Responses of Phyllosphere Microbiome to Ozone Stress: Abundance, Community Compositions and Functions
Abstract
:1. Introduction
2. Materials and Methods
3. Results
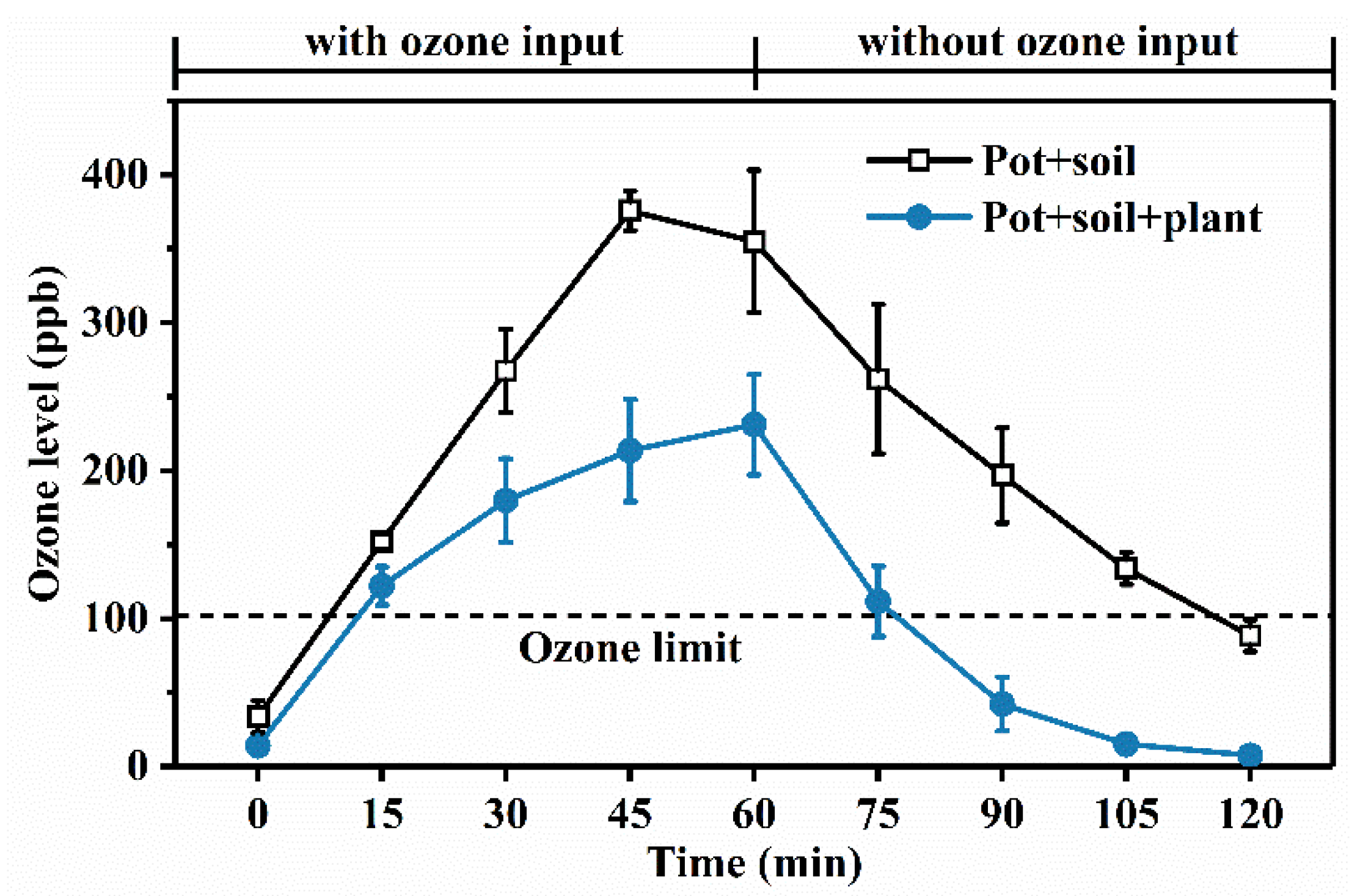
3.1. Ozone Phylloremediation
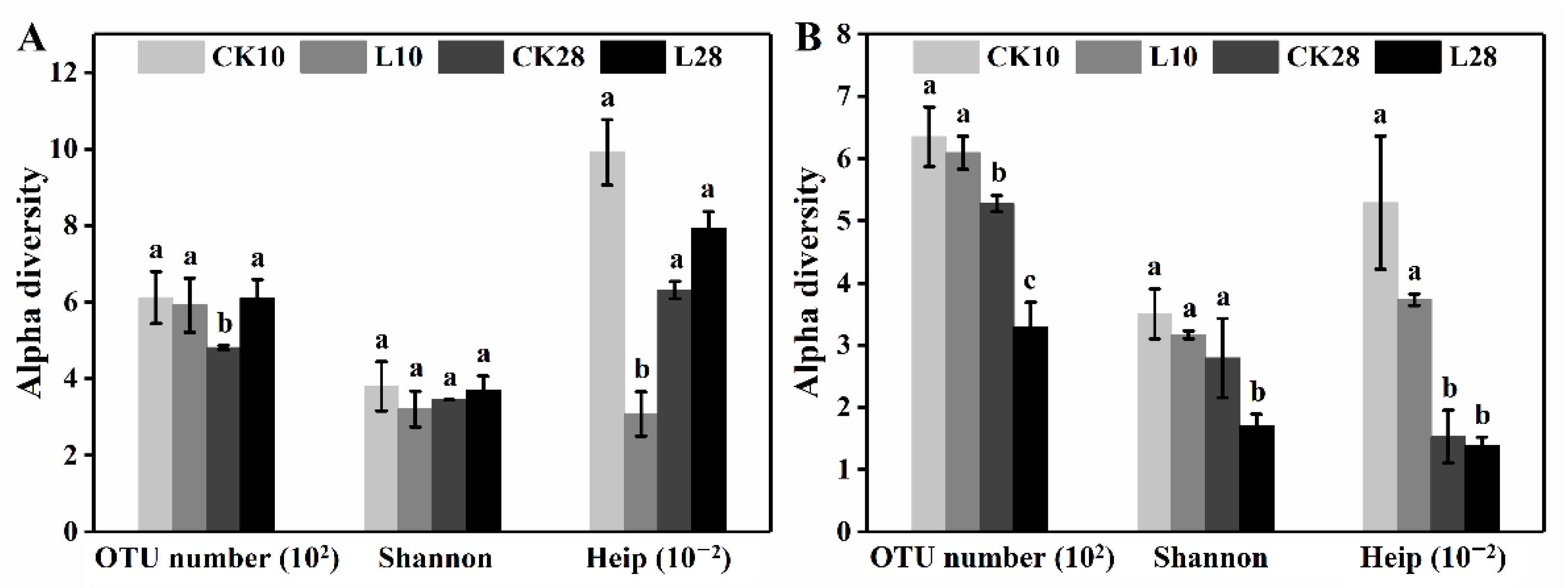
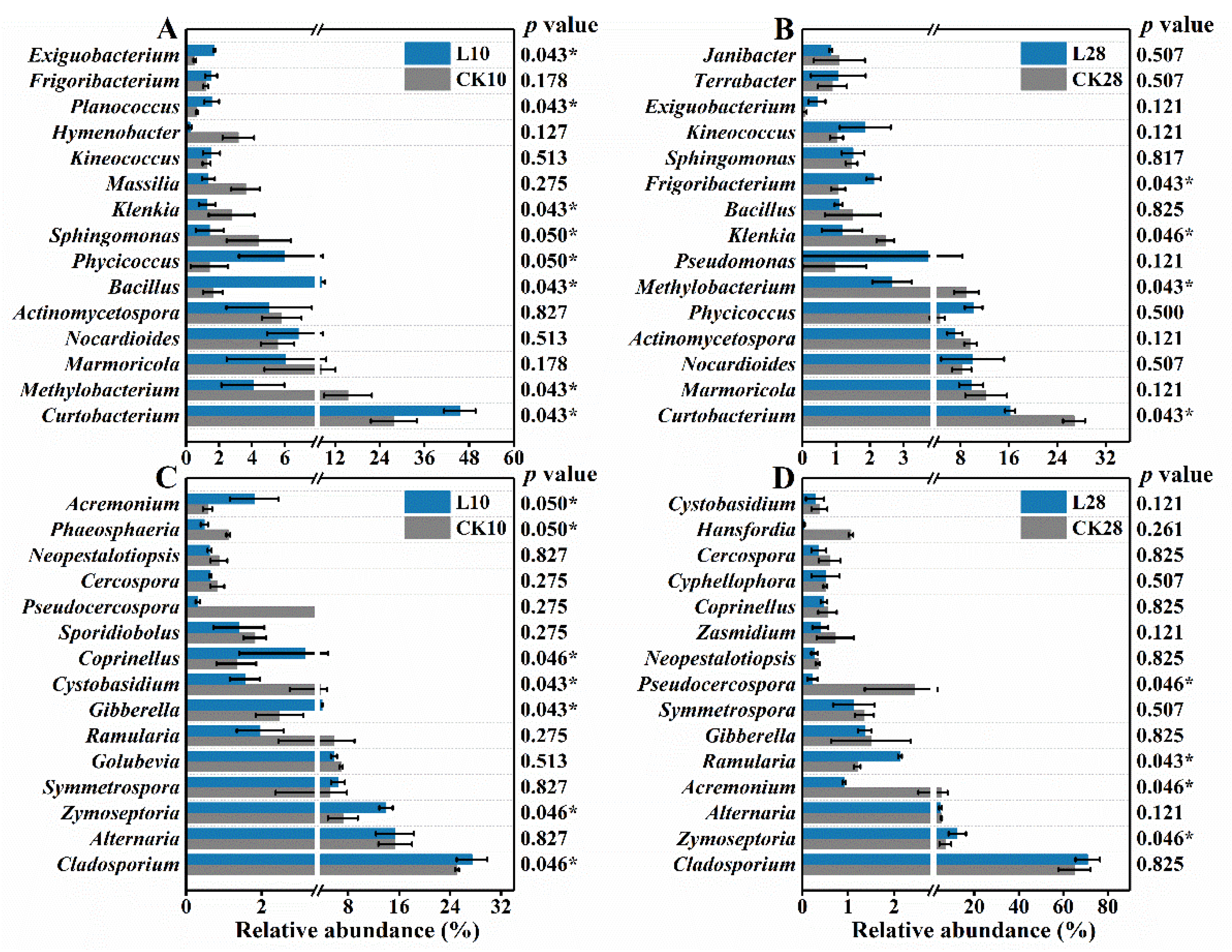
3.2. Low-Level Ozone Exposure
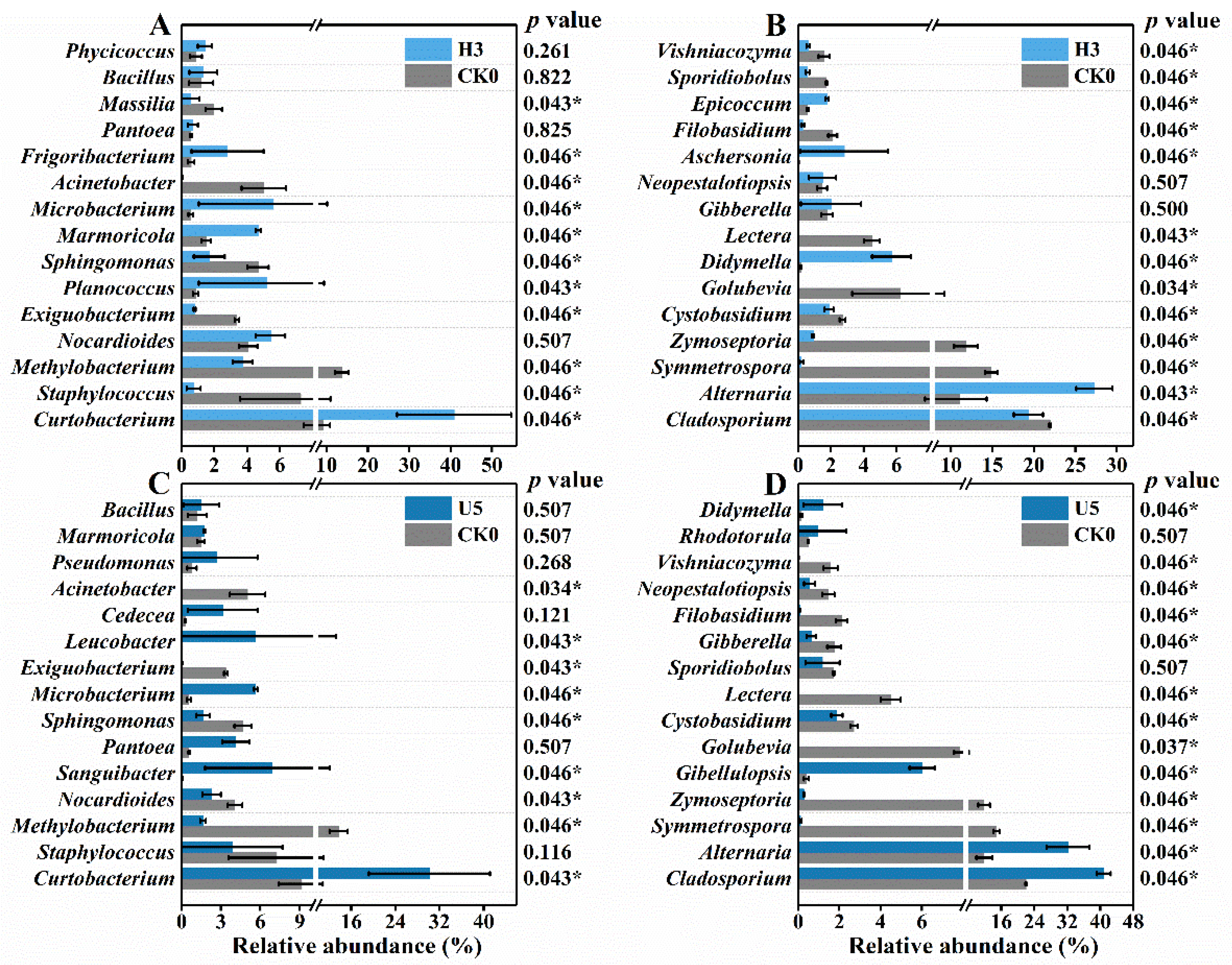
3.3. (Ultra)High-Level Ozone Exposure
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Phillips, D.L.; Johnson, M.G.; Tingey, D.T.; Storm, M.J. Elevated CO2 and O3 effects on fine-root survivorship in ponderosa pine mesocosms. Oecologia 2009, 160, 827–837. [Google Scholar] [CrossRef]
- Canella, R.; Borriello, R.; Cavicchio, C.; Cervellati, F.; Martini, M.; Muresan, X.; Valacchi, G. Tropospheric ozone effects on chlorine current in lung epithelial cells: An electrophysiological approach. Free Radic. Biol. Med. 2016, 96, S58–S59. [Google Scholar] [CrossRef]
- Monks, P.S.; Archibald, A.T.; Colette, A.; Cooper, O.; Coyle, M.; Derwent, R.; Fowler, D.; Granier, C.; Law, K.S.; Mills, G.E.; et al. Tropospheric ozone and its precursors from the urban to the global scale from air quality to short-lived climate forcer. Atmos. Chem. Phys. 2015, 15, 8889–8973. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Qiao, X.; Hopke, P.K.; Ying, Q.; Zhang, Y.; Zeng, Y.; Yuan, Y.; Tang, Y. Ozone pollution in the west China rain zone and its adjacent regions, Southwestern China: Concentrations, ecological risk, and Sources. Chemosphere 2020, 256, 127008. [Google Scholar] [CrossRef]
- Avnery, S.; Mauzerall, D.L.; Liu, J.; Horowitz, L.W. Global crop yield reductions due to surface ozone exposure: 1. Year 2000 crop production losses and economic damage. Atmos. Environ. 2011, 45, 2284–2296. [Google Scholar] [CrossRef]
