TKE1 | 3.11 ± 0.08 | N/D | N/D | 3.001 ± 0.05 | N/D |
| TKE2 | 2.31 ± 0.05 | 2.81 ± 0.01 | N/D | 0.69 ± 0.01 | N/D |
| TKE3 | 2.01 ± 0.02 | N/D | N/D | 2.05 ± 0.6 | 5.05 ± 0.95 |
| TKE4 | 2.42 ± 0.01 | 3.26 ± 0.04 | N/D | 2.55 ± 0.03 | N/D |
| TKR2 | 5.8 ± 0.03 | 4.64 ± 0.04 | 0.69 ± 0.03 | 2.18 ± 0.01 | N/D |
| TKR3 | 2.55 ± 0.02 | N/D | 3.3 ± 0.06 | 2.92 ± 0.05 | N/D |
| TKR4 | 2.72 ± 0.08 | 4.05 ± 0.09 | 0.98 ± 0.02 | 1.44 ± 0.01 | 6.72 ± 1.68 |
| TKR5 | 2.01 ± 0.05 | 0.74 ± 0.02 | 4.59 ± 0.08 | 1.97 ± 0.05 | N/D |
| TKR6 | N/D | 3.13 ± 0.08 | 0.98 ± 0.02 | 4.97 ± 0.06 | N/D |
| TKR7 | 0.7 ± 0.01 | N/D | 2.91 ± 0.05 | 1.85 ± 0.05 | 17.52 ± 1.17 |
| TKR8 | 2.8 ± 0.07 | N/D | N/D | 2.67 ± 0.03 | 6.77 ± 1.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, T.; Alzahrani, O.M.; Sohail, M.; Hasan, K.A.; Gulzar, S.; Rehman, A.U.; Mahmoud, S.F.; Alswat, A.S.; Abdel-Gawad, S.A. Enzyme Profiling and Identification of Endophytic and Rhizospheric Bacteria Isolated from Arthrocnemum macrostachyum. Microorganisms 2022, 10, 2112. https://doi.org/10.3390/microorganisms10112112
Khan T, Alzahrani OM, Sohail M, Hasan KA, Gulzar S, Rehman AU, Mahmoud SF, Alswat AS, Abdel-Gawad SA. Enzyme Profiling and Identification of Endophytic and Rhizospheric Bacteria Isolated from Arthrocnemum macrostachyum. Microorganisms. 2022; 10(11):2112. https://doi.org/10.3390/microorganisms10112112
Chicago/Turabian StyleKhan, Tooba, Othman M. Alzahrani, Muhammad Sohail, Khwaja Ali Hasan, Salman Gulzar, Ammad Ur Rehman, Samy F. Mahmoud, Amal S. Alswat, and Shebl Abdallah Abdel-Gawad. 2022. "Enzyme Profiling and Identification of Endophytic and Rhizospheric Bacteria Isolated from Arthrocnemum macrostachyum" Microorganisms 10, no. 11: 2112. https://doi.org/10.3390/microorganisms10112112





