Comparison of Fermentation Behaviors and Characteristics of Tomato Sour Soup between Natural Fermentation and Dominant Bacteria-Enhanced Fermentation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of the Starter
2.2. Sample Collection and Analysis
CTN Sample Collection
2.3. Extraction of Microbial Genome
2.4. Bioinformatics Analysis
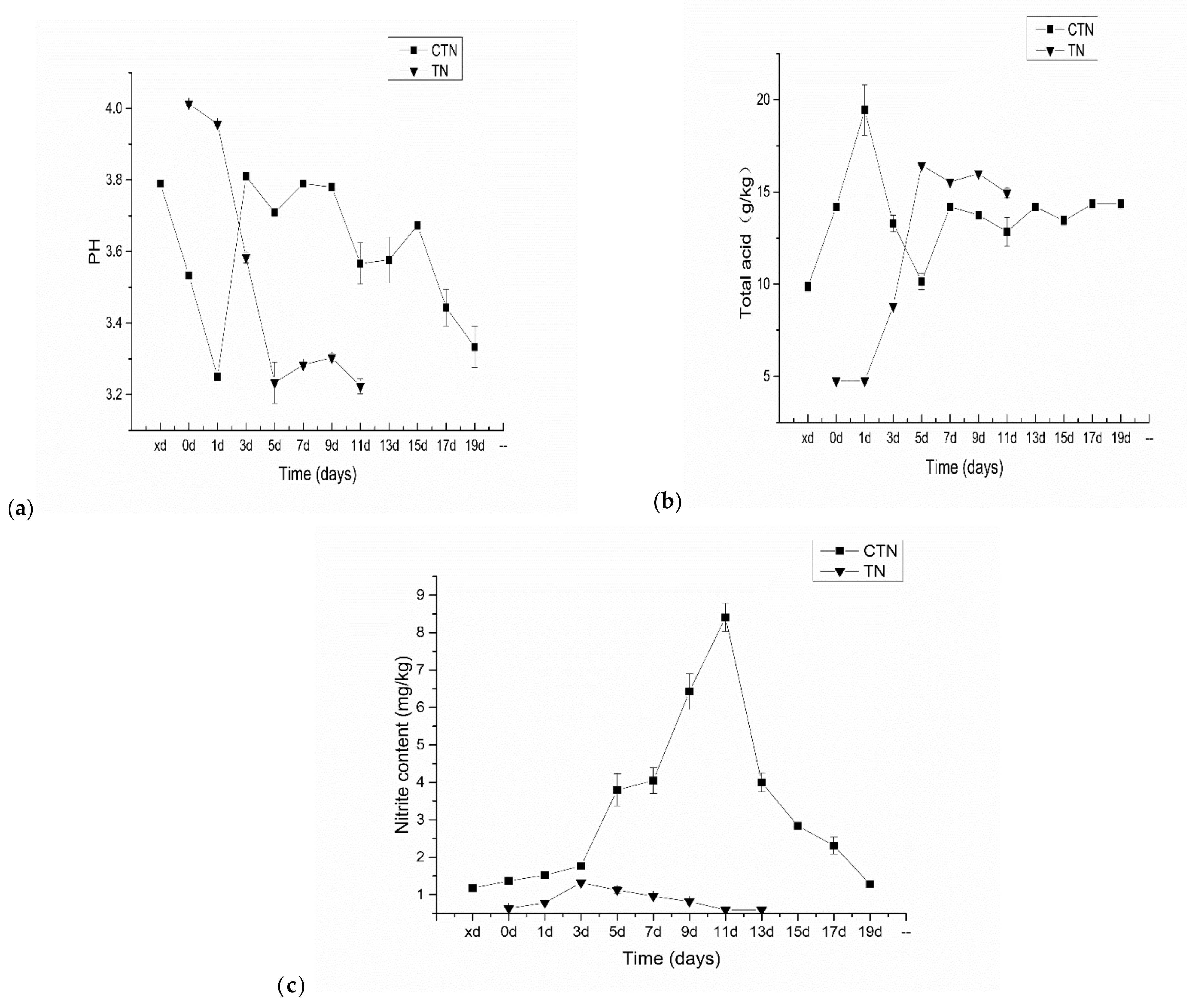
2.5. Analysis of the Physicochemical Indexes
HPLC Detection of Organic Acids
2.6. GC–IMS Analysis of VFCs
2.7. Statistical Analysis
3. Results and Discussion
3.1. Abundances of Bacterial and Fungal Sequences in CTN
3.2. Bacterial Microbiota in CTN Samples
3.3. Fungal Microbiota in CTN Samples
3.4. Physicochemical Indices Analysis of TN Fermentation Process
3.5. HPLC Detection of Organic Acids
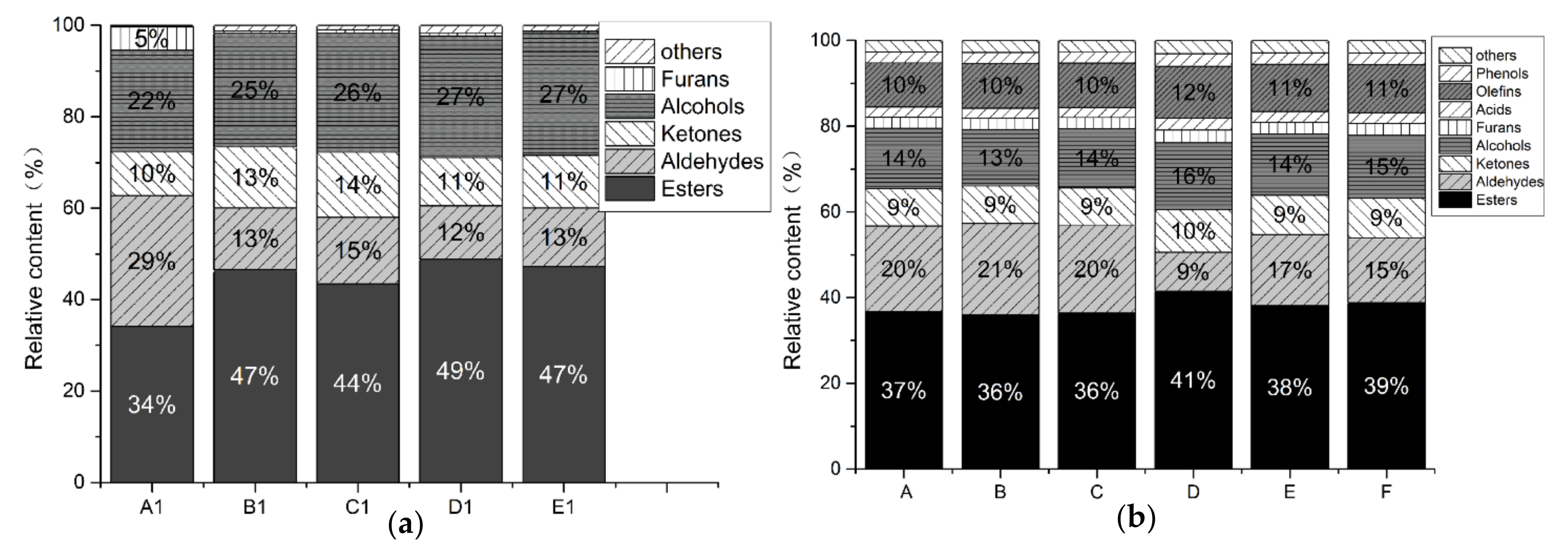
3.6. GC–IMS Analysis for VFCs
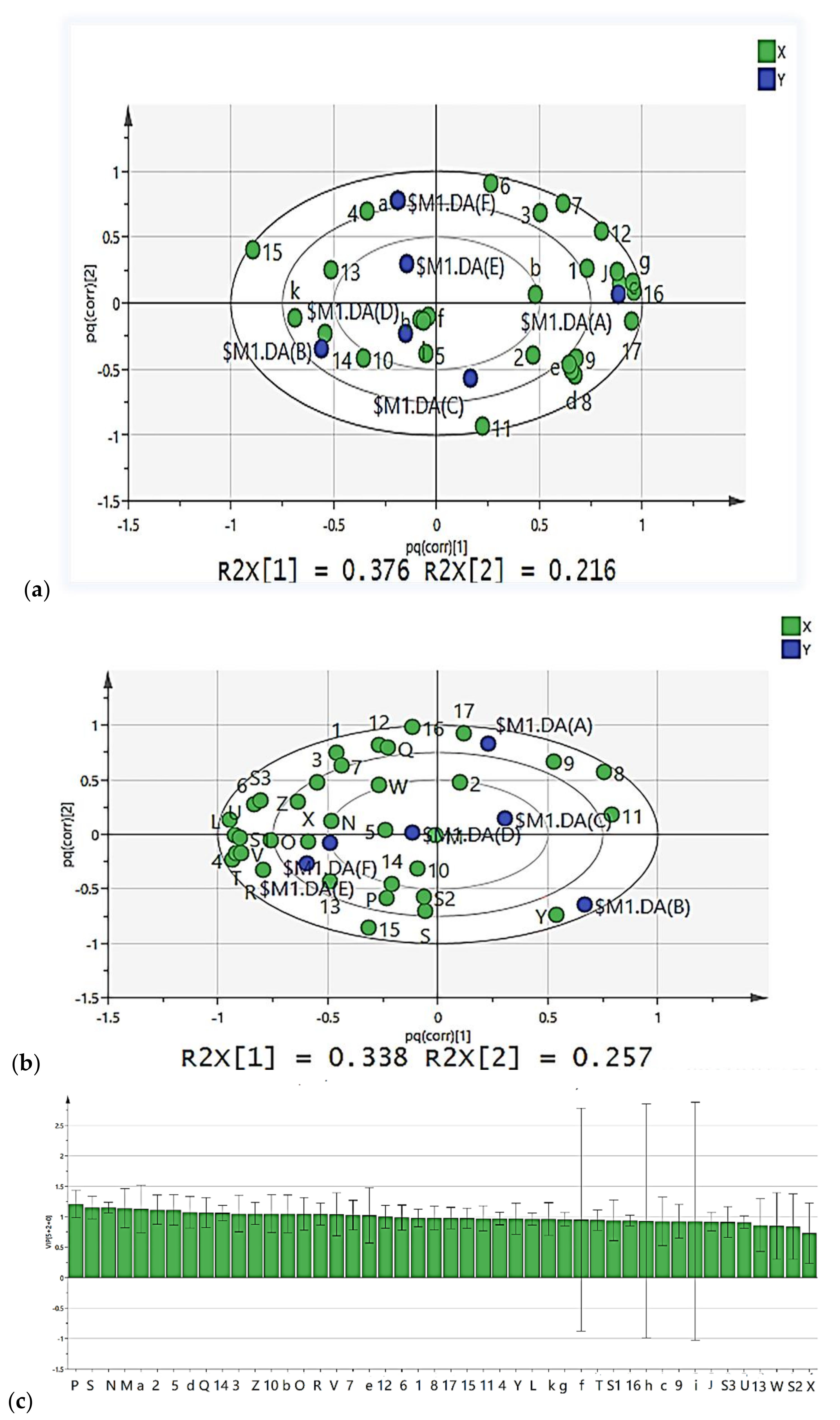
3.7. CTN Multi-Factor Analysis
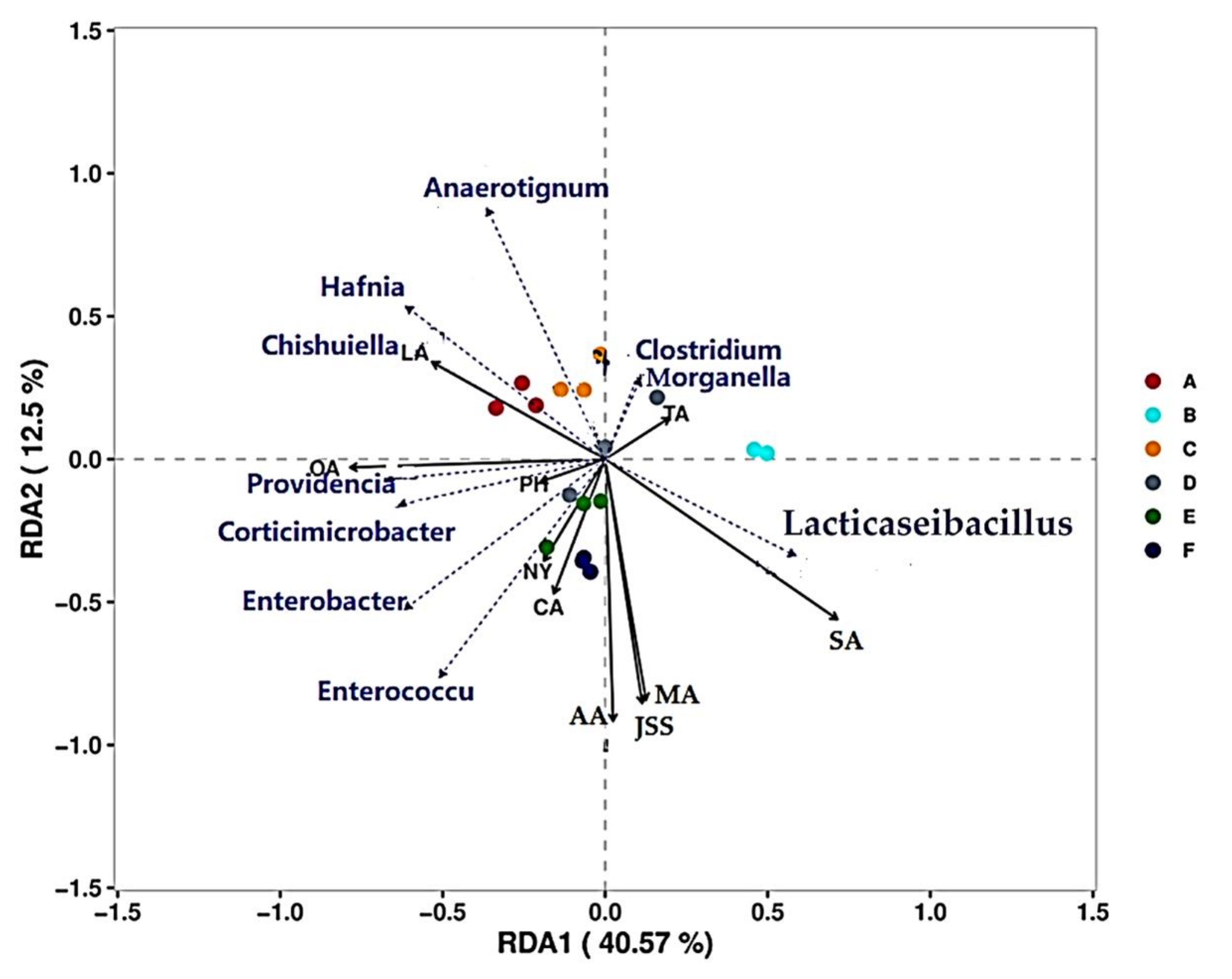
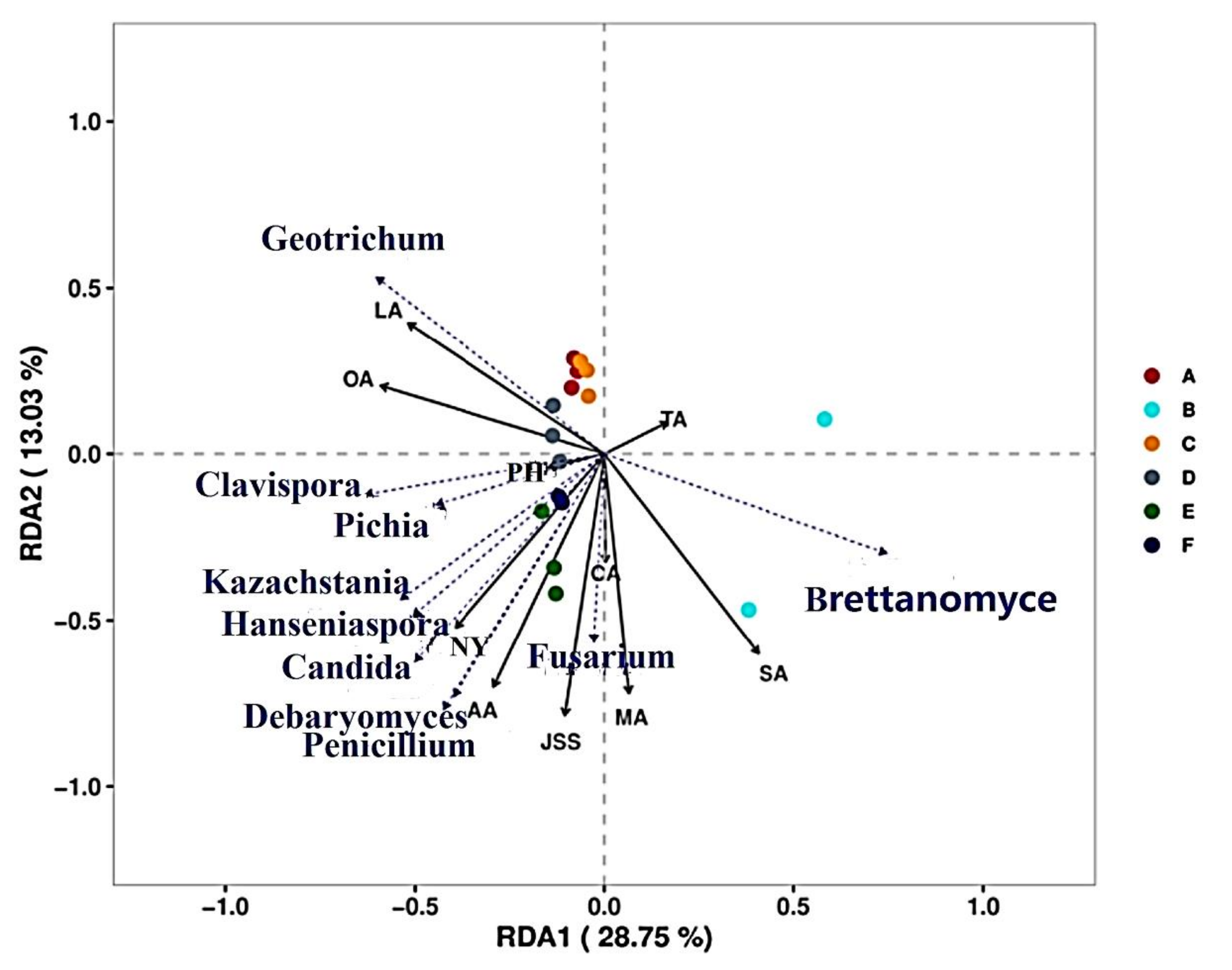
3.7.1. Correlation Analysis of Microorganisms and Physicochemical Indices during CTN Fermentation
3.7.2. Correlation Analysis between Microorganisms and Key VFCs during CTN Fermentation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, S.; Li, J.L.; Long, L.L.; Yang, T.T.; Wang, Y.; Li, G.F. Quality change of red sour soup during natural fermentation and lactic acid bacteria intensified fermentation. China Brew. 2020, 39, 75–78. [Google Scholar]
- Lu, Y.; Wang, N.L.; Li, X.; Qu, Z.H.; Yang, H.G.; Wang, H.Q. Analysis and Research on Main Nutrition and Functional Components of KaiLi Red Acid Soup. Food Res. Dev. 2019, 40, 163–166. [Google Scholar]
- Lu, Q.S.; Xu, L.; Mu, Q.; Wang, M.; Zhang, D.Y. Optimization of Extraction Process and Antioxidant Activity of Organic Acid from Kaili Red Sour Soup. Food Ind. 2019, 40, 89–94. [Google Scholar]
- Tiina, R.; Sari, V.; Kristhna, N.; Jukka, T.S. Lycopene, Atherosclerosis, and Coronary Heart Disease. Exp. Biol. Med. 2002, 227, 900–907. [Google Scholar]
- Zeng, R.M.; Han, L.; Huang, P. Research Progress and Industrialization Process on Guizhou Red Sour Soup Hotpot Seasoning. Food Ferment. Technol. 2014, 50, 72–75. [Google Scholar]
- Cong, S.; Li, Z.; Yu, L.; Liu, Y.; Hu, Y.; Bi, Y.; Cheng, M. Integrative proteomic and lipidomic analysis of Kaili Sour Soup-mediated attenuation of high-fat diet-induced nonalcoholic fatty liver disease in a rat model. Nutr. Metab. 2021, 18, 26. [Google Scholar] [CrossRef]
- Sylwia, P. Lycopene—A bioactive carotenoid offering multiple health benefits: A review. Int. J. Food Sci. Technol. 2020, 55, 11–32. [Google Scholar]
