The Emergence and Dynamics of Tick-Borne Encephalitis Virus in a New Endemic Region in Southern Germany
Abstract
:1. Introduction
2. Material and Methods
2.1. Case Definition and Case Data
2.2. Statistical Analysis
2.3. Identification of TBEV Natural Foci
2.4. Sampling and Processing of Ticks
2.5. Sequencing and Analysis of TBEV Genomes
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beaute, J.; Spiteri, G.; Warns-Petit, E.; Zeller, H. Tick-Borne Encephalitis in Europe, 2012 to 2016. Euro Surveill. 2018, 23, 1800201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demina, T.; Dzhioev, Y.; Verkhozina, M.; Kozlova, I.; Tkachev, S.; Plyusnin, A.; Doroshchenko, E.; Lisak, O.; Zlobin, V. Genotyping and characterization of the geographical distribution of tick-borne encephalitis virus variants with a set of molecular probes. J. Med. Virol. 2010, 82, 965–976. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Shang, G.; Lu, S.; Yang, J.; Xu, J. A New Subtype of Eastern Tick-Borne Encephalitis Virus Discovered in Qing-hai-Tibet Plateau, China. Emerg. Microbes Infect. 2018, 7, 1–9. [Google Scholar]
- Kutschera, L.S.; Wolfinger, M.T. Evolutionary traits of Tick-borne encephalitis virus: Pervasive non-coding RNA structure conservation and molecular epidemiology. Virus Evol. 2022, 8, veac051. [Google Scholar] [CrossRef]
- Kozlova, I.V.; Demina, T.V.; Tkachev, S.E.; Doroshchenko, E.K.; Lisak, O.V.; Verkhozina, M.M.; Karan, L.S.; Dzhioev, Y.P.; Paramonov, A.I.; Suntsova, O.V.; et al. Characteristics of the Baikal subtype of tick-borne encephalitis virus circulating in Eastern Siberia. Acta Biomed. Sci. 2018, 3, 53–60. [Google Scholar] [CrossRef] [Green Version]
- Heinze, D.M.; Gould, E.A.; Forrester, N.L. Revisiting the Clinal Concept of Evolution and Dispersal for the Tick-Borne Flaviviruses by Using Phylogenetic and Biogeographic Analyses. J. Virol. 2012, 86, 8663–8671. [Google Scholar] [CrossRef] [Green Version]
- Kovalev, S.Y.; Chernykh, D.N.; Kokorev, V.S.; Snitkovskaya, T.E.; Romanenko, V.V. Origin and Distribution of Tick-Borne Encephalitis Virus Strains of the Siberian Subtype in the Middle Urals, the North-West of Russia and the Baltic Countries. J. Gen. Virol. 2009, 90 Pt 12, 2884–2892. [Google Scholar] [CrossRef]
