Natural Infections of Potato Plants Grown from Minitubers with Blackleg-Causing Soft Rot Pectobacteriaceae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Sampling Plant Material
2.3. Pathogen Detection Procedure
2.4. Enrichment Procedure
2.5. DNA Extraction Procedure
2.6. Multiplex TaqMan Assay for Blackleg-Causing Pathogens
2.7. Species-Specific Simplex TaqMan Assays
2.8. Triplex TaqMan Assay for a Clade of Virulent P. brasiliense Strains
| Assay | Strain | Name | Sequence | Target | Reporter Dye | Addition | Reference |
|---|---|---|---|---|---|---|---|
| * and ** | Dickeya sp. | Dickeya Fw284 | tgtgcgttttcgggctactc | potassium transporter Kup | [12] | ||
| Dickeya Rv284 | ccctgtcttctgttatcaattcattaac | ||||||
| Dickeya P284 | aaccagaataaggccc | FAM | MGB-NFQ | ||||
| * and ** | Pectobacterium atrosepticum | ECA-CSL-1F | cggcatcataaaaacacgcc | unknown | [13] | ||
| ECA-CSL-89R | cctgtgtaatatccgaaaggtgg | ||||||
| ECA-CSL-36T-P | acattcaggctgatattccccctgc | FAM | ZEN/IBFQ | ||||
| * and ** | P. brasiliense | PcbrFw | tgcgggttctgcgtttc | araC | [14] | ||
| PcbrRv | tggcgcgttcgcaatat | ||||||
| PcbrP | caaggcacgatacg | FAM | MGB-NFQ | ||||
| * and ** | P. parmentieri | PwF1 | tctgttcaatgtcaacgcaggta | mdh | [14] | ||
| PwR1 | aggtaaccgcaatttgctcaa | ||||||
| PwP1 | tgtgcgcaacctg | FAM | MGB-NFQ | ||||
| * | Acidovorax cattleya | Acat 2-F | tgtagcgatccttcacaag | Unknown | [10] | ||
| Acat 2-R | tgtcgatagatgctcacaat | ||||||
| Acat 2-Pr | cttgctctgcttctctatcacg | HEX | |||||
| ** | D. dianthicola | DdiFw | gccgtatccatcatgcttacc | dnaX | [15] | ||
| DdiRv | aacgggcgatagtcgtcttg | ||||||
| DdiP | tttccggcactcgg | FAM | MGB-NFQ | ||||
| ** | D. chrysanthemi | Fw2 | cgatttcccggcaagtgt | dnaX | [15] | ||
| Rv2 | tggcaaaagggctgaattg | ||||||
| LNA probe 3 | cgccgTCActccc | FAM | LNA | ||||
| ** | D. zeae | Fw4 | tcccgcactaaagttgaaga | dnaX | [15] | ||
| R43 | gcgagctggcgcgtatt | ||||||
| probe | cgcgagactTACtggataacgt | FAM | LNA | ||||
| ** | D. solani | SOL-C-F | gcctacaccatcagggctat | Unknown | [16] | ||
| SOL-C-R | acactacagcgcgcataaac | ||||||
| SOL-C-P | ccaggccgtgctcgaaatcc | FAM | |||||
| ** | D. dadantii | Fw2 | cccggtttcgcaattcag | dnaX | [15] | ||
| Rv3 | gggcgtaggcaagacgacta | ||||||
| Probe | tttcgccAACaaacggg | FAM | LNA | ||||
| ** | D. dieffenbachiae | Fw2 | gaattgcgaaaccgggatta | dnaX | [15] | ||
| Rv1 | gatttcccggcaggtatcg | ||||||
| Probe | cggctaCACcctgc | FAM | LNA | ||||
| ** | D. fangzhongdai | DfF | cttcgccgcccaggtatttt | fusA | [17] | ||
| DfR | atcagggcgtgaccttcgtt | ||||||
| DfP | tgctgcagactcgatcaggttctga | FAM | ZEN/IBFQ | ||||
| *** | P. brasiliense (virulent group) | LZI_F1 | cggtaagttatgccgcatct | unpublished data | |||
| LZI_R1 | cactgatctctttcatttagccatatc | (Van der Lee, Van Gent, WUR) | |||||
| LZI_P1 | tggcattacagaattcattgccaac | Lysozyme inhibitor | FAM | ZEN/IBFQ | |||