- Fares, S.; Park, J.-H.; Ormeno, E.; Gentner, D.R.; McKay, M.; Loreto, F.; Karlik, J.; Goldstein, A.H. Ozone uptake by citrus trees exposed to a range of ozone concentrations. Atmos. Environ. 2010, 44, 3404–3412. [Google Scholar] [CrossRef]
- Loreto, F.; Fares, S. Is ozone flux inside leaves only a damage indicator? Clues from volatile isoprenoid studies. Plant Physiol. 2007, 143, 1096–1100. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, E.; Häikiö, E.; Sober, J.; Karnosky, D.F. Ozone-induced H2O2 accumulation in field-grown aspen and birch is linked to foliar ultrastructure and peroxisomal activity. New Phytol. 2004, 161, 791–799. [Google Scholar] [CrossRef]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef] [Green Version]
- Beattie, G.A.; Lindow, S.E. The secret life of foliar bacterial pathogens on leaves. Annu. Rev. Phytopathol. 1995, 33, 145–172. [Google Scholar] [CrossRef] [PubMed]
- Lindow, S.E.; Brandl, M.T. MINIREVIEW Microbiology of the Phyllosphere. Society 2003, 69, 1875–1883. [Google Scholar] [CrossRef]
- Rangel, K.; Cabral, F.O.; Lechuga, G.C.; Carvalh, J.P.R.S.; Villas-Bôas, M.H.S.; Midlej, V.; De-Simone, S.G. Detrimental effect of ozone on pathogenic bacteria. Microorganisms 2022, 10, 40. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Sun, M.Z.; Zhang, C.; Xin, Y. Antioxidant properties of Lactobacillus and its protecting effects to oxidative stress caco-2 cells. J. Anim. Plant Sci. 2014, 24, 1766–1771. [Google Scholar]
- Fenn, M.E.; Dunn, P.H.; Durall, D.M. Effects of Ozone and Sulfur Dioxide on Phyllosphere Fungi from Three Tree Species. Appl. Environ. Microbiol. 1989, 55, 412–418. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Yu, Y.; Tang, H.; Zu, Q.; Zhu, J.; Lin, X. The contrasting responses of soil microorganisms in two rice cultivars to elevated ground-level ozone. Environ. Pollut. 2015, 197, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tang, H.; Zhu, J.; Lin, X.; Feng, Y. Divergent responses of methanogenic archaeal communities in two rice cultivars to elevated ground-level O3. Environ. Pollut. 2016, 213, 127–134. [Google Scholar] [CrossRef]