- Li, J.; Gong, L.L.; Chen, F.Y.; Zhang, N.; Cui, L.Y. Analysis of the research status of Kaili Sour Decoction. Ind. Sci. Trib. 2020, 19, 55–57. [Google Scholar]
- He, Z.; Chen, H.; Wang, X.; Lin, X.; Ji, C.; Li, S.; Liang, H. Effects of different temperatures on bacterial diversity and volatile flavor compounds during the fermentation of suancai, a traditional fermented vegetable food from northeastern China. Lwt-Food Sci. Technol. 2020, 118, 108773. [Google Scholar] [CrossRef]
- Ping-Mei, Y.; Wen-Tong, X.; Sze-Sze, T.; Hui, Z.; Xiao-Hui, C. Effect of inoculating lactic acid bacteria starter cultures on the nitrite concentration of fermenting Chinese paocai. Food Control 2007, 19, 50–55. [Google Scholar]
- Pan, J.H.; Qin, L.K.; Wen, A.Y.; Yang, Z.J.; Liu, N. Analysis of nutritional quality and flavor characteristics of red acid soup in Guizhou province. China Condiment 2020, 45, 43–48. [Google Scholar]
- Lin, L.J.; Du Fang, M.; Zeng, J.; Liang, Z.J.; Zhang, X.Y.; Gao, X.Y. Deep insights into fungal diversity in traditional Chinese sour soup by Illumina MiSeq sequencing. Food Res. Int. 2020, 137, 109439. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Pan, J.; Miao, S.; Qin, L. Microbial community in Chinese traditional fermented acid rice soup (rice-acid) and its correlations with key organic acids and volatile compounds. Food Res. Int. 2020, 137, 109672. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Duan, F.; Tian, Q.; Zhong, D.; Wang, X.; Jia, L. Physiochemical, microbiological and flavor characteristics of traditional Chinese fermented food Kaili Red Sour Soup. LWT 2021, 142, 110933. [Google Scholar] [CrossRef]
- Xiong, K.; Han, F.; Wang, Z.; Ming, D.; Chen, Y.; Tang, Y.; Wang, Z. Screening of dominant strains in red sour soup from Miao nationality and the optimization of inoculating fermentation conditions. Food Sci. Nutr. 2020, 9, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Wu, J.; Chen, X.; Huang, L.; Zhang, X.; Gao, X. The Role of the Bacterial Community in Producing a Peculiar Smell in Chinese Fermented Sour Soup. Microorganisms 2020, 8, 1270. [Google Scholar] [CrossRef]
- Liang, H.; He, Z.; Wang, X.; Song, G.; Chen, H.; Lin, X.; Ji, C.; Zhang, S. Bacterial profiles and volatile flavor compounds in commercial Suancai with varying salt concentration from Northeastern China. Food Res. Int. 2020, 137, 09384. [Google Scholar] [CrossRef]
- Chen, Z.; Kang, J.; Zhang, Y.; Yi, X.; Pang, X.; Li, B.H.; Gao, X. Differences in the bacterial profiles and physicochemical between natural and inoculated fermentation of vegetables from Shanxi Province. Ann. Microbiol. 2020, 70, 66. [Google Scholar] [CrossRef]