| *** | P. brasiliense (virulent group) | TIR-F2 | agataaacaagcgagggttga | unpublished data | |||
| TIR-R2 | atctatctcccatttcacccaag | (Van der Lee, Van Gent, WUR) | |||||
| TIR-P2 | aaatacagcctccattagagtttccc | Toll-like receptor | Yakima Yellow | ZEN/IBFQ | |||
| *** | P. brasiliense (in triplex assay) | Pb1F | ccttaccaagaagatgtgtgttgc | [11] | |||
| Pb2R | cataaacccggcacgct | ||||||
| PbPr | caagcgcacctgttgatgtcatgagtg | 16–23 s intergenic spacer | Cy5 | TAO/IBRQ |
2.9. Pathogen Isolation
Confirmation of the Identity of Isolates
2.10. Tuber Maceration Assay
| Target | Sequence | Target Gene | bp | Comments |
|---|---|---|---|---|
| Generic Dickeya | GACCACTTTGCCGTTTTCCACCAACAGGTTAATTTTGCAGCCTGACCCACAGTAAGGGCAGACGGTAATTACTTTCTGCATGACATTGCTCTCCTTCATCGTACCGACGGCACGCTCAGAATTTCATTTCCGCGGCTTCATCGAGTGCTGCGCGCCGCTGTTTACGTTGGATCATTTCCTGAATGTCCTCCCGGCTGATCAG | formate dehydrogenase | 220 | |
| Pectobacterium parmentieri | ACCGGGTGTTGCTGTCGATCTGAGCCATATTCCTACAGCAGTGAAGATCAAAGGCTATAGCGGTGAAGACGCTAAACCAGCGCTTGCTGGTGCGGATATTGTGCTGATTTCCGCTGGCGTGGCACGTAAACCTGGTATGGATCGTTCCGATCTGTTCAATGTCAACGCAGGTATTGTGCGCAACCTGGTTGAGCAAATTGCGGTTACCTGCCCGAAAGCCTGCATCGGGATCATTACTAATCCCGTGAACACGACCGTCGCTATTGCAGCCGAAGTGCTG | malate dehydrogenase | 280 | |
| P. atrosepticum | AGGATTCAGTTAATAATGCAATGGAATAGCAATGTAATATCGAAATCATTGAACGCTTTTATAGAATAGAGAAGGATCGGCATCATAAAAACACGCCATTAATAAACACATCAACATTCAGGCTGATATTCCCCCTGCCTATTCCACCTTTCGGATATTACACAGGGTACTTCCCTTATTGCCTTCTATTAAATCAG | unknown | 210 | A = deletion in primer, CG = different from target sequence (AA). Adjusted to increase GC content. |
| P. brasiliense | CGTCAGGTGACCGGTGCCGGATGGGCGAGGTGAATCGTTCTGATTCGCGCCGAATACTGCCAGGCAACATCGGTGAGTGTTGGTCGGATTTCAGCCGTATTGATGAGGATCTGTCGCTGGCGCGTTCGCAATATCGCCCGTATCGTGCCTTGGTGGAAGAAACGCAGAACCCGCATTCGCAGCCGATGCTGATCATGACATTTGCACTG | unknown | 215 |
2.11. Field Bioassay
2.12. Statistics
3. Results
3.1. Development of a Generic Multiplex TaqMan Assay for Blackleg-Causing Bacteria
3.2. Survey 2019
3.3. Survey 2020
3.4. Characterization of the P. brasiliense Strains
3.5. Identification of Dickeya Species
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ma, B.; Hibbing, M.E.; Kim, H.S.; Reedy, R.M.; Yedidia, I.; Breuer, J.; Breuer, J.; Glasner, J.D.; Perna, N.T.; Kelman, A.; et al. Host range and molecular phylogenies of the soft rot enterobacterial genera Pectobacterium and Dickeya. Phytopathology 2007, 97, 1150–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toth, I.K.; Barny, M.-A.; Brurberg, M.B.; Condemine, G.; Czajkowski, R.; Elphinstone, J.G.; Helias, V.; Johnson, S.B.; Moleleki, L.N.; Pirhonen, M.; et al. Pectobacterium and Dickeya: Environment to Disease Development. In Plant Diseases Caused by Dickeya and Pectobacterium Species; Springer: Berlin/Heidelberg, Germany, 2021; pp. 39–84. [Google Scholar]