- Golovljova, I.; Katargina, O.; Geller, J.; Tallo, T.; Mittženkov, V.; Vene, S.; Nemirov, K.; Kutsenko, A.; Kilosanidze, G.; Vasilenko, V.; et al. Unique signature amino acid substitution in Baltic tick-borne encephalitis virus (TBEV) strains within the Siberian TBEV subtype. Int. J. Med. Microbiol. 2008, 298, 108–120. [Google Scholar] [CrossRef]
- Weidmann, M.; Růžek, D.; Křivanec, K.; Zöller, G.; Essbauer, S.; Pfeffer, M.; Zanotto, P.M.D.A.; Hufert, F.T.; Dobler, G. Relation of genetic phylogeny and geographical distance of tick-borne encephalitis virus in central Europe. J. Gen. Virol. 2011, 92, 1906–1916. [Google Scholar] [CrossRef]
- Weidmann, M.; Frey, S.; Freire, C.C.M.; Essbauer, S.; Ruzek, D.; Klempa, B.; Zubriková, D.; Vögerl, M.; Pfeffer, M.; Hufert, F.T.; et al. Molecular phylogeography of tick-borne encephalitis virus in central Europe. J. Gen. Virol. 2013, 94, 2129–2139. [Google Scholar] [CrossRef]
- Barzon, L. Ongoing and emerging arbovirus threats in Europe. J. Clin. Virol. 2018, 107, 38–47. [Google Scholar] [CrossRef]
- Atkinson, B.; Hewson, R. Emerging arboviruses of clinical importance in Central Asia. J. Gen. Virol. 2018, 99, 1172–1184. [Google Scholar] [CrossRef]
- Weststrate, A.C.; Knapen, D.; Laverman, G.D.; Schot, B.; Prick, J.J.; Spit, S.A.; Reimerink, J.; Rockx, B.; Geeraedts, F. Increasing Evidence of Tick-Borne Encephalitis (TBE) Virus Transmission, The Netherlands, June 2016. Euro Surveill. 2017, 22, 30482. [Google Scholar] [CrossRef]
- Holding, M.; Dowall, S.; Hewson, R. Detection of tick-borne encephalitis virus in the UK. Lancet 2020, 395, 411. [Google Scholar] [CrossRef] [Green Version]
- Holding, M.; Dowall, S.D.; Medlock, J.M.; Carter, D.P.; McGinley, L.; Curran-French, M.; Pullan, S.T.; Chamberlain, J.; Hansford, K.M.; Baylis, M.; et al. Detection of New Endemic Focus of Tick-Borne Encephalitis Virus (TBEV), Hampshire/Dorset Border, England, September 2019. Euro Surveill. 2019, 24, 1900658. [Google Scholar] [CrossRef] [Green Version]