- Yuecheng, M.; Xuliang, C.; Zhehang, S.; Yanhua, L.; Dongdong, C.; Sheng, F.; Jie, C. Exploring core microbiota responsible for the production of volatile flavor compounds during the traditional fermentation of Koumiss. LWT 2021, 135, 110049. [Google Scholar]
- He, W.; Chung, H.Y. Exploring core functional microbiota related with flavor compounds involved in the fermentation of a natural fermented plain sufu (Chinese fermented soybean curd). Food Microbiol. 2019, 90, 103408. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Hu, P.; Wang, J.L.; Liao, Q.W. Isolation of exopolysaccharides-producing lactic acid bacteria and its antioxidant properties. China Brew. 2015, 34, 37–42. [Google Scholar]
- Fangmin, D.; Xiaoyong, Z.; Huarong, G.; Jiajia, S.; Xiangyang, G. Dynamic Changes in the Bacterial Community During the Fermentation of Traditional Chinese Fish Sauce (TCFS) and Their Correlation with TCFS Quality. Microorganisms 2019, 7, 371. [Google Scholar]
- Zheng, S.S.; Hu, P.; Wang, X.Y.; Tian, Y.; Xu, L.; Ding, Z.H.; Yang, X.L.; Li, J.; Li, J.; Feng, D.D. Optimization of Tomato Sour Soup Fermented by Lactobacillus casei. Mod. Food Sci. Technol. 2020, 36, 218–226. [Google Scholar]
- Wang, C.; Zhang, Q.; He, L.; Li, C. Determination of the microbial communities of Guizhou Suantang, a traditional Chinese fermented sour soup, and correlation between the identified microorganisms and volatile compounds. Food Res. Int. 2020, 138, 109820. [Google Scholar] [CrossRef] [PubMed]
- Jeffrey, J.S.; Andrew, L.M. Human safety controversies surrounding nitrate and nitrite in the diet. Nitric Oxide 2012, 26, 259–266. [Google Scholar]
- Tian, Y. Study on the Correlation between the Dominant Microorganisms and Quality Characteristics of Guizhou Rice Sour Soup. Master’s Thesis, Guizhou University, Guiyang, China, 2020. [Google Scholar]
- Gou, M.; Hu, J.; Zhang, T.T.; Ju, N. The Succession of Microbial Community in Raw Milk during Cold Storage by Illumina MiSeq High-throughput Sequencing. J. Chin. Inst. Food Sci. Technol. 2021, 21, 313–319. [Google Scholar]
- Xu, X.; Wu, B.; Zhao, W.; Pang, X.; Lao, F.; Liao, X.; Wu, J. Correlation between autochthonous microbial communities and key odorants during the fermentation of red pepper (Capsicum annuum L.). Food Microbiol. 2020, 91, 103510. [Google Scholar] [CrossRef]
- Xiao, Y.; Xiong, T.; Peng, Z.; Liu, C.; Huang, T.; Yu, H.; Xie, M. Correlation between microbiota and flavours in fermentation of Chinese Sichuan Paocai. Food Res. Int. 2018, 114, 123–132. [Google Scholar] [CrossRef]
- Wu, J.; Tian, T.; Liu, Y.; Shi, Y.; Tao, D.; Wu, R.; Yue, X. The dynamic changes of chemical components and microbiota during the natural fermentation process in Da-Jiang, a Chinese popular traditional fermented condiment. Food Res. Int. 2018, 112, 457–467. [Google Scholar] [CrossRef]