- Van der Wolf, J.M.; Acuña, I.; De Boer, S.H.; Brurberg, M.B.; Cahill, G.; Charkowski, A.O.; Coutinho, T.; Davey, T.; Dees, M.W.; Degefu, Y. Diseases Caused by Pectobacterium and Dickeya species around the world. Chapter 7. In Plant Diseases Caused by Dickeya and Pectobacterium Species; Springer: Berlin/Heidelberg, Germany, 2021; pp. 215–261. [Google Scholar]
- Czajkowski, R.; Perombelon, M.C.M.; van Veen, J.A.; van der Wolf, J.M. Control of blackleg and tuber soft rot of potato caused by Pectobacterium and Dickeya species: A review. Plant Pathol. 2011, 60, 999–1013. [Google Scholar] [CrossRef]
- Van der Wolf, J.M.; De Boer, S.H.; Czajkowski, R.; Cahill, G.; Van Gijsegem, F.; Davey, T.; Dupuis, B.; Ellicott, J.; Jafra, S.; Kooman, M. Management of diseases caused by Pectobacterium and Dickeya species. Chapter 6. In Plant Diseases Caused by Dickeya and Pectobacterium Species; Springer: Berlin/Heidelberg, Germany, 2021; pp. 175–214. [Google Scholar]
- Boomsma, D.; Kastelein, P.; Van der Zouwen, P.S.; Krijger, M.; Förch, M.; Van der Wolf, J.; Czajkowski, C.; Wegierek, A.; Jafra, S.; Van den Bovenkamp, G.; et al. Het Project Deltaplan Erwinia Deel C—Pootaardappelen. Eindrapport van Het Onderzoek 209-2012; Nederlandse Aardappel Organisatie: Den Haag, The Nethelands, 2012; p. 104. [Google Scholar]
- Nykyri, J.; Fang, X.; Dorati, F.; Bakr, R.; Pasanen, M.; Niemi, O.; Palva, E.T.; Jackson, R.W.; Pirhonen, M. Evidence that nematodes may vector the soft rot-causing enterobacterial phytopathogens. Plant Pathol. 2014, 63, 747–757. [Google Scholar] [CrossRef]
- Kastelein, P.; Förch, M.; Krijger, M.; Van der Zouwen, P.; Van den Berg, W.; Van der Wolf, J. Systemic colonization of potato plants resulting from potato haulm inoculation with Dickeya solani or Pectobacterium parmentieri. Can. J. Plant Pathol. 2020, 43, 1–15. [Google Scholar] [CrossRef]
- Helias, V.; Hamon, P.; Huchet, E.; Van der Wolf, J.; Andrivon, D. Two new effective semiselective crystal violet pectate media for isolation of Pectobacterium and Dickeya. Plant Pathol. 2012, 61, 339–345. [Google Scholar] [CrossRef]
- Bonants, P.; Griekspoor, Y.; Houwers, I.; Krijger, M.; van der Zouwen, P.; van der Lee, T.A.; Van der Wolf, J. Development and evaluation of a triplex TaqMan assay and next-generation sequence analysis for improved detection of Xylella in plant material. Plant Dis. 2019, 103, 645–655. [Google Scholar] [CrossRef]
- Muzhinji, N.; Dube, J.P.; De Haan, E.; Woodhall, J.; Van der Waals, J.E. Development of a TaqMan PCR assay for specific detection and quantification of Pectobacterium brasiliense in potato tubers and soil. Eur. J. Plant Pathol. 2020, 158, 521–532. [Google Scholar] [CrossRef]
- Zijlstra, C.; Groenenboom–De Haas, L.; Krijger, M.; Verstappen, E.; Warris, S.; de Haan, E.; van der Wolf, J. Development and evaluation of two TaqMan assays for generic detection of Dickeya species. Eur. J. Plant Pathol. 2020, 156, 311–316. [Google Scholar] [CrossRef] [Green Version]
