Enzymatic Characterization of Unused Biomass Degradation Using the Clostridium cellulovorans Cellulosome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture Conditions
2.2. Preparation and Purification of the Cellulosome Fraction
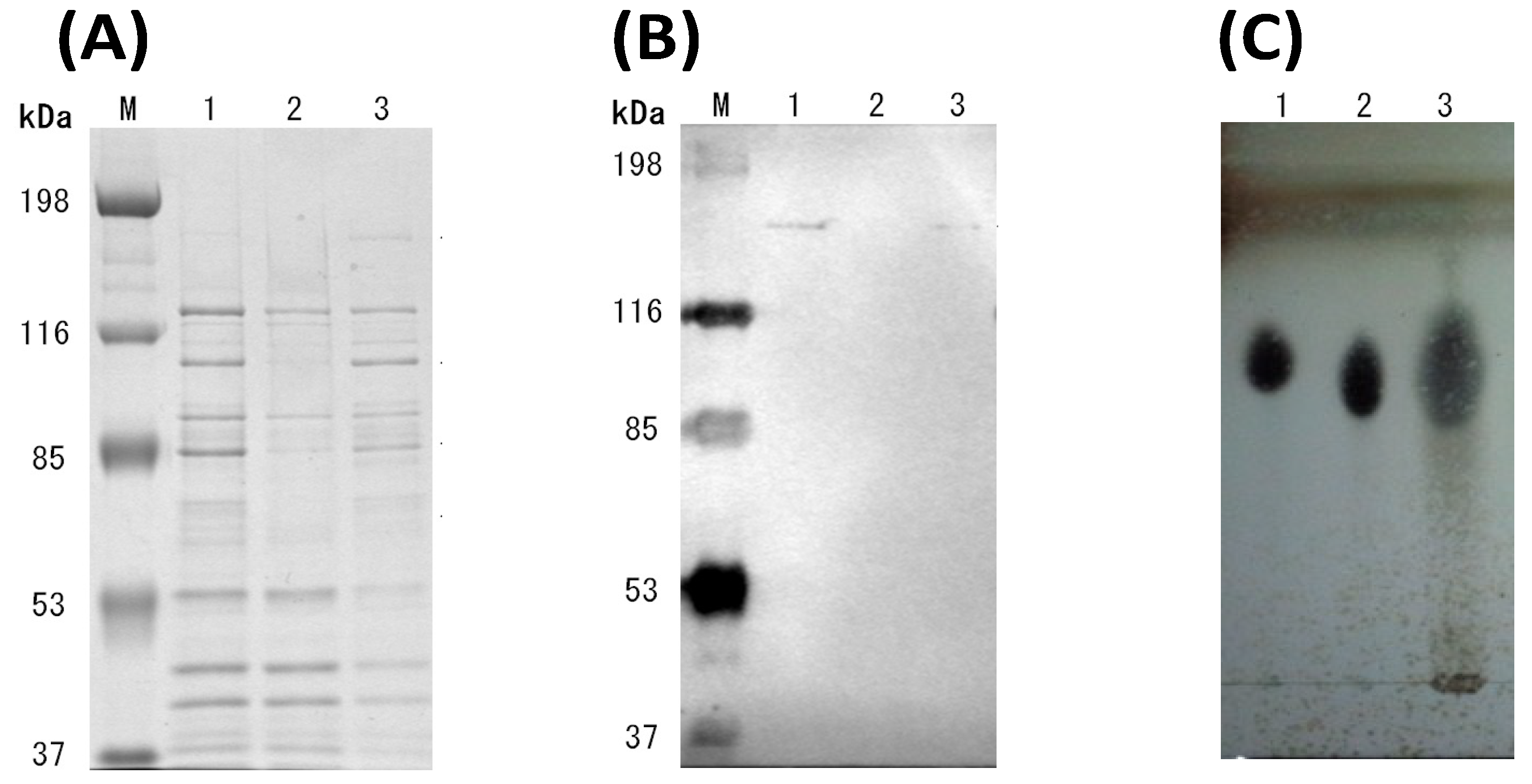
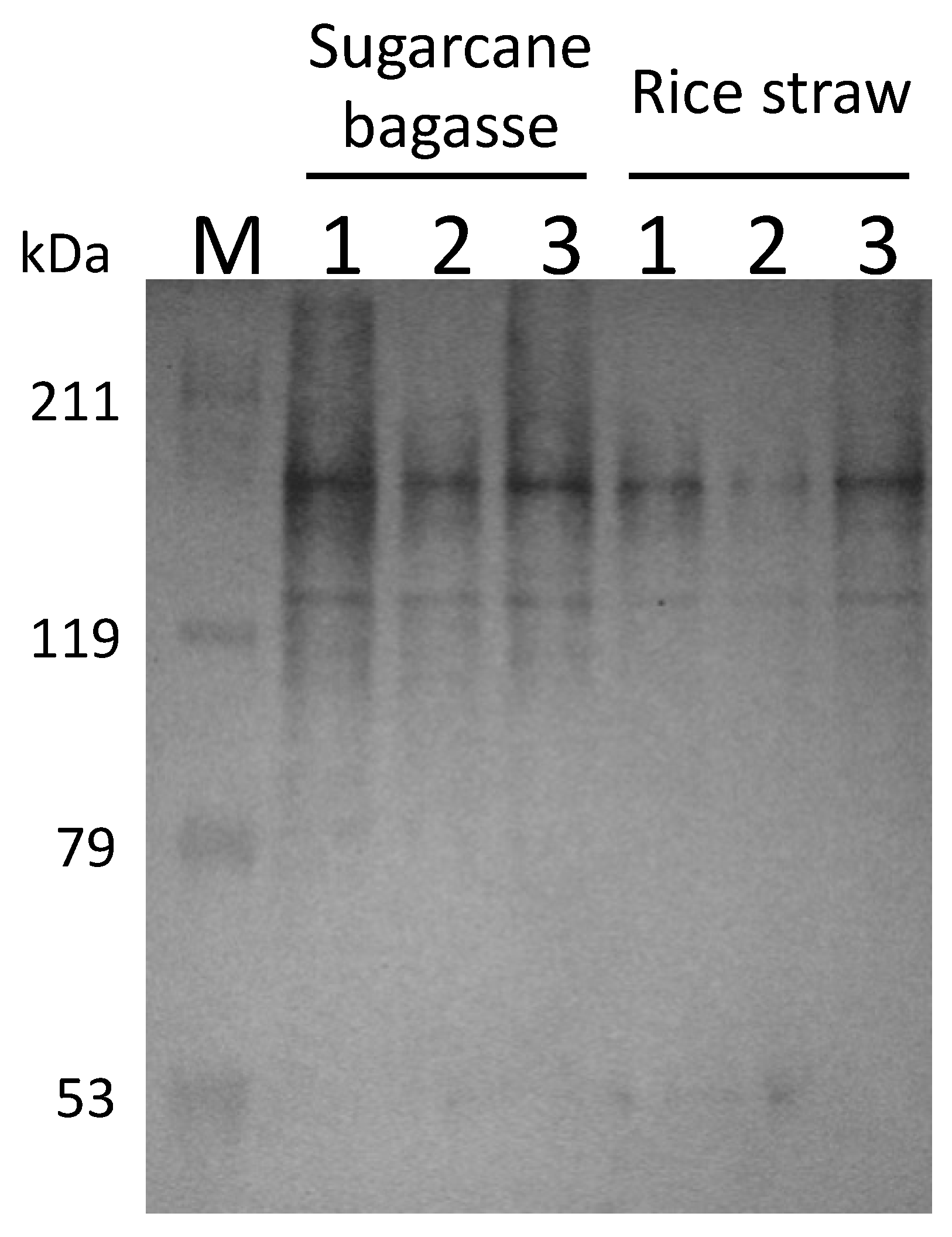
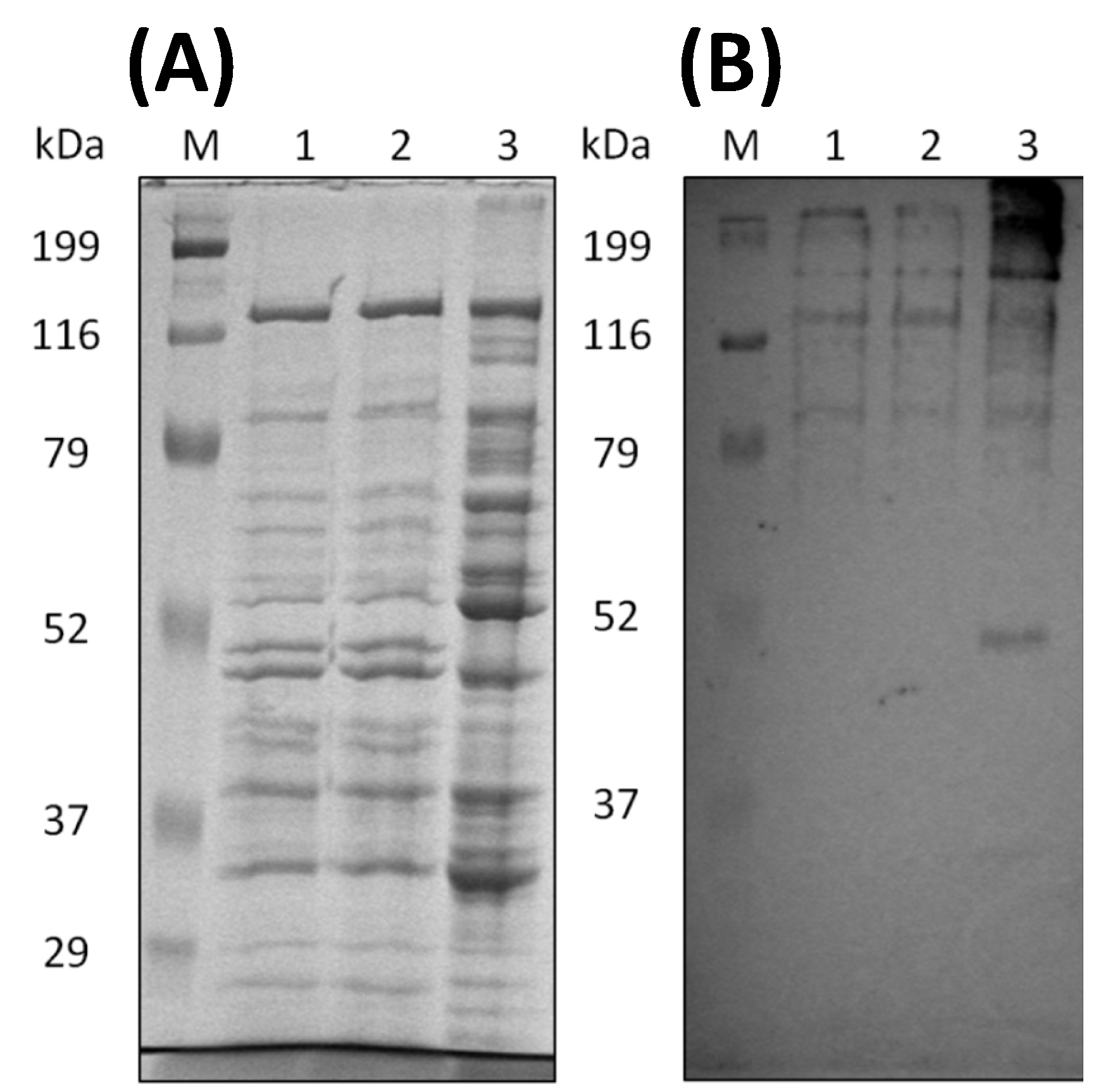
2.3. SDS-PAGE and Western Blot Analysis
2.4. Enzymatic Activity, Protein Assays and TLC Analysis
2.5. Alcohol Concentration
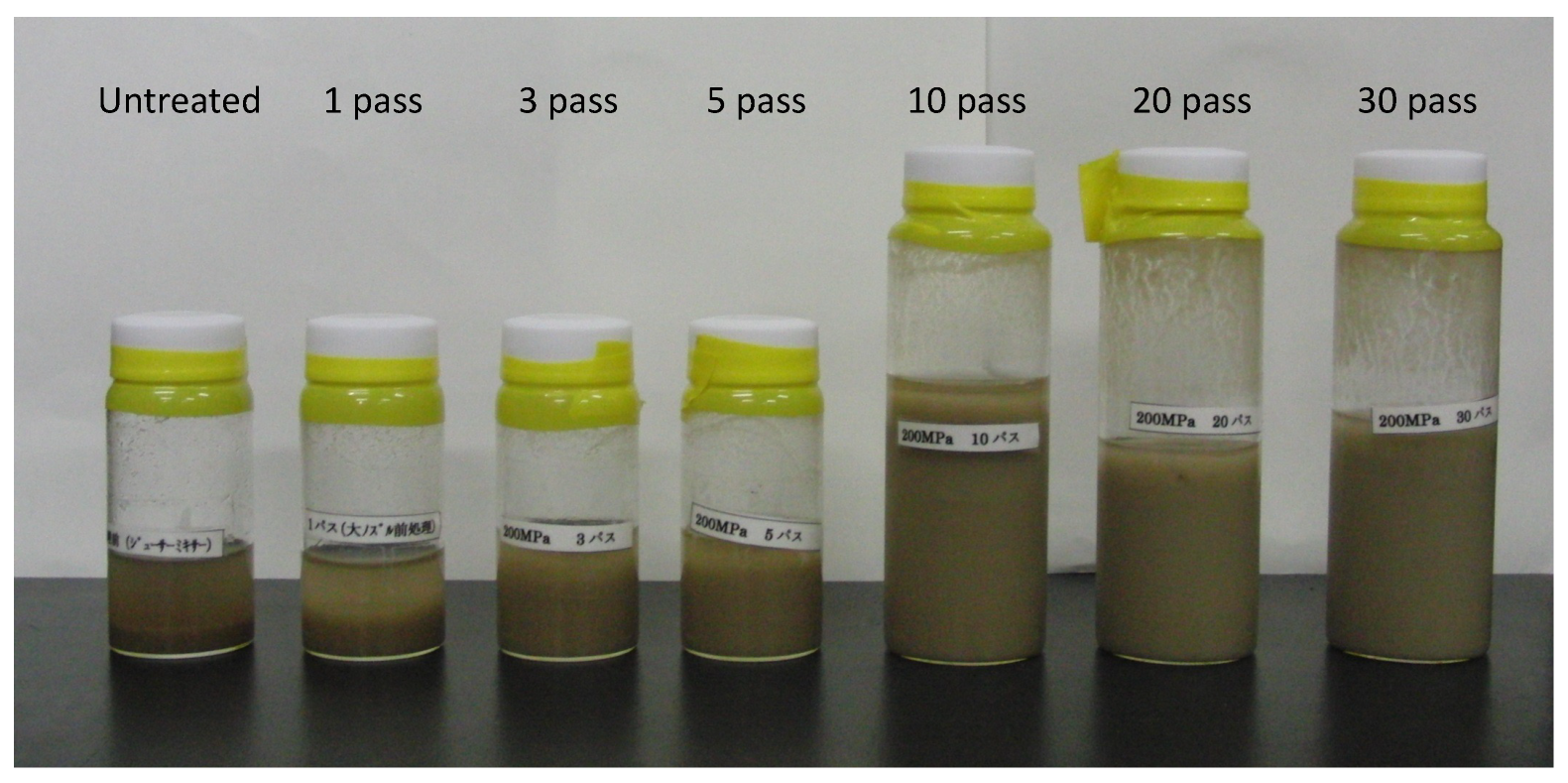
2.6. Pretreatment of Sugarcane Bagasse and Its Enzymatic Reaction
3. Results
3.1. Continuous Cultivation of C. cellulovorans with Shredded Paper
3.2. Cocultivation of C. cellulovorans and C. acetobutylicum
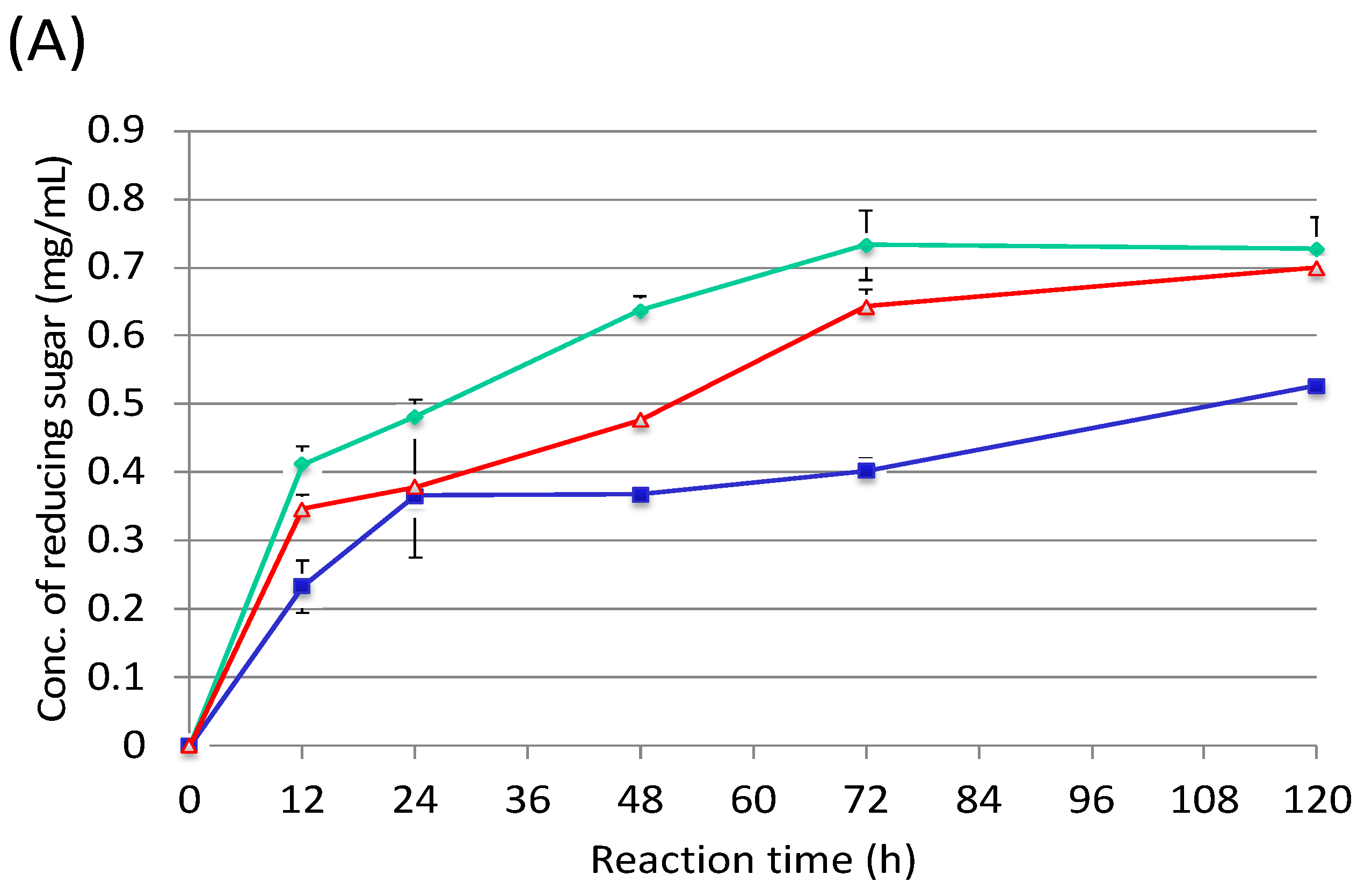
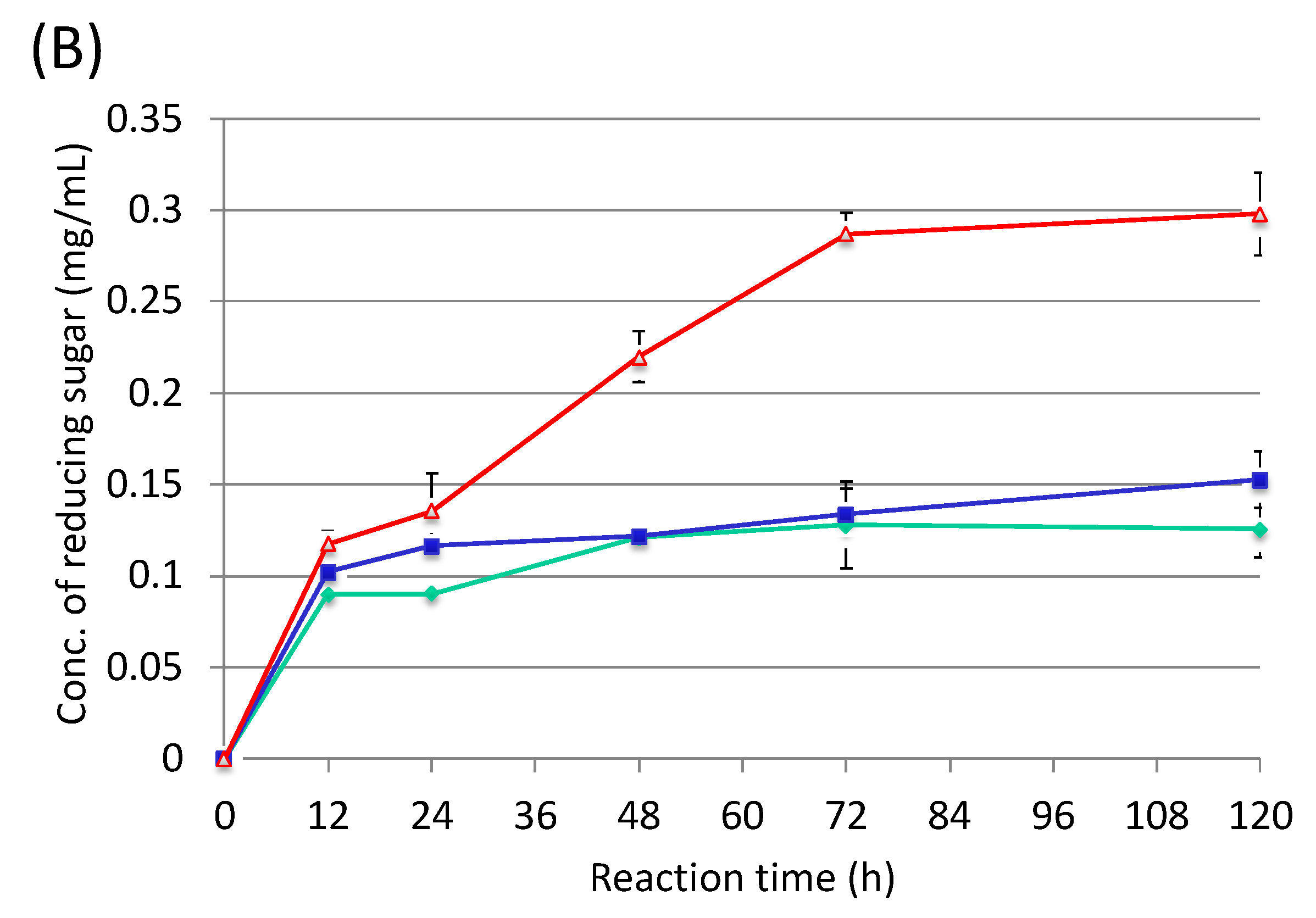
3.3. Cultivation of Cellulosic Biomass with C. cellulovorans
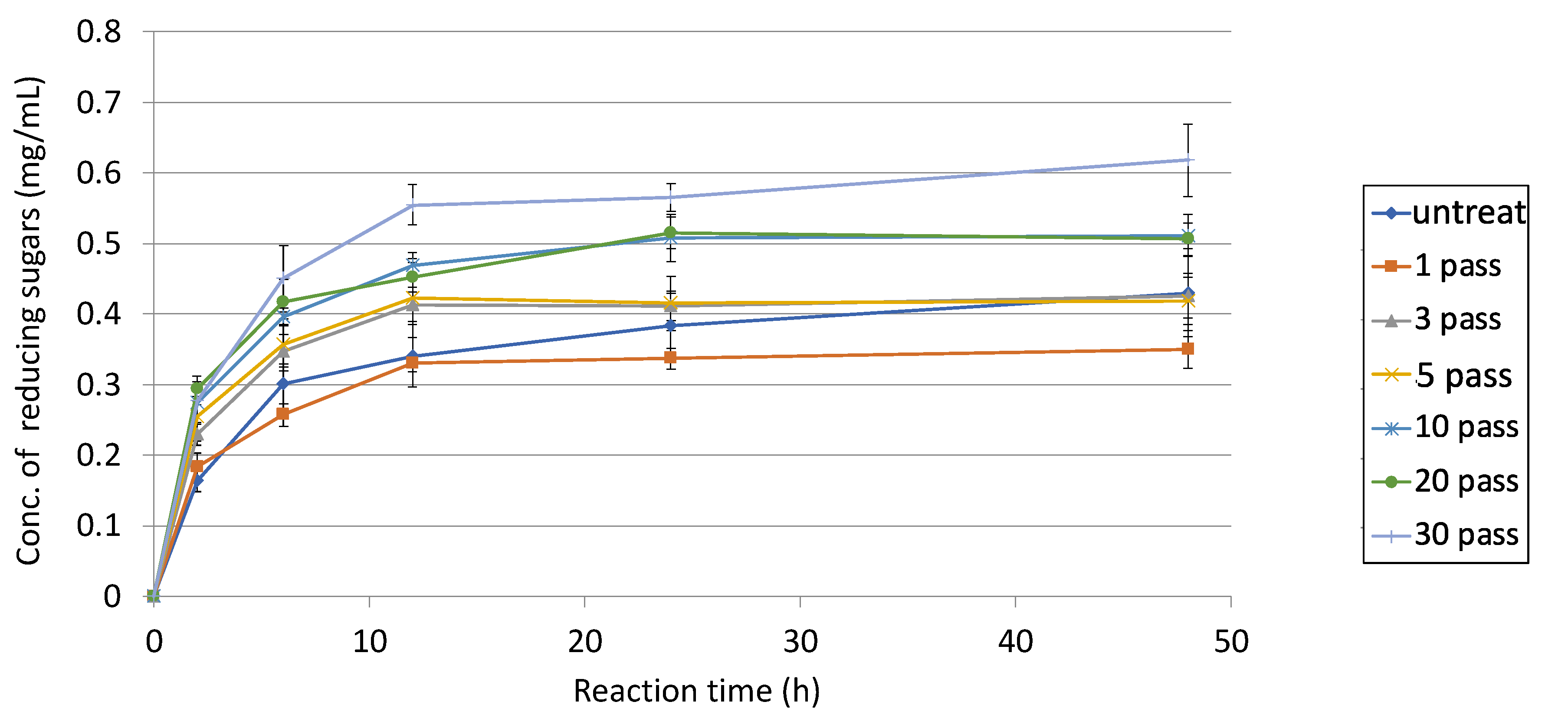
3.4. Futher Analysis of Sugarcane Bagasse Degradation
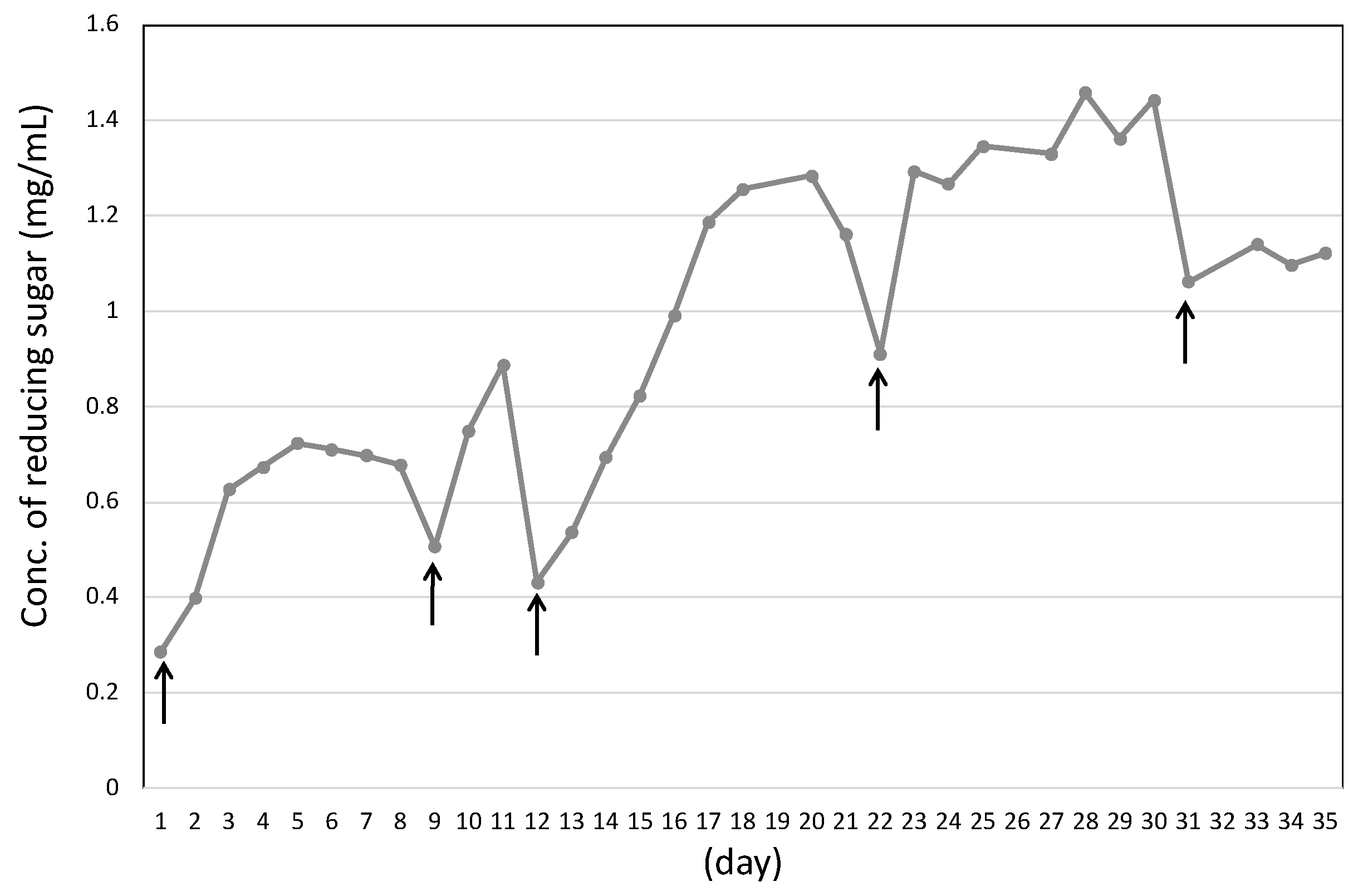
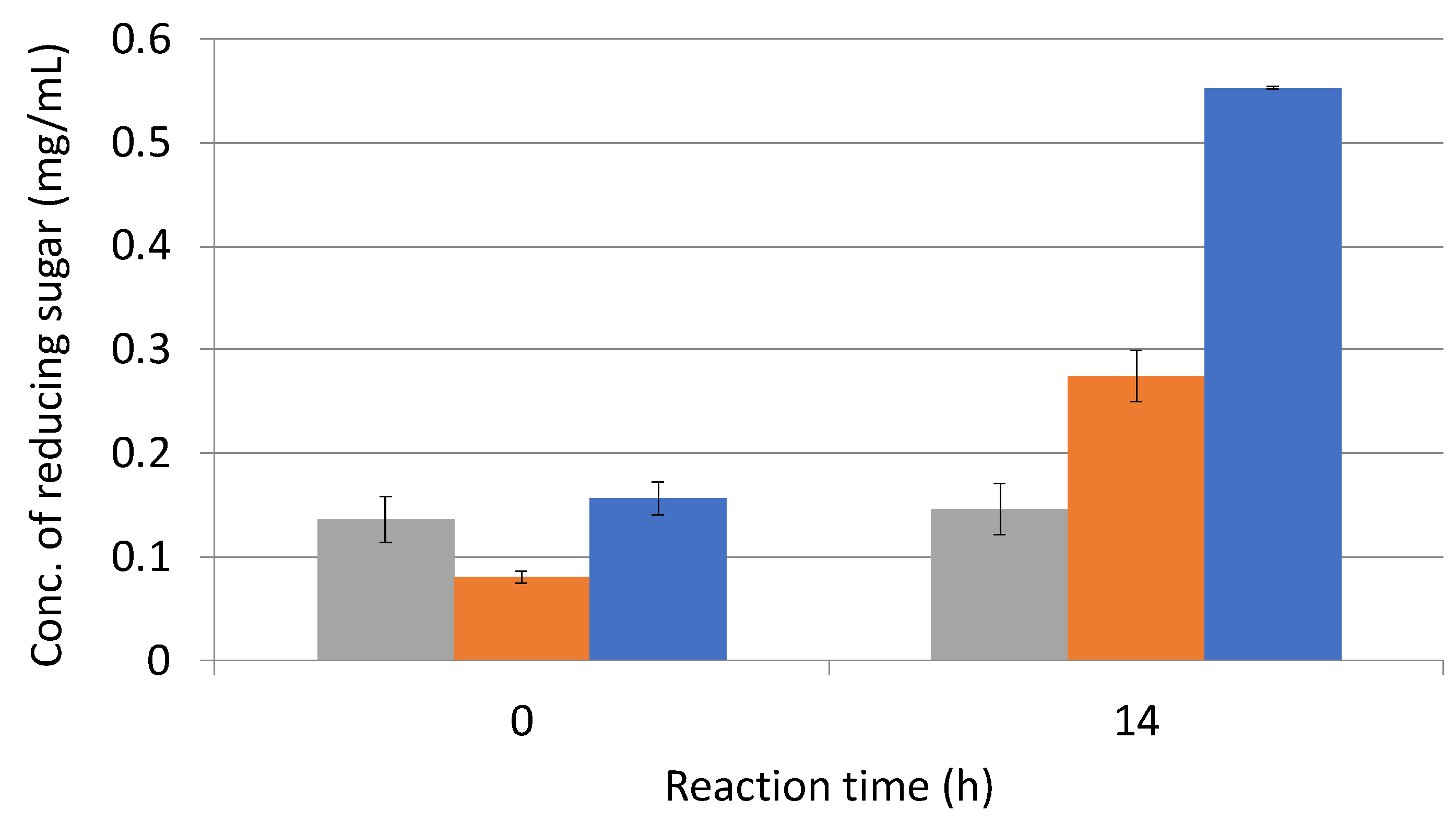
3.5. Effect of Enzymatic Reaction of Sugarcane Bagasse with L-Cysteine
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Olson, D.G.; McBride, J.E.; Joisha, A.; Lynd, L.R. Recent progress in consolidated bioprocessing. Curr. Opin. Biotechnol. 2012, 23, 396–405. [Google Scholar] [CrossRef] [PubMed]