- Wang, Q.Q.; Tian, J.X.; Pan, Z.D.; Du, J.; Xin, J.K.; Zhang, C.B. Analysis of dominant microflora associated with the unique flavor of Guizhou Kaili sour soup using Illumina MiSeq sequencing. Food Ferment. Ind. 2020, 46, 40–47. [Google Scholar]
- Cappello, M.S.; Zapparoli, G.; Logrieco, A.; Bartowsky, E.J. Linking wine lactic acid bacteria diversity with wine aroma and flavour. Int. J. Food Microbiol. 2017, 243, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, A.; Bartowsky, E.; Jiranek, V. Screening of Lactobacillus spp. and Pediococcus spp. for glycosidase activities that are important in oenology. J. Appl. Microbiol. 2005, 99, 1061–1069. [Google Scholar] [CrossRef]
- Wen-Ke, S.; Jia, W.; Fu-Sheng, C.; Xiu-Yan, Z. Effect of Issatchenkia terricola and Pichia kudriavzevii on wine flavor and quality through simultaneous and sequential co-fermentation with Saccharomyces cerevisiae. LWT 2019, 116, 108477. [Google Scholar]
- Wu, R.; Yu, M.; Liu, X.; Meng, L.; Wang, Q.; Xue, Y.; Wu, J.; Yue, X. Changes in flavour and microbial diversity during natural fermentation of suan-cai, a traditional food made in Northeast China. Int. J. Food Microbiol. 2015, 211, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Qisheng, Z.; Gong, C.; Wenxi, S.; Yong, W.; Wenxue, Z.; Yuanlong, C. Microbial safety and sensory quality of instant low-salt Chinese paocai. Food Control 2016, 59, 575–580. [Google Scholar]
- Li, W.Z.; Han, S.; Zi, Y.Y.; Bo, L.; Yan, Z.; Xin, T.W.; Chun, Y.Z. A New Method of Fermenting Northeast Sauerkraut Rapidly by Lactic Acid Bacteria. Adv. Mater. Res. 2015, 3702, 1858–1862. [Google Scholar]
- Zhang, J.; Yang, J.F.; Chen, J.; Zhu, Y.T.; Hu, K.; Ma, Q.; Zuo, Y. A novel propylene glycol alginate gel based colorimetric tube for rapid detection of nitrite in pickled vegetables. Food Chem. 2022, 373, 131678. [Google Scholar] [CrossRef]
- Yan, Y.M.; Lv, J.Z.; Wang, X.P. Research on pickle starter with the properties of degradation nitrite. China Condiment 2016, 41, 35–41. [Google Scholar]
- Tang, Q.L.; Yang, Z.N.; Wu, T.X. Preliminary study on the characteristic components in fermented sour soup of Miao people in Guizhou province. Sci. Technol. Food Ind. 2005, 9, 165–166. [Google Scholar]
- Zhang, D.Y. Research on the Optimization of Fermentation Process and Quality Control of the Red Sour Soup. Master’s Thesis, Guizhou University, Guiyang, China, 2018. [Google Scholar]
- Saerens, K.; Descamps, D.; Dewettinck, K. Release of short chain fatty acids from cream lipids by commercial lipases and esterases. Biotechnol. Lett. 2008, 30, 311–315. [Google Scholar] [CrossRef]