- Ueda, Y.; Frindte, K.; Knief, C.; Ashrafuzzaman, M.D.; Frei, M. Effects of elevated tropospheric ozone concentration on the bacterial community in the phyllosphere and rhizoplane of rice. PLoS ONE 2016, 11, e0163178. [Google Scholar] [CrossRef]
- Stevens, V.; Thijs, S.; Bongaerts, E.; Nawrot, T.; Marchal, W.; Van Hamme, J.; Vangronsveld, J. Ambient air pollution shapes bacterial and fungal ivy leaf communities. Microorganisms 2021, 9, 2088. [Google Scholar] [CrossRef]
- Bao, L.; Gu, L.; Sun, B.; Cai, W.; Zhang, S.; Zhuang, G.; Bai, Z.; Zhuang, X. Seasonal variation of epiphytic bacteria in the phyllosphere of Gingko biloba, Pinus bungeana and Sabina chinensis. FEMS Microbiol. Ecol. 2020, 96, fiaa017. [Google Scholar] [CrossRef]
- Voytas, D. Agarose Gel Electrophoresis. Curr. Protoc. Immunol. 1992, 2, 10.4.1–10.4.8. [Google Scholar] [CrossRef]
- Kembel, S.; O’Connor, T.; Arnold, H.; Hubbell, S.; Wright, S.J.; Green, J. Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc. Natl. Acad. Sci. USA 2014, 111, 13715–13720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaalid, R.; Kumar, S.; Nilsson, R.H.; Abarenkov, K.; Kirk, P.M.; Kauserud, H. ITS1 versus ITS2 as DNA metabarcodes for fungi. Mol. Ecol. Resour. 2013, 13, 218–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, M.; Sun, B.; Li, R.; Zhang, Z.; Bai, Z.; Zhuang, X. Dynamic succession patterns and interactions of phyllospheric microorganisms during NOx exposure. J. Hazard. Mater. 2022, 430, 128371. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Aßhauer, K.P.; Wemheuer, B.; Daniel, R.; Meinicke, P. Tax4Fun: Predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 2015, 31, 2882–2884. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Mahilang, M.; Deb, M.K.; Pervez, S. Biogenic secondary organic aerosols: A review on formation mechanism, analytical challenges and environmental impacts. Chemosphere 2021, 262, 127771. [Google Scholar] [CrossRef] [PubMed]