- Humphris, S.N.; Cahill, G.; Elphinstone, J.G.; Kelly, R.; Parkinson, N.M.; Pritchard, L.; Saddler, G.S. Detection of the bacterial potato pathogens Pectobacterium and Dickeya spp. using conventional and real-time PCR. In Plant Pathology: Techniques and Protocols, 2nd ed.; Lacomme, C., Ed.; Humana Press: New York, NY, USA, 2015; Volume 1302, pp. 1–6. [Google Scholar]
- Van der Wolf, J.M.; de Haan, E.G.; Kastelein, P.; Krijger, M.; de Haas, B.H.; Velvis, H.; Mendes, O.; Kooman-Gersmann, M.; van der Zouwen, P.S. Virulence of Pectobacterium carotovorum subsp. brasiliense on potato compared with that of other Pectobacterium and Dickeya species under climatic conditions prevailing in the Netherlands. Plant Pathol. 2017, 66, 571–583. [Google Scholar]
- Van der Wolf, J.M.; de Haas, B.H.; van Hoof, R.; de Haan, E.G.; van den Bovenkamp, G.W. Development and evaluation of Taqman assays for the differentiation of Dickeya (sub)species. Eur. J. Plant Pathol. 2014, 138, 695–709. [Google Scholar] [CrossRef]
- Pritchard, L.; Humphris, S.; Saddler, G.S.; Parkinson, N.M.; Bertrand, V.; Elphinstone, J.G. Detection of phytopathogens of the genus Dickeya using a PCR primer prediction pipeline for draft bacterial genome sequences: Dickeya diagnostics from draft bacterial genome sequences. Plant Pathol. 2013, 62, 587–596. [Google Scholar] [CrossRef]
- Tian, Y.; Zhao, Y.; Yuan, X.; Yi, J.; Fan, J.; Xu, Z.; Hu, B.; De Boer, S.H.; Li, X. Dickeya fangzhongdai sp. nov. a plant-pathogenic bacterium isolated from pear trees (Pyrus pyrifolia). Int. J. Syst. Evol. Microbiol. 2016, 66, 2831–2835. [Google Scholar] [CrossRef]
- Cigna, J.; Dewaegeneire, P.; Beury, A.; Gobert, V.; Faure, D. A gapA PCR-sequencing assay for identifying the Dickeya and Pectobacterium potato pathogens. Plant Dis. 2017, 101, 1278–1282. [Google Scholar] [CrossRef] [Green Version]
- Czajkowski, R.; de Boer, W.J.; Velvis, H.; van der Wolf, J.M. Systemic colonization of potato plants by a soilborne, green fluorescent protein-tagged strain of Dickeya sp biovar 3. Phytopathology 2010, 100, 134–142. [Google Scholar] [CrossRef] [Green Version]
- De Haan, E.G.; Dekker-Nooren, T.C.E.M.; van den Bovenkamp, G.W.; Speksnijder, A.G.C.L.; van der Zouwen, P.S.; van der Wolf, J.M. Pectobacterium carotovorum subsp carotovorum can cause potato blackleg in temperate climates. Eur. J. Plant Pathol. 2008, 122, 561–569. [Google Scholar] [CrossRef]
- De Boer, S.H. Relative incidence of Erwinia carotovora subsp. atroseptica in stolon end and peridermal tissue of potato tubers in Canada. Plant Disease 2002, 86, 960–964. [Google Scholar] [PubMed] [Green Version]
- Pazdernik, N.; Prediger, E. qPCR Probes-Selecting the Best Reporter Dye and Quencher. Available online: https://www.idtdna.com/pages/education/decoded/article/qpcr-probes-selecting-the-best-reporter-dye-and-quencher (accessed on 29 June 2022).