- Tomita, H.; Okazaki, F.; Tamaru, Y. Direct IBE fermentation from mandarin orange wastes by combination of Clostridium Cellulomonas and Clostridium beijerinckii. AMB Express 2019, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Tomita, H.; Okazaki, F.; Tamaru, Y. Biomethane production from sugar beet pulp under cocultivation with Clostridium cellulovorans and methanogens. AMB Express 2019, 9, 28. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Sun, J.; Chang, J.-S.; Shukla, P. Engineering microbes for direct fermentation of cellulose to bioethanol. Crit. Rev. Biotechnol. 2018, 38, 1089–1105. [Google Scholar] [CrossRef]
- Cho, W.; Jeon, S.D.; Shim, H.J.; Doi, R.H.; Han, S.O. Cellulosomic profiling produced by Clostridium cellulovorans during growth on different carbon sources explored by the cohesin marker. J. Biotechnol. 2010, 145, 233–239. [Google Scholar] [CrossRef]
- Morisaka, H.; Matsui, K.; Tatsukami, Y.; Kuroda, K.; Miyake, H.; Tamaru, Y.; Ueda, M. Profile of native cellulosomal proteins of Clostridium cellulovorans adapted to various carbon sources. AMB Express 2012, 2, 37. [Google Scholar] [CrossRef] [Green Version]
- Matsui, K.; Bae, J.; Esaka, K.; Morisaka, H.; Kuroda, K.; Ueda, M. Exoproteome profiles of Clostridium cellulovorans grown on various carbon sources. Appl. Environ. Microbiol. 2013, 79, 6576–6584. [Google Scholar] [CrossRef] [Green Version]
- Esaka, E.; Aburaya, S.; Morisaka, H.; Kuroda, K.; Ueda, M. Exoproteome analysis of Clostridium cellulovorans in natural soft-biomass degradation. AMB Express 2015, 5, 2. [Google Scholar] [CrossRef] [Green Version]
- Murashima, K.; Kosugi, A.; Doi, R.H. Synergistic effects on crystalline cellulose degradation between cellulosomal cellulases from Clostridium cellulovorans. J. Bacteriol. 2002, 184, 5088–5095. [Google Scholar] [CrossRef] [Green Version]