- Kanerva, T.; Palojärvi, A.; Rämö, K.; Manninen, S. Changes in soil microbial community structure under elevated tropospheric O3 and CO2. Soil Biol. Biochem. 2008, 40, 2502–2510. [Google Scholar] [CrossRef]
- Phillips, R.L.; Zak, D.R.; Holmes, W.E.; White, D.C. Microbial community composition and function beneath temperate trees exposed to elevated atmospheric carbon dioxide and ozone. Oecologia 2002, 131, 236–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osdaghi, E.; Pakdaman Sardrood, B.; Bavi, M.; Akbari Oghaz, N.; Kimiaei, S.; Hadian, S. First report of Curtobacterium flaccumfaciens pv. flaccumfaciens causing cowpea bacterial wilt in Iran. J. Phytopathol. 2015, 163, 653–656. [Google Scholar] [CrossRef]
- Raupach, G.S.; Kloepper, J.W. Mixtures of plant growth-promoting rhizobacteria enhance biological control of multiple cucumber pathogens. Phytopathology 1998, 88, 1158–1164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buczolits, S.; Denner, E.B.M.; Vybiral, D.; Wieser, M.; Kämpfer, P.; Busse, H.-J. Classification of three airborne bacteria and proposal of Hymenobacter aerophilus sp. nov. Int. J. Syst. Evol. Microbiol. 2002, 52, 445–456. [Google Scholar] [CrossRef]
- Muangchinda, C.; Srisuwankarn, P.; Boubpha, S.; Chavanich, S.; Pinyakong, O. The effect of bioaugmentation with Exiguobacterium sp. AO-11 on crude oil removal and the bacterial community in sediment microcosms, and the development of a liquid ready-to-use inoculum. Chemosphere 2020, 250, 126303. [Google Scholar] [CrossRef]
- Chen, M.-S.; Chen, X.-H.; Yan, X.-R.; Li, F.-N.; Tuo, L. Phycicoccus mangrovi sp. nov., a novel endophytic actinobacterium isolated from bark of Sonneratia apetala. Syst. Appl. Microbiol. 2021, 44, 126275. [Google Scholar] [CrossRef]
