Road Salt versus Urban Snow Effects on Lake Microbial Communities
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Properties of the Lake Water and Urban Snow
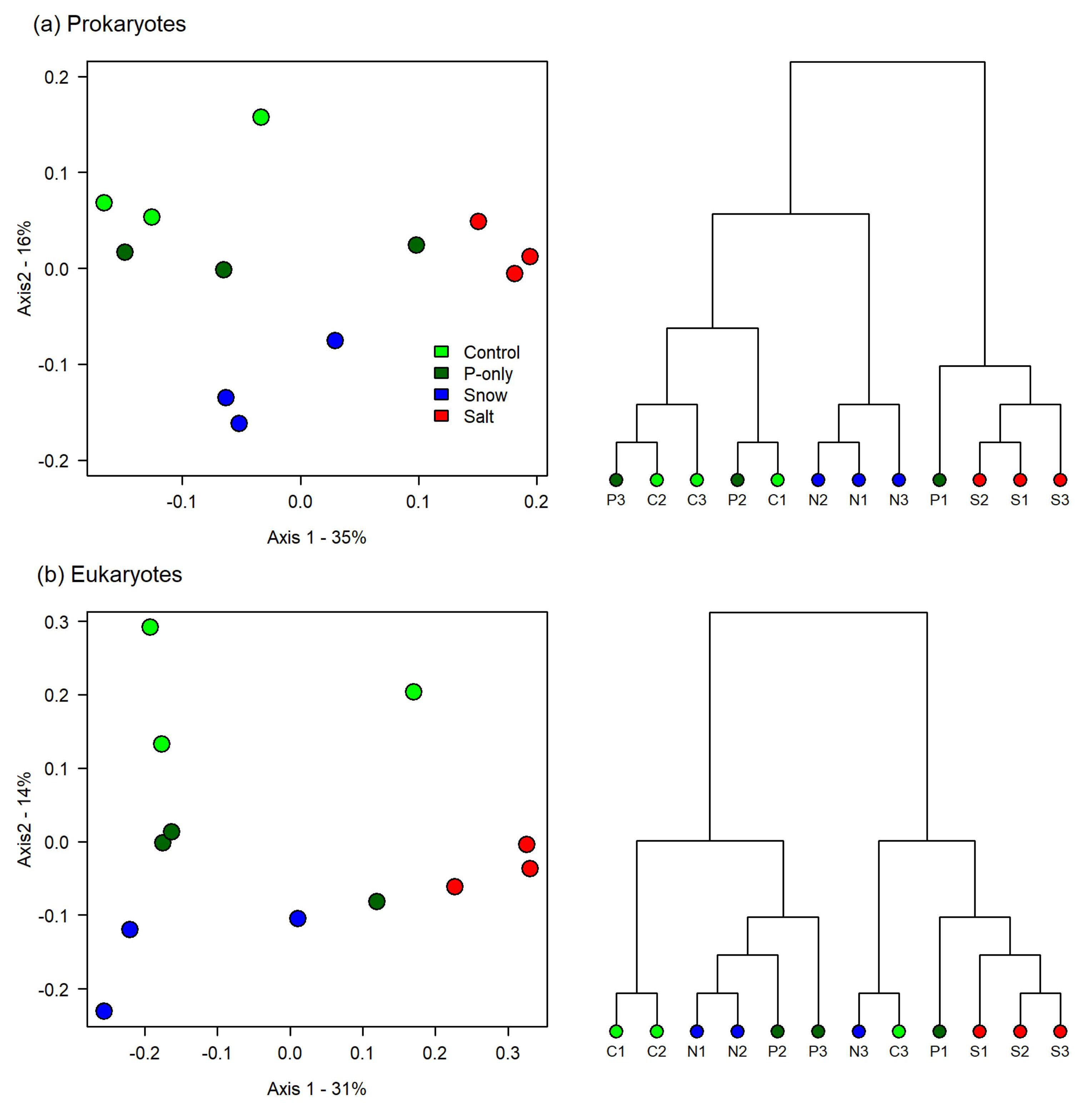
3.2. Microbial Plankton Responses
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dugan, H.A.; Skaff, N.K.; Doubek, J.P.; Bartlett, S.L.; Burke, S.M.; Krivak-Tetley, F.E.; Summers, J.C.; Hanson, P.C.; Weathers, K.C. Lakes at risk of chloride contamination. Environ. Sci. Technol. 2020, 54, 6639–6650. [Google Scholar] [CrossRef] [PubMed]
- Hintz, W.D.; Relyea, R.A. A review of the species, community, and ecosystem impacts of road salt salinisation in fresh waters. Freshw. Biol. 2019, 64, 1084–1097. [Google Scholar] [CrossRef] [Green Version]
- Moffett, E.R.; Baker, H.K.; Bonadonna, C.C.; Shurin, J.B.; Symons, C.C. Cascading effects of freshwater salinization on plankton communities in the Sierra Nevada. Limnol. Oceanogr. Lett. 2022, in press. [Google Scholar] [CrossRef]
- Greco, D.A.; Arnott, S.E.; Fournier, I.B.; Schamp, B.S. Effects of chloride and nutrients on freshwater plankton communities. Limnol. Oceanogr. Lett. 2022; in press. [Google Scholar] [CrossRef]
- Bartlett, A.J.; Rochfort, Q.; Brown, L.R.; Marsalek, J. Causes of toxicity to Hyalella azteca in a stormwater management facility receiving highway runoff and snowmelt. Part I: Polycyclic aromatic hydrocarbons and metals. Sci. Total Environ. 2012, 414, 227–237. [Google Scholar] [CrossRef]
- Bartlett, A.J.; Rochfort, Q.; Brown, L.R.; Marsalek, J. Causes of toxicity to Hyalella azteca in a stormwater management facility receiving highway runoff and snowmelt. Part II: Salts, nutrients, and water quality. Sci. Total Environ. 2012, 414, 238–247. [Google Scholar] [CrossRef]
- Fournier, I.B.; Galvez-Cloutier, R.; Vincent, W.F. Roadside snowmelt: A management target to reduce lake and river contamination. Inland Waters 2022, 12, 47–60. [Google Scholar] [CrossRef]
- Ruth, O. The effects of de-icing in Helsinki urban streams, Southern Finland. Water Sci. Technol. 2003, 48, 33–43. [Google Scholar] [CrossRef] [Green Version]
- Wetzel, R.B.; Likens, G.E. Limnological Analyses, 3rd ed.; Springer: New York, NY, USA, 2000. [Google Scholar]