- Picard, C.; Fioramonti, J.; Francois, A.; Robinson, T.; Neant, F.; Matuchansky, C. Review article: Bifidobacteria as probiotic agents—Physiological effects and clinical benefits. Aliment Pharm. 2005, 22, 495–512. [Google Scholar] [CrossRef]
- Shang, J.T.; Wang, X.J.; Wang, J.H. Effect of degradation of nitrite by organic acids in pickles. FoodMach 2018, 34, 73–78. [Google Scholar]
- Khairy, H.L.; Saadoon, A.F.; Zzaman, W.; Yang, T.A.; Easa, A.M. Identification of flavor compounds in rambutan seed fat and its mixture with cocoa butter determined by SPME-GCMS. J. King Saud Univ.-Sci. 2017, 30, 2316–2323. [Google Scholar] [CrossRef]
- Harada, R.; Yuzuki, M.; Ito, K.; Shiga, K.; Bamba, T.; Fukusaki, E. Microbe participation in aroma production during soy sauce fermentation. J. Biosci. Bioeng. 2018, 125, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.; Fan, W.; Xu, Y. Changes in Volatile Compounds of Chinese Rice Wine Wheat Qu during Fermentation and Storage. J. Inst. Brew. 2009, 115, 300–307. [Google Scholar] [CrossRef]
- Cai-Lin, K.; Ai-Hua, L.; Jun, S.; Xing-Chen, W.; Chao-Qi, C.; Yong-Sheng, T. Flavor modification of dry red wine from Chinese spine grape by mixed fermentation with Pichia fermentans and S. cerevisiae. LWT 2019, 109, 83–92. [Google Scholar]
- Zhao, C.; Su, W.; Mu, Y.; Jiang, L.; Mu, Y. Correlations between microbiota with physicochemical properties and volatile flavor components in black glutinous rice wine fermentation. Food Res. Int. 2020, 138, 109800. [Google Scholar] [CrossRef]
- Kennes, C.; Veiga, M.C.; Dubourguier, H.C.; Touzel, J.P.; Albagnac, G.; Naveau, H.; Nyns, E.J. Trophic relationships between Saccharomyces cerevisiae and Lactobacillus plantarum and their metabolism of glucose and citrate. Appl. Environ. Microbiol. 1991, 57, 1046–1051. [Google Scholar] [CrossRef] [Green Version]

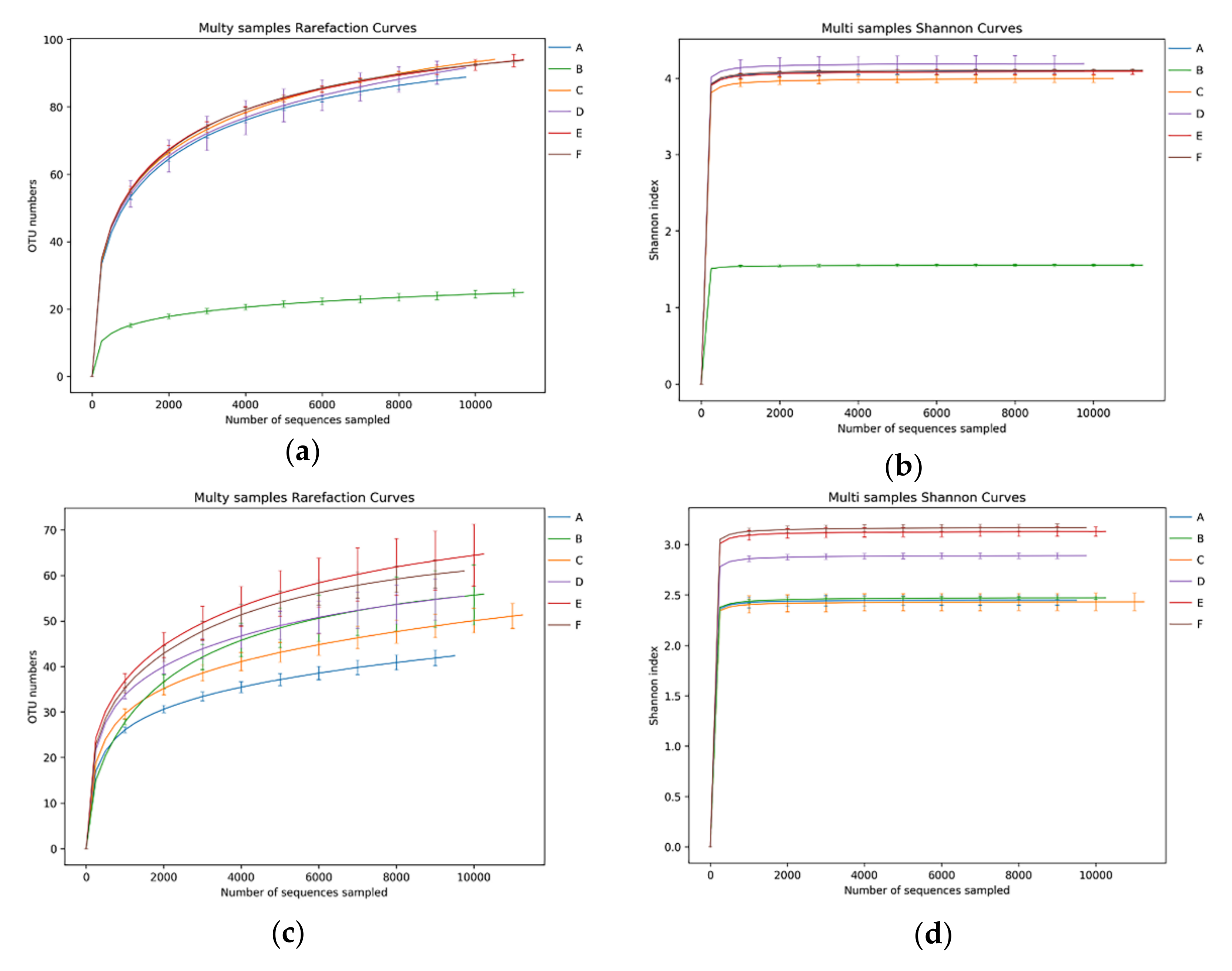
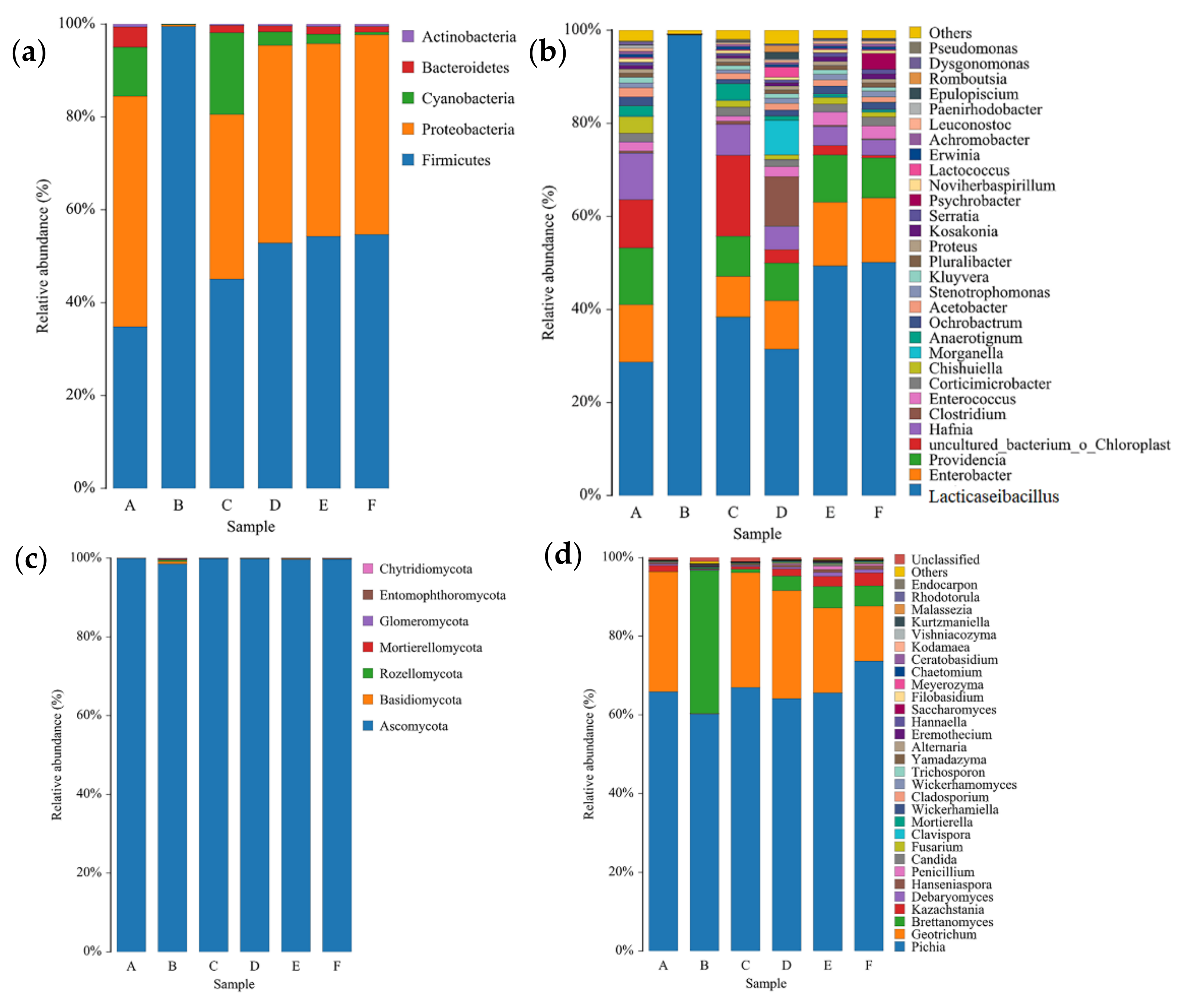
| Sample | Richness Index | Diversity Index | Coverage | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ACE | Chao 1 | Simpson | Shannon | Bacteria | Fungi | |||||
| Bacteria | Fungi | Bacteria | Fungi | Bacteria | Fungi | Bacteria | Fungi | |||
| A | 96.41 ± 3.93 b | 56.72 ± 9.07 a | 94.87 ± 3.45 b | 52.19 ± 6.54 a | 0.9 ± 0.003 a | 0.70 ± 0.02 d | 4.09 ± 0.05 ab | 2.45 ± 009 c | 0.999 | 0.9992 |
| B | 29.87 ± 1.97 c | 60.55 ± 14.77 a | 27.32 ± 1.36 c | 59.50 ± 13.45 a | 0.45 ± 0.01 b | 0.74 ± 0.01 c | 1.55 ± 0.03 c | 2.47 ± 0.03 c | 0.9997 | 0.9994 |
| C | 106.3 ± 4.7 ab | 71.69 ± 10.81 a | 105.51 ± 5.20 ab | 63.95 ± 6.65 a | 0.89 ± 0.01 a | 0.67 ± 0.03 e | 4.00 ± 0.08 b | 2.43 ± 0.15 c | 0.9987 | 0.999 |