- Bensch, K.; Braun, U.; Groenewald, J.Z.; Crous, P.W. The genus Cladosporium. Stud. Mycol. 2012, 72, 1–401. [Google Scholar] [CrossRef] [Green Version]
- Köhl, J.; Scheer, C.; Holb, I.J.; Masny, S.; Molhoek, W. Toward an integrated use of biological control by Cladosporium cladosporioides H39 in apple scab (Venturia inaequalis) management. Plant Dis. 2014, 99, 535–543. [Google Scholar] [CrossRef] [Green Version]
- Fones, H.; Gurr, S. The impact of Septoria tritici Blotch disease on wheat: An EU perspective. Fungal Genet. Biol. 2015, 79, 3–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russ, L.; Lombaers-van der Plas, C.; Castillo-Russi, J.D.; Zijlstra, C.; Köhl, J. Deciphering the modes of action of Golubevia sp., an antagonist against the causal agent of powdery mildew in wheat, using an mRNA-based systems approach. Biol. Control 2021, 152, 104446. [Google Scholar] [CrossRef]
- Liang, C.; Jayawardena, R.S.; Zhang, W.; Wang, X.; Liu, M.; Liu, L.; Zang, C.; Xu, X.; Hyde, K.D.; Yan, J.; et al. Identification and characterization of Pseudocercospora species causing grapevine leaf spot in China. J. Phytopathol. 2016, 164, 75–85. [Google Scholar] [CrossRef]
- Hepperly, P.R. Hansfordia sp.: A parasitic pathogen of dematiaceous plant pathogenic fungi in Puerto Rico. J. Agric. Univ. Puerto Rico 1969, 70, 113–119. [Google Scholar] [CrossRef]
- Akbar, A.; Medina, A.; Magan, N. Potential control of mycotoxigenic fungi and ochratoxin a in stored coffee using gaseous ozone treatment. Microorganisms 2020, 8, 1462. [Google Scholar] [CrossRef]
- Porto, Y.D.; Trombete, F.M.; Freitas-Silva, O.; de Castro, I.M.; Direito, G.M.; Ascheri, J.L.R. Gaseous ozonation to reduce aflatoxins levels and microbial contamination in corn grits. Microorganisms 2019, 7, 220. [Google Scholar] [CrossRef] [Green Version]