- Koukiekolo, R.; Cho, H.Y.; Kosugi, A.; Inui, M.; Yukawa, H.; Doi, R.H. Degradation of corn fiber by Clostridium cellulovorans cellulases and hemicellulases and contribution of scaffolding protein CbpA. Appl. Environ. Microbiol. 2005, 71, 3504–3511. [Google Scholar] [CrossRef]
- Cha, J.; Matsuoka, S.; Chan, H.; Yukawa, H.; Inui, M.; Doi, R.H. Effect of multiple copies of cohesins on cellulase and hemicellulase activities of Clostridium cellulovorans mini-cellulosomes. J. Microbiol. Biotechnol. 2007, 17, 1782–1788. [Google Scholar]
- Dredge, R.; Radloff, S.E.; van Dyk, J.S.; Pletschke, B.I. Lime pretreatment of sugar beet pulp and evaluation of synergy between ArfA, ManA and XynA from Clostridium cellulovorans on the pretreated substrate. 3 Biotech. 2011, 1, 151–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olver, B.; Van Dyk, J.S.; Beukes, N.; Pletschke, B.I. Synergy between EngE, XynA and ManA from Clostridium cellulovorans on corn stalk, grass and pineapple pulp substrates. 3 Biotech. 2011, 1, 187–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, S.D.; Lee, J.E.; Kim, S.J.; Park, S.H.; Choi, G.W.; Han, S.O. Unique contribution of the cell wall-binding endoglucanase G to the cellulolytic complex in Clostridium cellulovorans. Appl. Environ. Microbiol. 2013, 79, 5942–5948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, S.D.; Kim, S.J.; Park, S.H.; Choi, G.W.; Han, S.O. Hydrolytic effects of scaffolding proteins CbpB and CbpC on crystalline cellulose mediated by the major cellulolytic complex from Clostridium cellulovorans. Bioresour. Technol. 2015, 191, 505–511. [Google Scholar] [CrossRef]