| D | 121.17 ± 26.52 a | 64.22 ± 12.29 a | 122.89 ± 15.01 a | 66.86 ± 12.92 a | 0.91 ± 0.01 a | 0.77 ± 0.01 b | 4.19 ± 0.19 a | 2.89 ± 0.05 b | 0.9983 | 0.9991 |
| E | 103.8 ± 8.53 ab | 73.198 ± 15.50 a | 114.32 ± 19.22 ab | 72.53 ± 18.22 a | 0.90 ± 00.003 a | 0.79 ± 0.01 ab | 4.09 ± 0.05 ab | 3.13 ± 0.08 a | 0.9988 | 0.9991 |
| F | 100.64 ± 4.47 ab | 66.52 ± 6.88 a | 100.56 ± 5.39 b | 64.86 ± 7.67 a | 0.90 ± 0.002 a | 0.82 ± 0.01 a | 4.11 ± 0.02 ab | 3.17 ± 0.06 a | 0.999 | 0.9992 |
| Category | Oxalic Acid | Tartaric Acid | Malic Acid | Lactic Acid | Acetic Acid | Citric Acid | Succinic Acid | Propionic Acid | Fumaric Acid |
|---|---|---|---|---|---|---|---|---|---|
| A (mg·mL−1) | 0.37 ± 0.05 d | 0.84 ± 0.01 cd | 1.22 ± 0.05 f | 27.09 ± 0.06 ef | 5.93 ± 0.06 d | 4.33 ± 0.11 f | 9.03 ± 0.05 g | 0.2 ± 0.12 f | 0 |
| B (mg·mL−1) | 0.15 ± 0.06 f | 1.02 ± 0.03 bc | 2.49 ± 0.01 d | 17.22 ± 0.04 h | 6.71 ± 0.05 cd | 3.45 ± 0.06 h | 16.23 ± 0.17 b | 1.3 ± 0.05 b | 0 |
| C (mg·mL−1) | 0.18 ± 0.05 ef | 0.63 ± 0.04 de | 0.53 ± 0.05 h | 44.28 ± 0.51 a | 6.09 ± 0.05 d | 1.88 ± 0.05 j | 12.93 ± 0.05 f | 0.67 ± 0.12 e | 0 |
| D (mg·mL−1) | 0.22 ± 0.05 e | 1.04 ± 0.02 bc | 0.36 ± 0.02 i | 25.08 ± 0.12 g | 7.37 ± 0.02 c | 2.74 ± 0.05 i | 14.29 ± 0.11 e | 0.93 ± 0.06 d | 0 |
| E (mg·mL−1) | 0.23 ± 0.04 e | 1.33 ± 0.03 a | 3.66 ± 0.02 c | 27.18 ± 0.12 ef | 8.31 ± 0.57 b | 3.82 ± 0.12 g | 14.52 ± 0.17 d | 1.06 ± 0.06 c | 0 |
| F (mg·mL−1) | 0.24 ± 0.01 e | 1.21 ± 0.05 ab | 4.1 ± 0.06 b | 28.72 ± 0.12 d | 9.18 ± 0.58 a | 3.92 ± 0.05 g | 15.26 ± 0.12 c | 1.14 ± 0.017 c | - |
| A1 (mg·mL−1) | 0.37 ± 0.01 d | 0.71 ± 0.52 de | 4.85 ± 0.10 a | 3.12 ± 0.06 j | 3.62 ± 0.05 e | 7.5 ± 0.02 e | 0.85 ± 0.01 l | 0 | 0.71 ± 0.01 b |
| B1 (mg·mL−1) | 1.22 ± 0.01 b | 0.47 ± 0.01 e | 1.56 ± 0.03 e | 5.5 ± 0.05 i | 1.3 ± 0.58 f | 10.38 ± 0.23 d | 2.25 ± 0.02 k | 0 | 0.33 ± 0.03 c |
| C1 (mg·mL−1) | 1.4 ± 0.01 a | 0.41 ± 0.01 e | 1.57 ± 0.09 e | 26.57 ± 0.58 f | 3.88 ± 0.01 e | 11.98 ± 0.58 c | 3.54 ± 0.14 j | 0 | 0.02 ± 0.01 d |
| D1 (mg·mL−1) | 0.38 ± 0.06 d | 0.49 ± 0.03 e | 0.95 ± 0.03 g | 30.09 ± 0.57 c | 6.66 ± 0.58 cd | 12.51 ± 0.05 b | 6.68 ± 0.05 i | 0 | 0.03 ± 0.01 d |
| E1 (mg·mL−1) | 0.78 ± 0.01 c | 0.49 ± 0.02 e | 0.45 ± 0.06 hi | 31.26 ± 0.58 b | 6.2 ± 0.57 d | 12.85 ± 0.05 a | 7.42 ± 0.05 h | 0 | 0.02 ± 0.01 d |
| F1 (mg·mL−1) | 0.23 ± 0.01 e | 0.62 ± 0.06 de | 1.2 ± 0.08 f | 30.75 ± 0.57 e | 6.52 ± 0.57 d | 7.28 ± 0.17 e | 18.54 ± 0.12 a | 1.44 ± 0.05 a | 1.95 ± 0.01 a |
| Compound | Retention Index | Retention Time/s | Migration Time/ms | F | D1 |
|---|---|---|---|---|---|