- Jonkheer, E.M.; Brankovics, B.; Houwers, I.M.; van der Wolf, J.M.; Bonants, P.J.; Vreeburg, R.A.; Bollema, R.; de Haan, J.R.; Berke, L.; Smit, S. The Pectobacterium pangenome, with a focus on Pectobacterium brasiliense, shows a robust core and extensive exchange of genes from a shared gene pool. BMC Genom. 2021, 22, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Sławiak, M.; van Beckhoven, J.R.; Speksnijder, A.G.; Czajkowski, R.; Grabe, G.; van der Wolf, J.M. Biochemical and genetical analysis reveal a new clade of biovar 3 Dickeya spp. strains isolated from potato in Europe. Eur. J. Plant Pathol. 2009, 125, 245–261. [Google Scholar] [CrossRef]
- McNally, R.; Curland, R.; Webster, B.; Robinson, A.; Ishimaru, C. First report of stem rot on potato caused by Dickeya chrysanthemi in Minnesota. Plant Dis. 2018, 102, 238. [Google Scholar] [CrossRef]
- Pédron, J.; Schaerer, S.; Kellenberger, I.; Van Gijsegem, F. Early Emergence of Dickeya solani revealed by analysis of Dickeya diversity of potato blackleg and soft rot causing pathogens in Switzerland. Microorganisms 2021, 9, 1187. [Google Scholar] [CrossRef]
- Cother, E.J.; Powell, V. Physiological and pathological characteristics of Erwinia chrysanthemi isolates from potato tubers. J. Appl. Bacteriol. 1983, 54, 37–44. [Google Scholar] [CrossRef]
- Suharjo, R.; Sawada, H.; Takikawa, Y. Phylogenetic study of Japanese Dickeya spp. and development of new rapid identification methods using PCR–RFLP. J. Gen. Plant Pathol. 2014, 80, 237–254. [Google Scholar] [CrossRef]
- Velvis, H.; Van der Wolf, J.M. Project Bacterievrije Pootgoedteelt—Een Uitdaging! 2005–2008. Eindrapport van het Onderzoek. 2008. Available online: https://kennisakker.nl/storage/2444/Eindrapport_Erwiniaproject_2005-08.pdf,31pages (accessed on 1 September 2022).