- Yamamoto, K.; Tamaru, Y. Synergistic properties of cellulases from Clostridium cellulovorans in the presence of cellobiose. AMB Express 2016, 6, 1. [Google Scholar] [CrossRef] [Green Version]
- Arai, T.; Kosugi, A.; Chan, H.; Koukiekolo, R.; Yukawa, H.; Inui, M.; Doi, R.H. Properties of cellulosomal family 9 cellulases from Clostridium cellulovorans. Appl. Microbiol. Biotechnol. 2006, 71, 654–660. [Google Scholar] [CrossRef]
- Tamaru, Y.; Ui, S.; Murashima, K.; Kosugi, A.; Chan, H.; Doi, R.H.; Liu, B. Formation of protoplasts from cultured tobacco cells and Arabidopsis thaliana by the action of cellulosomes and pectate lyase from Clostridium cellulovorans. Appl. Environ. Microbiol. 2002, 68, 2614–2618. [Google Scholar] [CrossRef] [Green Version]
- Tomita, H.; Tamaru, Y. The second-generation biomethane from mandarin orange peel under cocultivation with methanogens and the armed Clostridium cellulovorans. Fermentation 2019, 5, 95. [Google Scholar] [CrossRef] [Green Version]
- Resch, M.G.; Donohoe, B.S.; Baker, J.O.; Decker, S.R.; Bayer, E.A.; Beckhamde, G.T.; Himmel, M.E. Fungal cellulases and complexed cellulosomal enzymes exhibit synergistic mechanisms in cellulose deconstruction. Energy Environ. Sci. 2013, 6, 1858–1867. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Tamaru, Y.; Doi, R.H. Three surface layer homology domains at the N terminus of the Clostridium cellulovorans major cellulosomal subunit EngE. J. Bacteriol. 