| Esters | |||||
| Methyl Salicylate | 1235.7 | 688.365 | 1.20588 | 424.79 | – |
| Butyl butanoate | 1004.3 | 356.079 | 1.81327 | 280.95 | – |
| Pentyl acetate | 912.8 | 274.089 | 1.76114 | 239.64 | 154.88 |
| Isoamyl acetate | 875 | 247.755 | 1.75345 | 224.98 | 6023.73 |
| Ethyl 2-methylbutanoate | 848.8 | 234.134 | 1.66369 | 216.25 | – |
| Ethyl 2-hydroxypropanoate | 818.5 | 218.374 | 1.54705 | 206.15 | – |
| Ethyl butanoate | 792.4 | 204.821 | 1.56923 | 197.52 | 105.65 |
| Butyl acetate | 806.1 | 211.905 | 1.63724 | 202.08 | – |
| Methyl 2-methylbutanoate | 769.8 | 194.685 | 1.54033 | 190.52 | – |
| 2-Methylpropyl acetate | 764 | 192.337 | 1.62702 | 188.86 | – |
| Ethyl 2-methylpropanoate | 748.9 | 186.175 | 1.58294 | 184.37 | 268.66 |
| Ethyl propanoate | 705.9 | 168.646 | 1.46174 | 171.59 | – |
| Ethyl Acetate | 596.7 | 137.37 | 1.34657 | 143.45 | 13898.89 |
| Isopropyl acetate | 654.9 | 152.715 | 1.48967 | 158.07 | – |
| Propyl butanoate | 898.9 | 262.349 | 1.69733 | 233.57 | – |
| Methyl hexanoate | 923.8 | 283.382 | 1.6923 | 244.39 | – |
| Ethyl heptanoate | 1100.3 | 493.969 | 1.92705 | 340.83 | – |
| Ethyl hexanoate-M | 1005.5 | 357.86 | 1.33 | – | 334.68 |
| Ethyl hexanoate-D | 1006.3 | 359.03 | 1.81 | – | 1157.28 |
| Methyl hexanoate | 923.9 | 283.5 | 1.69 | – | 265.26 |
| Ethyl butanoate | 793.3 | 205.29 | 1.57 | – | 5383.76 |
| 2-Methylpropyl acetate | 768 | 193.94 | 1.63 | – | 1753.84 |
| Propyl acetate | 706.3 | 168.82 | 1.47 | – | 1665.82 |
| Methyl acetate | 552.8 | 125.76 | 1.19 | – | 1256.93 |
| Hexyl acetate | 1016.4 | 373.57 | 1.91 | – | 591.28 |
| Olefins | |||||
| Limonene | 1031.8 | 395.617 | 1.21701 | 297.79 | 307.08 |
| alpha-Terpinene | 1025.4 | 386.493 | 1.22695 | 293.83 | – |
| alpha-Pinene | 946.6 | 302.684 | 1.22113 | 254.07 | – |
| Aldehydes | |||||
| 2-Decenal | 1267 | 733.342 | 1.47569 | 444.44 | – |
| (E)-2-Heptenal | 959.3 | 313.499 | 1.27664 | 259.65 | – |
| Benzaldehyde | 961.5 | 315.324 | 1.47823 | 260.7 | – |
| 3-Methylbutanal | 653.1 | 152.236 | 1.41542 | 157.58 | 308.79 |
| 2-Methylbutanal | 679.4 | 159.186 | 1.39664 | 164.15 | – |
| Butanal | 579.2 | 132.737 | 1.304 | 139.05 | – |
| (E)-2-Hexenal | 864.7 | 242.411 | 1.52651 | 221.46 | 2919.78 |
| Vanillin | 1398.9 | 922.715 | 1.26926 | 526.49 | – |
| Nonanal | 1110.2 | 508.25 | 1.48 | – | 115.96 |
| (E)-2-Octenal-M | 1058.6 | 434.04 | 1.34 | – | 488.64 |
| (E)-2-Octenal-D | 1058.6 | 434.04 | 1.83 | – | 117.27 |
| (E)-2-Heptenal-D | 955 | 309.85 | 1.68 | – | 58.54 |
| (Z)-2-Hexenal | 844.9 | 232.09 | 1.53 | – | 887.34 |
| Furfural | 828.5 | 223.58 | 1.34 | – | 399.64 |
| (E)-2-Pentenal | 749.4 | 186.37 | 1.37 | – | 244.06 |