- Heinz, F.X.; Stiasny, K.; Holzmann, H.; Kundi, M.; Sixl, W.; Wenk, M.; Kainz, W.; Essl, A.; Kunz, C. Emergence of tick-borne encephalitis in new endemic areas in Austria: 42 years of surveillance. Euro Surveill. 2015, 20, 21077. [Google Scholar] [CrossRef] [Green Version]
- Dobler, G.; Essbauer, S.; Terzioglu, R.; Thomas, A.; Wolfel, R. Prevalence of Tick-Borne Encephalitis Virus and Rickettsiae in Ticks of the District Burgenland, Austria. Wien Klin. Wochenschr. 2008, 120 (Suppl. 4), 45–48. [Google Scholar] [CrossRef]
- Schwaiger, M.; Cassinotti, P. Development of a quantitative real-time RT-PCR assay with internal control for the laboratory detection of tick borne encephalitis virus (TBEV) RNA. J. Clin. Virol. 2002, 27, 136–145. [Google Scholar] [CrossRef]
- Kupča, A.M.; Essbauer, S.; Zoeller, G.; de Mendonça, P.G.; Brey, R.; Rinder, M.; Pfister, K.; Spiegel, M.; Doerrbecker, B.; Pfeffer, M.; et al. Isolation and molecular characterization of a tick-borne encephalitis virus strain from a new tick-borne encephalitis focus with severe cases in Bavaria, Germany. Ticks Tick-Borne Dis. 2010, 1, 44–51. [Google Scholar] [CrossRef]
- Kosakovsky Pond, S.L.; Posada, D.; Gravenor, M.B.; Woelk, C.H.; Frost, S.D. Automated Phylogenetic Detection of Re-combination Using a Genetic Algorithm. Mol. Biol. Evol. 2006, 23, 1891–1901. [Google Scholar] [CrossRef] [Green Version]
- Weaver, S.; Shank, S.D.; Spielman, S.J.; Li, M.; Muse, S.V.; Pond, S.L.K. Datamonkey 2.0: A Modern Web Application for Characterizing Selective and Other Evolutionary Processes. Mol. Biol. Evol. 2018, 35, 773–777. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Sagulenko, P.; Puller, V.; Neher, R.A. TreeTime: Maximum-likelihood phylodynamic analysis. Virus Evol. 2018, 4, vex042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritz, R.; Blazevic, J.; Taucher, C.; Pangerl, K.; Heinz, F.X.; Stiasny, K. The Unique Transmembrane Hairpin of Flavivirus Fusion Protein E Is Essential for Membrane Fusion. J. Virol. 2011, 85, 4377–4385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rey, F.A.; Heinz, F.X.; Mandl, C.; Kunz, C.; Harrison, S.C. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 1995, 375, 291–298. [Google Scholar] [CrossRef]