1999, 181, 3270–3276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glasgow, E.M.; Kemna, E.I.; Bingman, C.A.; Ing, N.; Deng, K.; Bianchetti, C.M.; Takasuka, T.E.; Northen, T.R.; Fox, B.G. A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis. J. Biol. Chem. 2020, 295, 17752–17769. [Google Scholar] [CrossRef]
- Chandel, A.K.; Garlapati, V.K.; Singh, A.K.; Antunes, F.A.F.; da Silva, S.S. The path forward for lignocellulose biorefineries: Bottle-necks, solutions, and perspective on commercialization. Bioresour. Technol. 2018, 264, 370–381. [Google Scholar] [CrossRef] [PubMed]
- Oke, M.A.; Annuar, M.S.M.; Simarani, K. Mixed feedstock approach to lignocellulosic ethanol production-prospects and limitations. Bioenerg. Res. 2016, 9, 1189–1203. [Google Scholar] [CrossRef] [Green Version]
- Tamaru, Y.; Miyake, H.; Kuroda, K.; Ueda, M.; Doi, R.H. Comparative genomics of the mesophilic cellulosome-producing Clostridium cellulovorans and its application to biofuel production via consolidated bioprocessing. Environ. Technol. 2010, 31, 889–903. [Google Scholar] [CrossRef]
- Tamaru, Y.; Miyake, H.; Kuroda, K.; Nakanishi, A.; Matsushima, C.; Doi, R.H.; Ueda, M. Comparison of the mesophilic cellulosome-producing Clostridium cellulovorans genome with other cellulosome-related clostridial genomes. Micro. Biotechnol. 2011, 4, 64–73. [Google Scholar] [CrossRef] [Green Version]