- Ul-Hassan, A.; Wellington, E.M. Actinobacteria, 3rd ed.; Schaechter, M., Ed.; Academic Press: Oxford, UK, 2009; pp. 25–44. ISBN 978-0-12-373944-5. [Google Scholar]
- Chen, C.; Wang, X.; Wang, J. Phytoremediation of cadmium-contaminated soil by Sorghum bicolor and the variation of microbial community. Chemosphere 2019, 235, 985–994. [Google Scholar] [CrossRef]
- Zhang, Y.-Z.; Jiang, D.-Y.; Zhang, C.; Yang, K.; Wang, H.-F.; Xia, X.-W.; Ding, W.-J. Pathological impact on the phyllosphere microbiota of Artemisia argyi by haze. J. Microbiol. Biotechnol. 2021, 31, 510–519. [Google Scholar] [CrossRef]
- Zhao, D.; Liu, G.; Wang, X.; Daraz, U.; Sun, Q. Abundance of human pathogen genes in the phyllosphere of four landscape plants. J. Environ. Manag. 2020, 255, 109933. [Google Scholar] [CrossRef]
- Zeng, J.; Dou, J.; Gao, L.; Xiang, Y.; Huang, J.; Ding, S.; Chen, J.; Zeng, Q.; Luo, Z.; Tan, W.; et al. Topical ozone therapy restores microbiome diversity in atopic dermatitis. Int. Immunopharmacol. 2020, 80, 106191. [Google Scholar] [CrossRef]
- Aveskamp, M.M.; de Gruyter, J.; Woudenberg, J.H.C.; Verkley, G.J.M.; Crous, P.W. Highlights of the Didymellaceae: A polyphasic approach to characterise Phoma and related pleosporalean genera. Stud. Mycol. 2010, 65, 1–60. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Song, X.; Zhang, H. Isolation and characterization of Aschersonia placenta from citrus orchards and its pathogenicity towards Dialeurodes citri (Ashmead). J. Invertebr. Pathol. 2013, 112, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Braga, R.M.; Padilla, G.; Araújo, W.L. The biotechnological potential of Epicoccum spp.: Diversity of secondary metabolites. Crit. Rev. Microbiol. 2018, 44, 759–778. [Google Scholar] [CrossRef]