- Bestehorn, M.; Weigold, S.; Kern, W.V.; Chitimia-Dobler, L.; Mackenstedt, U.; Dobler, G.; Borde, J.P. Phylogenetics of tick-borne encephalitis virus in endemic foci in the upper Rhine region in France and Germany. PLoS ONE 2018, 13, e0204790. [Google Scholar] [CrossRef]
- Formanova, P.; Cerny, J.; Bolfikova, B.C.; Valdes, J.J.; Kozlova, I.; Dzhioev, Y.; Ruzek, D. Full Genome Sequences and Molecular Characterization of Tick-Borne Encephalitis Virus Strains Isolated from Human Patients. Ticks Tick Borne Dis. 2015, 6, 38–46. [Google Scholar] [CrossRef]
- Gäumann, R.; Růžek, D.; Mühlemann, K.; Strasser, M.; Beuret, C.M. Phylogenetic and virulence analysis of tick-borne encephalitis virus field isolates from Switzerland. J. Med. Virol. 2011, 83, 853–863. [Google Scholar] [CrossRef]
- Gaäumann, R.; Muúhlemann, K.; Strasser, M.; Beuret, C.M. High-Throughput Procedure for Tick Surveys of Tick-Borne Encephalitis Virus and Its Application in a National Surveillance Study in Switzerland. Appl. Environ. Microbiol. 2010, 76, 4241–4249. [Google Scholar] [CrossRef] [Green Version]
- Ecker, M.; Allison, S.L.; Meixner, T.; Heinz, F.X. Sequence analysis and genetic classification of tick-borne encephalitis viruses from Europe and Asia. J. Gen. Virol. 1999, 80, 179–185. [Google Scholar] [CrossRef]
- Borde, J.P.; Kaier, K.; Hehn, P.; Matzarakis, A.; Frey, S.; Bestehorn, M.; Dobler, G.; Chitimia-Dobler, L. The Complex Interplay of Climate, TBEV Vector Dynamics and TBEV Infection Rates in Ticks-Monitoring a Natural TBEV Focus in Germany, 2009–2018. PLoS ONE 2021, 16, e0244668. [Google Scholar] [CrossRef]
- Hannoun, C.; Chatelain, J.; Krams, S.; Guillon, J.C. Isolation, in Alsace, of the tick encephalitis virus (arbovirus, group B). Comptes Rendus Hebd. Seances L’Academie Sci. Ser. D Sci. Nat. 1971, 272, 766–768. [Google Scholar]
- Jääskeläinen, A.; Tonteri, E.; Pieninkeroinen, I.; Sironen, T.; Voutilainen, L.; Kuusi, M.; Vaheri, A.; Vapalahti, O. Siberian subtype tick-borne encephalitis virus in Ixodes ricinus in a newly emerged focus, Finland. Ticks Tick-Borne Dis. 2016, 7, 216–223. [Google Scholar] [CrossRef]