- Aburaya, S.; Aoki, W.; Kuroda, K.; Minakuchi, H.; Ueda, M. Temporal proteome dynamics of Clostridium cellulovorans cultured with major plant cell wall polysaccharides. BMC Microbiol. 2019, 19, 118. [Google Scholar] [CrossRef] [Green Version]
- Usaia, G.; Cirrincionea, S.; Reb, A.; Manfredic, M.; Pagnanid, A.; Pessionea, E.; Mazzoli, R. Clostridium cellulovorans metabolism of cellulose as studied by comparative proteomic approach. J. Proteom. 2020, 216, 103667. [Google Scholar] [CrossRef]
- Rabemanolontsoa, H.; Kuninori, Y.; Saka, S. High conversion efficiency of Japanese cedar hydrolyzates into acetic acid by co-culture of Clostridium thermoaceticum and Clostridium thermocellum. J. Chem. Technol. Biotechnol. 2016, 91, 1040–1047. [Google Scholar] [CrossRef]
- Xu, L.; Tschirner, U. Immobilized anaerobic fermentation for bio-fuel production by Clostridium co-culture. Bioprocess Biosyst. Eng. 2014, 37, 1551–1559. [Google Scholar]
- Oliva-Rodríguez, A.G.; Quintero, J.; Medina-Morales, M.A.; Morales-Martínez, T.K.; Rodríguez-De la Garza, J.A.; Moreno-Dávila, M.; Aroca, G.; González, L.J.R. Clostridium strain selection for co-culture with Bacillus subtilis for butanol production from agave hydrolysates. Bioresour. Technol. 2019, 275, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Qi, G.; Xiong, L.; Luo, M.; Huang, Q.; Huang, C.; Li, H.; Chen, X.; Chen, X. Solvents production from cassava by co-culture of Clostridium acetobutylicum and Saccharomyces cerevisiae. J. Environ. Chem. Eng. 2018, 6, 128–133. [Google Scholar] [CrossRef]