- Behrendt, U.; Ulrich, A.; Schumann, P. Leucobacter tardus sp. nov., isolated from the phyllosphere of Solanum tuberosum L. Int. J. Syst. Evol. Microbiol. 2008, 58, 2574–2578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, J.; Liu, M.; Gu, J.; Liu, Q.; Zhao, L.; Ma, Y.; Wei, D. Metagenomic sequencing reveals microbial gene catalogue of phosphinothricin-utilized soils in South China. Gene 2019, 711, 143942. [Google Scholar] [CrossRef]
- Li, J.; Xu, Y.; Song, Q.; Yang, J.; Xie, L.; Yu, S.; Zheng, L. Polycyclic aromatic hydrocarbon and n-alkane pollution characteristics and structural and functional perturbations to the microbial community: A case-study of historically petroleum-contaminated soil. Environ. Sci. Pollut. Res. 2021, 28, 10589–10602. [Google Scholar] [CrossRef]
- van der Geer, P. Signal Transduction, 2nd ed.; Maloy, S., Hughes, K.B.T.-B.E., Eds.; Academic Press: San Diego, CA, USA, 2013; pp. 436–439. ISBN 978-0-08-096156-9. [Google Scholar]
- Liu, C.; Sun, D.; Zhu, J.; Liu, W. Two-component signal transduction systems: A major strategy for connecting input stimuli to biofilm formation. Front. Microbiol. 2018, 9, 3279. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Song, M.; Wei, X.; Zhang, H.; Bai, Z.; Zhuang, X. Responses of Phyllosphere Microbiome to Ozone Stress: Abundance, Community Compositions and Functions. Microorganisms 2022, 10, 680. https://doi.org/10.3390/microorganisms10040680
Liu J, Song M, Wei X, Zhang H, Bai Z, Zhuang X. Responses of Phyllosphere Microbiome to Ozone Stress: Abundance, Community Compositions and Functions. Microorganisms. 2022; 10(4):680. https://doi.org/10.3390/microorganisms10040680
Chicago/Turabian StyleLiu, Jiayu, Manjiao Song, Xinyuan Wei, Huanzhen Zhang, Zhihui Bai, and Xuliang Zhuang. 2022. "Responses of Phyllosphere Microbiome to Ozone Stress: Abundance, Community Compositions and Functions" Microorganisms 10, no. 4: 680. https://doi.org/10.3390/microorganisms10040680