- Suzuki, Y. Multiple transmissions of tick-borne encephalitis virus between Japan and Russia. Genes Genet. Syst. 2007, 82, 187–195. [Google Scholar] [CrossRef] [Green Version]
- Aspöck, H. The synecological system of arboviruses and its possible modification by man. Zent. Bakteriol. Parasitenkd. Infekt. Hyg. Abt. Med. Bakteriol. Virusforsch. Parasitol. Orig. 1970, 213, 434–454. [Google Scholar]
- Nah, K.; Bede-Fazekas, Á.; Trájer, A.J.; Wu, J. The potential impact of climate change on the transmission risk of tick-borne encephalitis in Hungary. BMC Infect. Dis. 2020, 20, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Zeman, P. Prolongation of Tick-Borne Encephalitis Cycles in Warmer Climatic Conditions. Int. J. Environ. Res. Public Health 2019, 16, 4532. [Google Scholar] [CrossRef] [Green Version]
- Leibovici, D.G.; Bylund, H.; Björkman, C.; Tokarevich, N.; Thierfelder, T.; Evengård, B.; Quegan, S. Associating Land Cover Changes with Patterns of Incidences of Climate-Sensitive Infections: An Example on Tick-Borne Diseases in the Nordic Area. Int. J. Environ. Res. Public Health 2021, 18, 10963. [Google Scholar] [CrossRef]
- Randolph, S.E.; Eden-Tbd Sub-Project Team. Human Activities Predominate in Determining Changing Incidence of Tick-Borne Encephalitis in Europe. Euro Surveill. 2010, 15, 24–31. [Google Scholar] [CrossRef] [Green Version]
- Stefanoff, P.; Rosinska, M.; Samuels, S.; White, D.J.; Morse, D.L.; Randolph, S.E. A National Case-Control Study Identifies Human Socio-Economic Status and Activities as Risk Factors for Tick-Borne Encephalitis in Poland. PLoS ONE 2012, 7, e45511. [Google Scholar] [CrossRef]
- Brugger, K.; Walter, M.; Chitimia-Dobler, L.; Dobler, G.; Rubel, F. Seasonal cycles of the TBE and Lyme borreliosis vector Ixodes ricinus modelled with time-lagged and interval-averaged predictors. Exp. Appl. Acarol. 2017, 73, 439–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kříž, B.; Kott, I.; Daniel, M.; Vráblík, T.; Beneš, Č. Impact of climate changes on the incidence of tick-borne encephalitis in the Czech Republic in 1982–2011. Epidemiol. Mikrobiol. Imunol. Cas. Spol. Epidemiol. A Mikrobiol. Ceske Lek. Spol. J.E. Purkyne 2015, 64, 24–32. [Google Scholar]