| Propanal | 523.5 | 118.04 | 1.15 | – | 1403.93 |
| Pentanal | 702.3 | 167.22 | 1.41 | – | 895.79 |
| Ketones | |||||
| 6-Methyl-5-hepten-2-one | 993.1 | 342.088 | 1.18294 | 274.31 | 4247.15 |
| 3-Pentanone | 691.6 | 162.854 | 1.34156 | 167.31 | 438.19 |
| 2-Pentanone | 695.4 | 164.38 | 1.37 | – | 2387.85 |
| 2-Butanone | 572.5 | 130.984 | 1.25643 | 137.36 | |
| Alcohols | |||||
| 2-Octanol | 1004.7 | 356.687 | 1.45694 | 281.03 | – |
| (E)-3-hexen-1-ol | 835.7 | 227.325 | 1.54284 | 211.85 | – |
| 3-Methylpentanol | 840.2 | 229.658 | 1.60397 | 213.37 | – |
| 2-Hexanol | 803.7 | 210.673 | 1.58697 | 201.26 | 1985.79 |
| 3-Methyl-1-butanol | 727.7 | 177.527 | 1.47801 | 178.04 | – |
| Propanol | 543 | 123.195 | 1.27084 | 129.99 | 487.59 |
| 2-Propanol | 510.4 | 114.57 | 1.20354 | 121.8 | 4498.07 |
| Linalool | 1104.7 | 500.263 | 1.2256 | 343.2 | |
| Ethanol | 466.1 | 102.865 | 1.14036 | 110.69 | 8048.69 |
| 1-Hexanol | 875.8 | 248.17 | 1.64 | – | 1985.79 |
| 2-Methyl-1-butanol | 751.7 | 187.32 | 1.49 | – | 113.88 |
| 2-Pentenal | 738.2 | 181.83 | 1.36 | – | 381.44 |
| Isopentanol | 728.6 | 177.9 | 1.5 | – | 1914.83 |
| Isohexanol | 845.2 | 232.27 | 1.62 | – | 316.96 |
| 1-Pentanol | 761 | 191.09 | 1.51 | – | 71.46 |
| Acids | |||||
| Hexanoic acid | 938.5 | 295.882 | 1.66882 | 250.8 | |
| Propanoic acid | 714.4 | 172.15 | 1.26 | – | 92.86 |
| Phenols | |||||
| Methional | 986 | 336.005 | 1.48249 | 271.35 | – |
| Furans | |||||
| 2-Ethylfuran | 682.9 | 160.13 | 1.32 | – | 356.27 |
| 2-Pentylfuran | 997.1 | 345.738 | 1.2596 | 276.18 | 90.3 |
| others | |||||
| 2-Ethyl pyrazine | 924.6 | 284.1 | 1.52 | – | 317.48 |
| 2-Acetylthiazole | 1015.3 | 371.894 | 1.49811 | 287.65 | 821.19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Wang, X.; Wu, W.; Jiang, J.; Feng, D.; Shi, Y.; Hu, P. Comparison of Fermentation Behaviors and Characteristics of Tomato Sour Soup between Natural Fermentation and Dominant Bacteria-Enhanced Fermentation. Microorganisms 2022, 10, 640. https://doi.org/10.3390/microorganisms10030640
Li J, Wang X, Wu W, Jiang J, Feng D, Shi Y, Hu P. Comparison of Fermentation Behaviors and Characteristics of Tomato Sour Soup between Natural Fermentation and Dominant Bacteria-Enhanced Fermentation. Microorganisms. 2022; 10(3):640. https://doi.org/10.3390/microorganisms10030640
Chicago/Turabian StyleLi, Juan, Xiaoyu Wang, Wenyan Wu, Jingzhu Jiang, Dandan Feng, Yuanyuan Shi, and Ping Hu. 2022. "Comparison of Fermentation Behaviors and Characteristics of Tomato Sour Soup between Natural Fermentation and Dominant Bacteria-Enhanced Fermentation" Microorganisms 10, no. 3: 640. https://doi.org/10.3390/microorganisms10030640