- Raut, M.P.; Pham, T.K.; Gomez, L.D.; Dimitriou, I.; Wright, P.C. Alcoholic fermentation of thermochemical and biological hydrolysates derived from miscanthus biomass by Clostridium acetobutylicum ATCC 824. Biomass Bioenergy 2019, 130, 105382. [Google Scholar] [CrossRef]
- Flythe, M.D.; Elía, N.M.; Schmal, M.B.; Nokes, S.E. Switchgrass (Panicum virgatum) fermentation by Clostridium thermocellum and Clostridium beijerinckii sequential culture: Effect of feedstock particle size on gas production. Adv. Microbiol. 2015, 5, 311–316. [Google Scholar] [CrossRef] [Green Version]
- Pang, J.; Hao, M.; Shi, Y.; Li, Y.; Zhu, M.; Hu, J.; Liu, J.; Zhang, Q.; Liu, Z. Enhancing the ethanol yield from salix using a Clostridium thermocellum and Thermoanaerobacterium thermosaccharolyticum co-culture system. Bioresources 2018, 13, 5377–5393. [Google Scholar] [CrossRef]
- Du, Y.; Zou, W.; Zhang, K.; Ye, G.; Yang, J. Advances and applications of Clostridium co-culture systems in biotechnology. Front. Microbiol. 2020, 11, 560223. [Google Scholar] [CrossRef]
- Wen, Z.; Ledesma-Amaro, R.; Lin, J.; Jiang, Y.; Yang, S. Improved n-Butanol Production from Clostridium cellulovorans by integrated metabolic and evolutionary engineering. Appl. Environ. Microbiol. 2019, 85, e02560-18. [Google Scholar] [CrossRef] [Green Version]
- Singhvi, M.; Kim, B.S. Green hydrogen production through consolidated bioprocessing of lignocellulosic biomass using nanobiotechnology approach. Bioresour. Technol. 2022, 365, 128108. [Google Scholar] [CrossRef]
- Lipkiewicz, A.; Małachowska, E.; Dubowik, M.; Piotr Przybysz, P. Impact of shredding degree on papermaking potential of recycled waste. Sci. Rep. 2021, 11, 17528. [Google Scholar] [CrossRef]