- Daniel, M.; Danielová, V.; Fialová, A.; Malý, M.; Kříž, B.; Nuttall, P.A. Increased Relative Risk of Tick-Borne Encephalitis in Warmer Weather. Front. Cell. Infect. Microbiol. 2018, 8, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gondard, M.; Michelet, L.; Nisavanh, A.; Devillers, E.; Delannoy, S.; Fach, P.; Aspan, A.; Ullman, K.; Chirico, J.; Hoffmann, B.; et al. Prevalence of Tick-Borne Viruses in Ixodes ricinus Assessed by High-Throughput Real-Time PCR. Pathog. Dis. 2018, 76, fty083. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, J.H.-O.; Golovljova, I.; Vene, S.; Jaenson, T.G. Prevalence of tick-borne encephalitis virus in Ixodes ricinus ticks in northern Europe with particular reference to Southern Sweden. Parasites Vectors 2014, 7, 102. [Google Scholar] [CrossRef] [Green Version]
- Drelich, A.; Andreassen, Å.; Vainio, K.; Kruszyński, P.; Wąsik, T.J. Prevalence of tick-borne encephalitis virus in a highly urbanized and low risk area in Southern Poland. Ticks Tick-Borne Dis. 2014, 5, 663–667. [Google Scholar] [CrossRef]
- Zubrikova, D.; Wittmann, M.; Honig, V.; Svec, P.; Vichova, B.; Essbauer, S.; Dobler, G.; Grubhoffer, L.; Pfister, K. Prevalence of Tick-Borne Encephalitis Virus and Borrelia Burgdorferi Sensu Lato in Ixodes ricinus Ticks in Lower Bavaria and Upper Palatinate, Germany. Ticks Tick Borne Dis. 2020, 11, 101375. [Google Scholar] [CrossRef]
- Bertrand, Y.J.; Johansson, M.; Norberg, P. Revisiting Recombination Signal in the Tick-Borne Encephalitis Virus: A Sim-ulation Approach. PLoS ONE 2016, 11, e0164435. [Google Scholar] [CrossRef] [Green Version]
- Uzcategui, N.Y.; Sironen, T.; Golovljova, I.; Jaaskelainen, A.E.; Valimaa, H.; Lundkvist, A.; Plyusnin, A.; Vaheri, A.; Vapalahti, O. Rate of Evolution and Molecular Epidemiology of Tick-Borne Encephalitis Virus in Europe, Including Two Isolations from the Same Focus 44 Years Apart. J. Gen. Virol. 2012, 93 Pt 4, 786–796. [Google Scholar] [CrossRef]