- Elphinstone, J.; Pérombelon, M. Contamination of potatoes by Erwinia carotovora during grading. Plant Pathol. 1986, 35, 25–33. [Google Scholar] [CrossRef]
- Li, X.S.; Yuan, K.X.; Cullis, J.; Lévesque, C.A.; Chen, W.; Lewis, C.T.; De Boer, S.H. Draft genome sequences for Canadian isolates of Pectobacterium carotovorum subsp. brasiliense with weak virulence on potato. Genome Announc. 2015, 3, e00240-15. [Google Scholar]
- Moleleki, L.N.; Onkendi, E.M.; Mongae, A.; Kubheka, G.C. Characterisation of Pectobacterium wasabiae causing blackleg and soft rot diseases in South Africa. Eur. J. Plant Pathol. 2013, 135, 279–288. [Google Scholar] [CrossRef]
- Ge, T.; Ekbataniamiri, F.; Johnson, S.B.; Larkin, R.P.; Hao, J. Interaction between Dickeya dianthicola and Pectobacterium parmentieri in Potato Infection under Field Conditions. Microorganisms 2021, 9, 316. [Google Scholar] [CrossRef]
- Yin, G.; Xu, Y. Study on the occurrence and control of the bleeding canker of pear. Xuzhou Hortic 1973, 1, 15–19. [Google Scholar]
- Alič, Š.; Van Gijsegem, F.; Pedron, J.; Ravnikar, M.; Dreo, T. Diversity within the novel Dickeya fangzhongdai sp. isolated from infected orchids, water and pears. Plant Pathol. 2018, 67, 1612–1620. [Google Scholar] [CrossRef]
- Van der Wolf, J.M.; Nijhuis, E.H.; Kowalewska, M.J.; Saddler, G.S.; Parkinson, N.; Elphinstone, J.G.; Pritchard, L.; Toth, I.K.; Lojkowska, E.; Potrykus, M. Dickeya solani sp. nov. a pectinolytic plant-pathogenic bacterium isolated from potato (Solanum tuberosum). Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 3, 768–774. [Google Scholar] [CrossRef]