- Homaei, A.A.; Sajedi, R.H.; Sariri, R.; Seyfzadeh, S.; Stevanato, R. Cysteine enhances activity and stability of immobilized papain. Amino Acids 2010, 38, 937–942. [Google Scholar] [CrossRef] [PubMed]

| Bacterial Cultivation | Concentration (mM) | |
|---|---|---|
| Ethanol | n-Butanol | |
| C. cellulovorans *1 | 1.50 | 0 |
| C. acetobutylicum *1 | 0 | 0 |
| Cocultivation *2 | 1.89 | 1.67 |
| Negative control | 0 | 0 |
| Enzyme Solution | Degradation Activity with Pretreated Sugarcane Bagasse (mU/mL) | Final Protein Concentration (mg/mL) | Specific Activity (mU/mg) |
|---|---|---|---|
| E1 | 6.90 | 0.247 | 27.9 |
| E2 | 9.44 | 0.247 | 38.2 |
| E3 | 10.8 | 0.385 | 28.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eljonaid, M.Y.; Tomita, H.; Okazaki, F.; Tamaru, Y. Enzymatic Characterization of Unused Biomass Degradation Using the Clostridium cellulovorans Cellulosome. Microorganisms 2022, 10, 2514. https://doi.org/10.3390/microorganisms10122514
Eljonaid MY, Tomita H, Okazaki F, Tamaru Y. Enzymatic Characterization of Unused Biomass Degradation Using the Clostridium cellulovorans Cellulosome. Microorganisms. 2022; 10(12):2514. https://doi.org/10.3390/microorganisms10122514
Chicago/Turabian StyleEljonaid, Mohamed Yahia, Hisao Tomita, Fumiyoshi Okazaki, and Yutaka Tamaru. 2022. "Enzymatic Characterization of Unused Biomass Degradation Using the Clostridium cellulovorans Cellulosome" Microorganisms 10, no. 12: 2514. https://doi.org/10.3390/microorganisms10122514