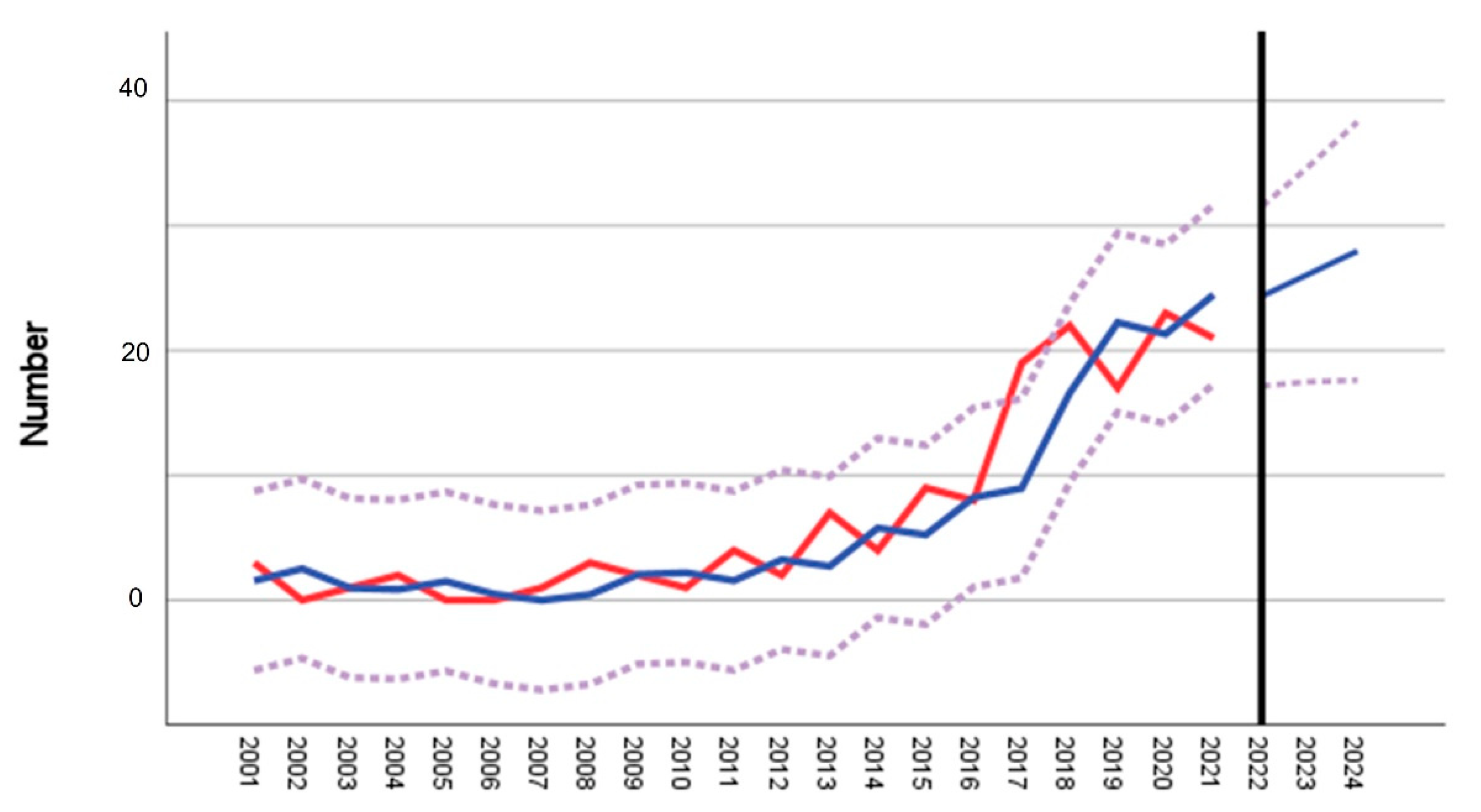
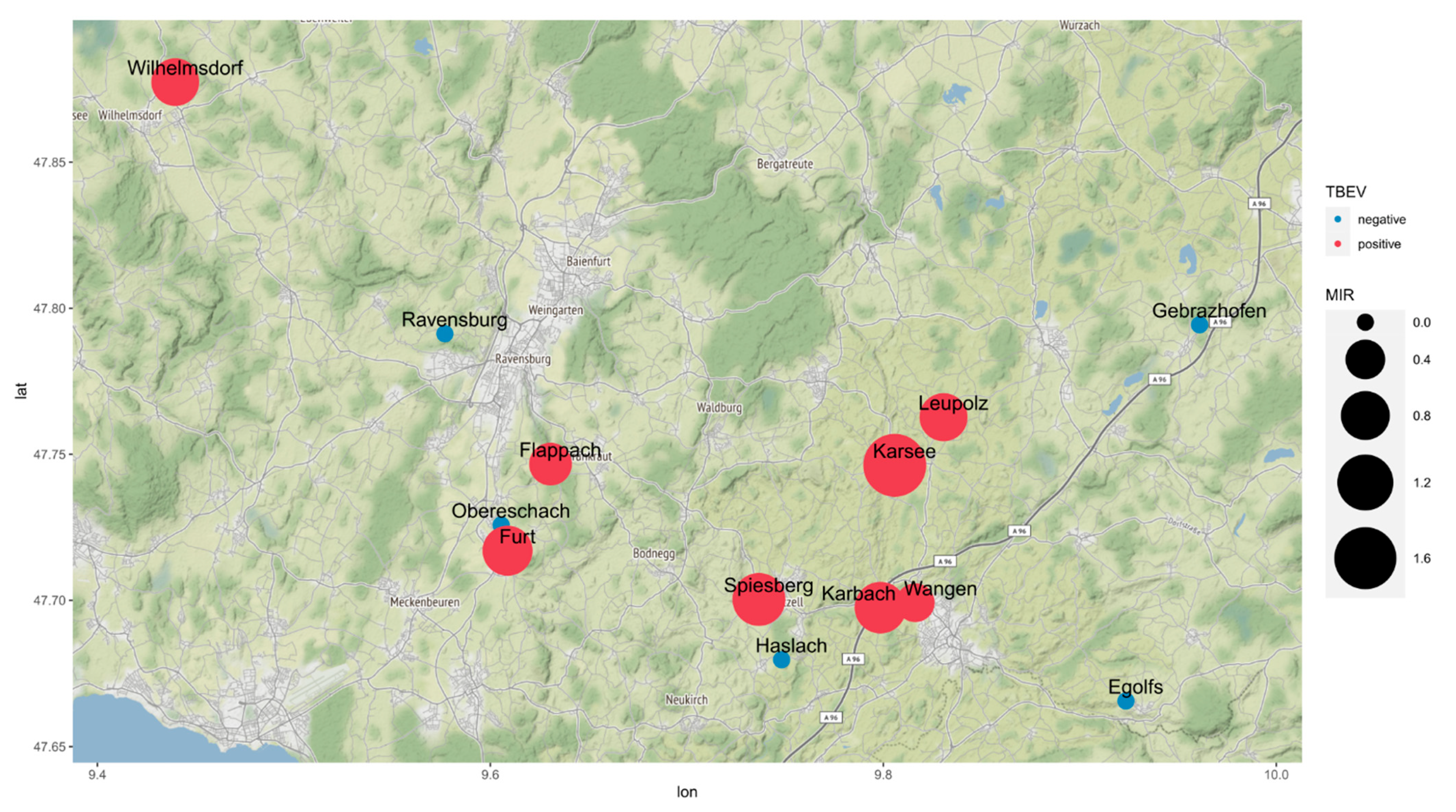
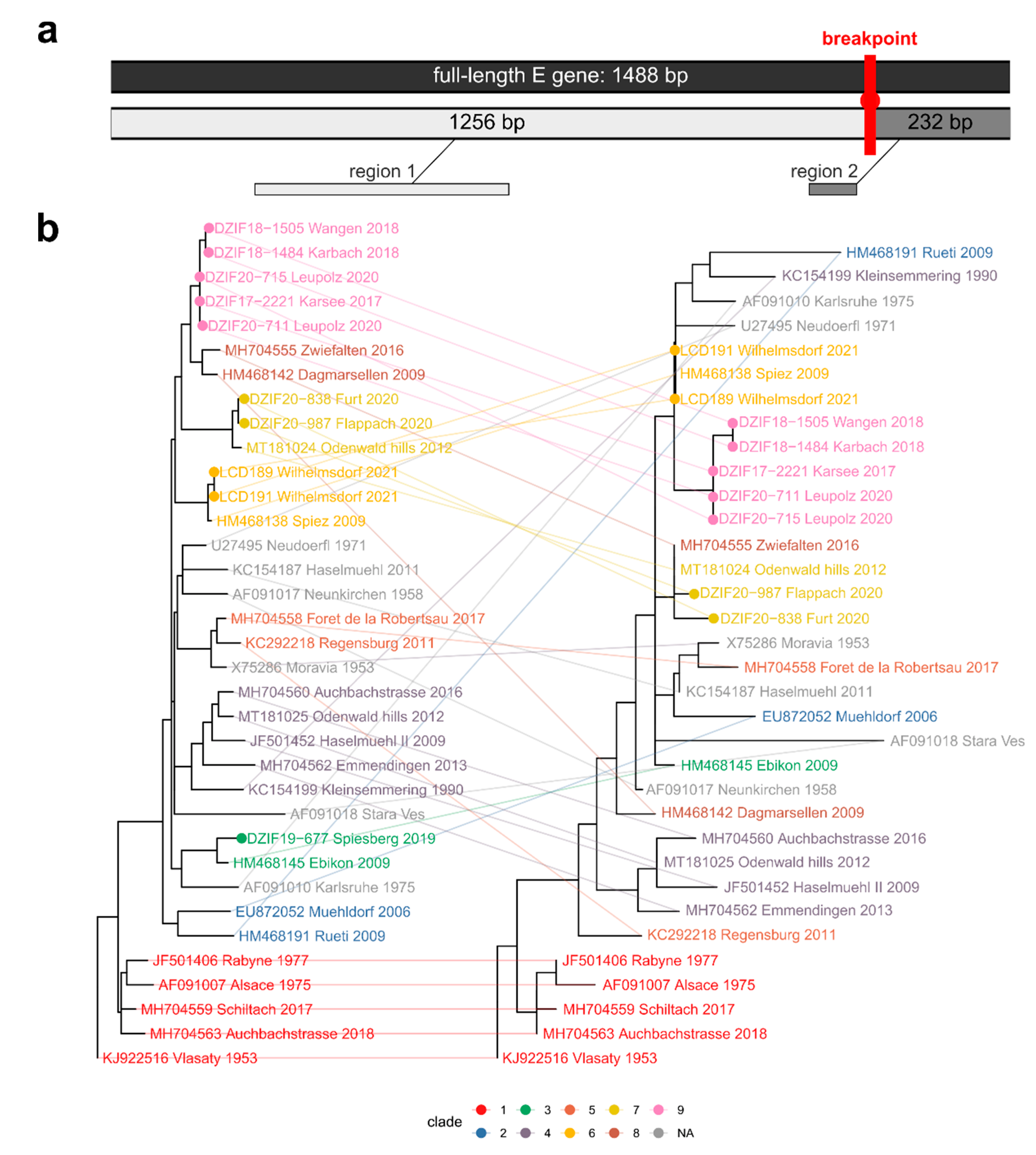
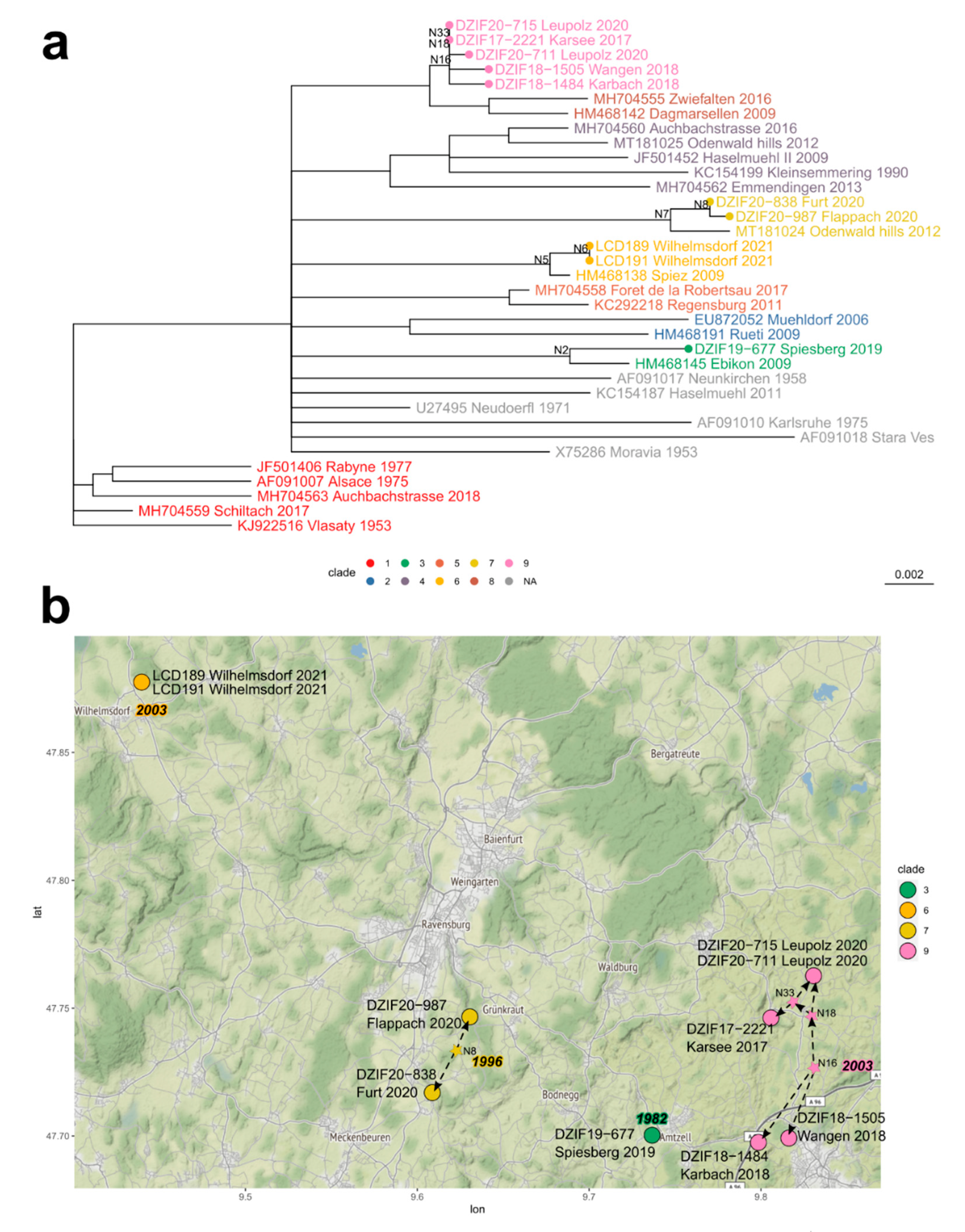
| District ID (Figure 1b) | District | Cases 2001–2012 | Mean 2001–2012 | Cases 2017–2020 | Mean 2017–2020 | Fold Increase | t-Test p | Time Series/Forecast Model 2001–2021 R2 |
|---|---|---|---|---|---|---|---|---|
| A | Ravensburg | 19 | 1.6 | 81 | 20.3 | 13.0 | <0.001 *** | 0.819 |
| B | Bodensee | 60 | 5.0 | 31 | 7.8 | 1.6 | 0.027 * | 0.79 |
| C | Reutlingen | 30 | 2.5 | 24 | 6 | 2.4 | 0.009 ** | 0.034 |
| D | Sigmaringen | 25 | 2.1 | 33 | 8.3 | 3.9 | 0.02 * | 0.102 |
| E | Tübingen | 48 | 4.0 | 31 (17) + | 7.8 (4.3) + | 0.5 (0.9) + | n.d. | n.d. |
| F | Zollernalb | 81 | 7.8 | 53 | 13.3 | 1.7 | 0.011 * | 0.027 |
| G | Oberallgäu | 4 | 0.3 | 19 | 4.75 | 15.8 | <0.001 *** | 0.321 |
| H | Unterallgäu | 11 | 0.9 | 23 | 5.8 | 6.4 | 0.071 | −0.014 |
| I | Lindau | 4 | 0.3 | 18 | 4.5 | 15 | <0.001 *** | 0.589 |
| J | Biberach | 7 | 0.6 | 18 | 4.5 | 7.7 | 0.027 * | 0.103 |
| Location | Altitude (m above s.l.) | Sampling Date | Males | Females | Nymphs | Total MIR (%) |
|---|---|---|---|---|---|---|
| Karsee | 635 | 26 September 2017 | 32 (1) | 20 | 8 | 1.7 |
| Wangen | 573 | 27 June 2018 | 24 (1) | 14 | 233 | 0.37 |
| Karbach | 579 | 27 June 2018 | 14 | 15 (1) | 77 | 0.94 |
| Ravensburg | 510 | 16 July 2018 | 8 | 7 | 150 | 0 |
| Gebrazhofen | 679 | 16 July 2018 | 26 | 19 | 169 | 0 |
| Spiesberg | 539 | 03 July 2019 | 10 | 19 (1) | 168 (1) | 1.0 |
| Haslach | 667 | 03 July 2019 | 27 | 23 | 285 | 0 |
| Egolfs | 646 | 28 May 2020 | 51 | 47 | 229 | 0 |
| Leupolz | 601 | 23 July 2020 | 100 (2) | 79 (2) | 346 | 0.76 |
| Obereschach | 485 | 19 August 2020 | 2 | 5 | 39 | 0 |
| Furt | 496 | 19 August 2020 | 6 | 0 | 110 (1) | 0.86 |
| Flappach | 556 | 14 September 2020 | 35 | 30 (1) | 127 | 0.52 |
| Wilhelmsdorf | 679 | 2018/2019 | 31 (1) | 14 | 356 (2) | 0.75 |
| Total | 366 (5) | 292 (5) | 2297 (4) | 0.47 |
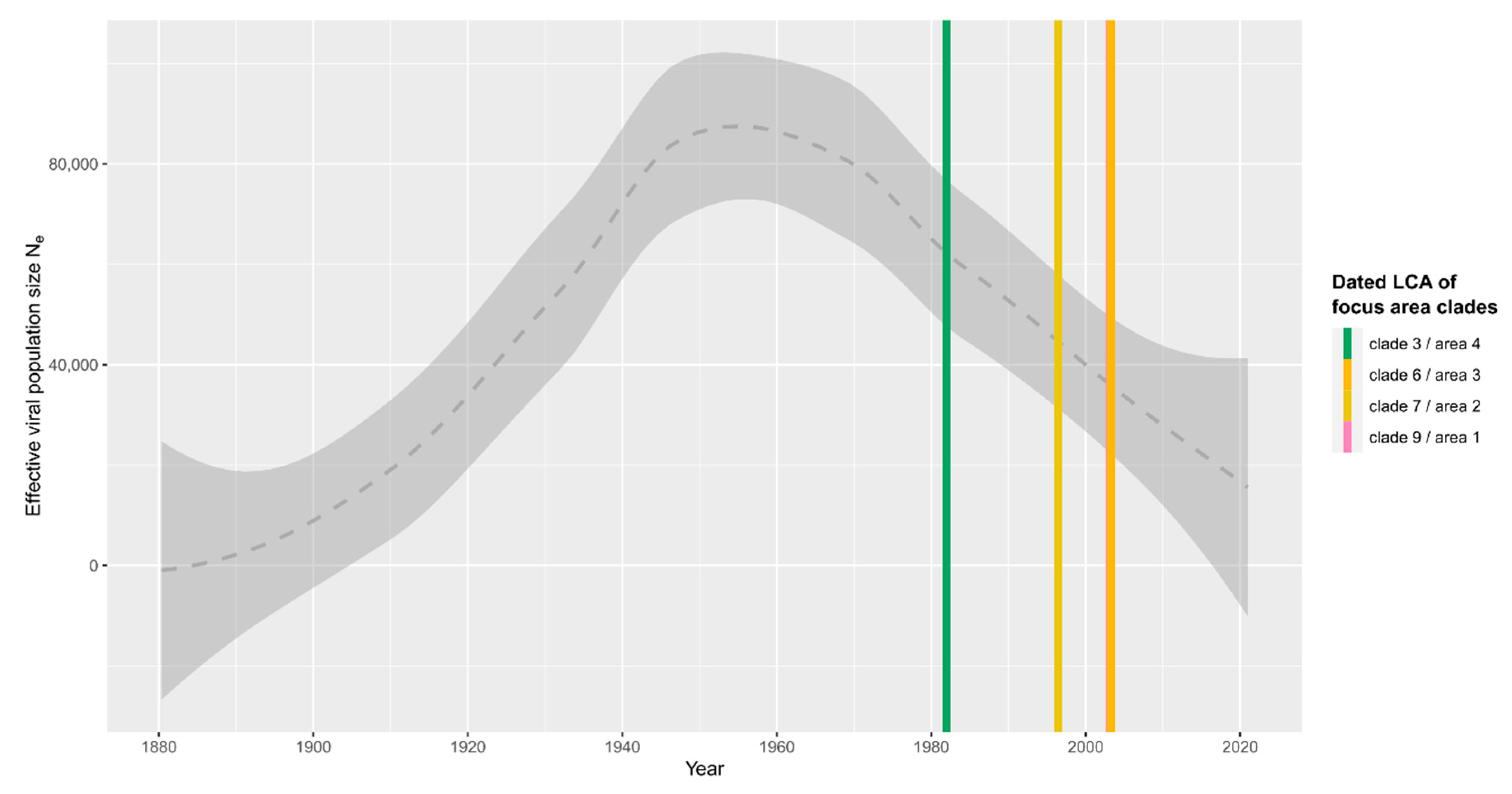
| Focus Area | Year | 90% Confidence Interval | ||||