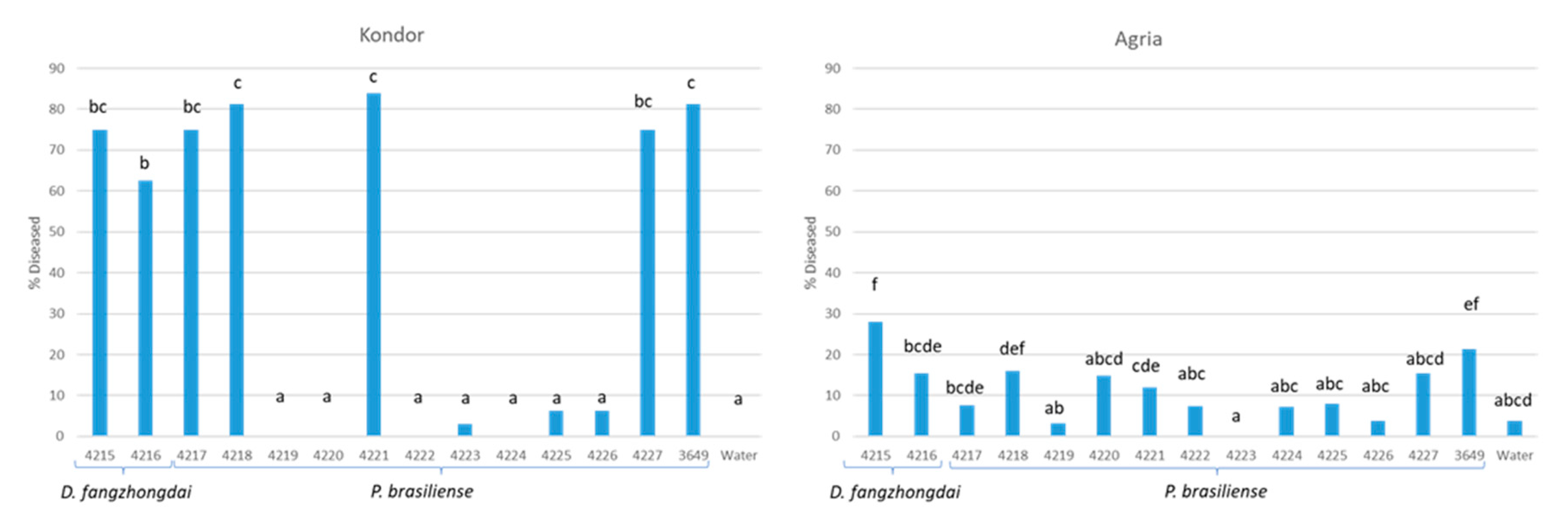
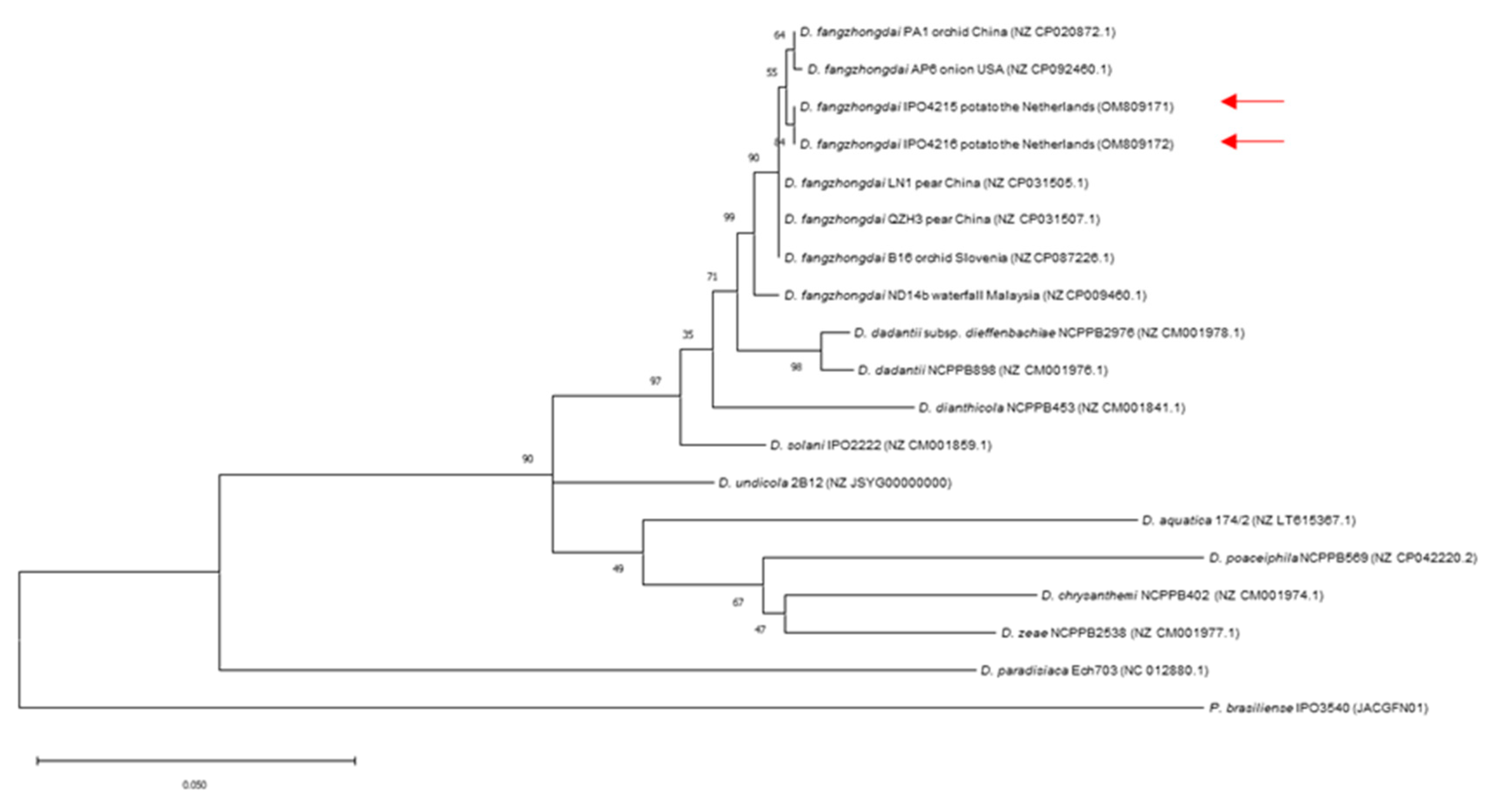
| Grower | Region | Sampling Date | |
|---|---|---|---|
| 2019 | 2020 | ||
| A | Noordoostpolder | 25 June | 17 June |
| B | Noordoostpolder | 27 June | 25 June |
| C | Noord-Holland | 4 July | 2 July |
| D | Noord-Holland | 9 July | 15 July |
| E | Noord-Holland | 17 July | 16 July |
| TaqMan Assay (Ct-Values) | |||||||
|---|---|---|---|---|---|---|---|
| Sample | Dickeya sp. | D. solani | D. dianthicola | D. dadantii | D. dieffenbachiae | D. chrysanthemi | D. zeae |
| 979 | 30 | - | - | - | - | 30.4 | - |
| 1018 | 30.4 | - | - | - | - | - | 31.3 |
| 1081 | 24 | - | - | - | - | 24.5 | - |
| 1141 | 23.9 | - | - | - | - | 24.4 | - |
| 1198 | 20.5 | - | - | - | - | - | 21.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van der Wolf, J.; Krijger, M.; Mendes, O.; Kurm, V.; Gros, J. Natural Infections of Potato Plants Grown from Minitubers with Blackleg-Causing Soft Rot Pectobacteriaceae. Microorganisms 2022, 10, 2504. https://doi.org/10.3390/microorganisms10122504
van der Wolf J, Krijger M, Mendes O, Kurm V, Gros J. Natural Infections of Potato Plants Grown from Minitubers with Blackleg-Causing Soft Rot Pectobacteriaceae. Microorganisms. 2022; 10(12):2504. https://doi.org/10.3390/microorganisms10122504
Chicago/Turabian Stylevan der Wolf, Jan, Marjon Krijger, Odette Mendes, Viola Kurm, and Jack Gros. 2022. "Natural Infections of Potato Plants Grown from Minitubers with Blackleg-Causing Soft Rot Pectobacteriaceae" Microorganisms 10, no. 12: 2504. https://doi.org/10.3390/microorganisms10122504