|---|---|---|---|---|---|---|
| Clade | Node/Taxon | Estimate | Min | Max | Location | |
| 1 | 9 | N16 | 2003 | 1989 | 2008 | |
| DZIF18-1484 | 2018 | 2018 | 2018 | Karbach | ||
| DZIF18-1505 | 2018 | 2018 | 2018 | Wangen | ||
| N18 | 2006 | 1999 | 2015 | |||
| DZIF20-711 | 2020 | 2020 | 2020 | Leupolz | ||
| N33 | 2014 | 2007 | 2017 | |||
| DZIF17-2221 | 2017 | 2017 | 2017 | Karsee | ||
| DZIF20-715 | 2020 | 2020 | 2020 | Leupolz | ||
| 2 | 7 | N7 | 1996 | 1983 | 2006 | |
| MT181024 | 2012 | 2012 | 2012 | Odenwald | ||
| N8 | 2014 | 2004 | 2018 | |||
| DZIF20-987 | 2020 | 2020 | 2020 | Flappach | ||
| DZIF20-838 | 2020 | 2020 | 2020 | Furt | ||
| 3 | 6 | N5 | 2003 | 1987 | 2007 | |
| HM468138 | 2009 | 2009 | 2009 | Spiez | ||
| N6 | 2017 | 2006 | 2020 | |||
| LCD191 | 2021 | 2021 | 2021 | Wilhelmsdorf | ||
| LCD189 | 2021 | 2021 | 2021 | Wilhelmsdorf | ||
| 4 | 3 | N2 | 1982 | 1966 | 1998 | |
| HM468145 | 2009 | 2009 | 2009 | Ebikon | ||
| DZIF19-677 | 2019 | 2019 | 2019 | Spiesberg | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lang, D.; Chitimia-Dobler, L.; Bestehorn-Willmann, M.; Lindau, A.; Drehmann, M.; Stroppel, G.; Hengge, H.; Mackenstedt, U.; Kaier, K.; Dobler, G.; et al. The Emergence and Dynamics of Tick-Borne Encephalitis Virus in a New Endemic Region in Southern Germany. Microorganisms 2022, 10, 2125. https://doi.org/10.3390/microorganisms10112125
Lang D, Chitimia-Dobler L, Bestehorn-Willmann M, Lindau A, Drehmann M, Stroppel G, Hengge H, Mackenstedt U, Kaier K, Dobler G, et al. The Emergence and Dynamics of Tick-Borne Encephalitis Virus in a New Endemic Region in Southern Germany. Microorganisms. 2022; 10(11):2125. https://doi.org/10.3390/microorganisms10112125
Chicago/Turabian StyleLang, Daniel, Lidia Chitimia-Dobler, Malena Bestehorn-Willmann, Alexander Lindau, Marco Drehmann, Gabriele Stroppel, Helga Hengge, Ute Mackenstedt, Klaus Kaier, Gerhard Dobler, and et al. 2022. "The Emergence and Dynamics of Tick-Borne Encephalitis Virus in a New Endemic Region in Southern Germany" Microorganisms 10, no. 11: 2125. https://doi.org/10.3390/microorganisms10112125