- Boxall, A.B.A.; Maltby, L. The effects of motorway runoff on freshwater ecosystems: 3. Toxicant confirmation. Arch. Environ. Contam. Toxicol. 1997, 33, 9–16. [Google Scholar] [CrossRef]
- Comte, J.; Culley, A.I.; Lovejoy, C.; Vincent, W.F. Microbial connectivity and sorting in a High Arctic watershed. ISME J. 2018, 12, 2988–3000. [Google Scholar] [CrossRef] [Green Version]
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Comeau, A.M.; Li, W.K.W.; Tremblay, J.-É.; Carmack, E.C.; Lovejoy, C. Arctic Ocean microbial community structure before and after the 2007 record sea ice minimum. PLoS ONE 2011, 6, e27492. [Google Scholar] [CrossRef]
- Fournier, I.B.; Lovejoy, C.; Vincent, W.F. Changes in the community structure of under-ice and open-water microbiomes in urban lakes exposed to road salts. Front. Microbiol. 2021, 12, 660719. [Google Scholar] [CrossRef]
- Bushnell, B.; Rood, J.; Singer, E. BBMerge-Accurate paired shotgun read merging via overlap. PLoS ONE 2017, 12, e0185056. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Edgar, E.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Guillou, L.; Bachar, D.; Audic, S.; Bass, D.; Berney, C.; Bittner, L.; Boutte, C.; Burgaud, G.; de Vargas, C.; Decelle, J.; et al. The Protist Ribosomal Reference database (PR2): A catalog of unicellular eukaryote Small Sub-Unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2013, 41, D597–D604. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Guiry, M.D. AlgaeBase; National University of Ireland: Galway, Ireland, 2022; Available online: https://www.algaebase.org/ (accessed on 4 April 2022).
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5–6. 2019. Available online: https://CRAN.R-project.org/package=vegan (accessed on 4 April 2022).
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: http://www.R-project.org/ (accessed on 4 April 2022).
- Azevedo, J.S.N.; Correia, A.; Henriques, I. Molecular analysis of the diversity of genus Psychrobacter present within a temperate estuary. FEMS Microbiol. Ecol. 2013, 84, 451–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, A.H.; Yan, N.D. Food quantity affects the sensitivity of Daphnia to road salt. Environ. Sci. Technol. 2015, 49, 4673–4680. [Google Scholar] [CrossRef]
- Park, M.-H.; Park, C.-H.; Sim, Y.B.; Hwang, S.-J. Response of Scenedesmus quadricauda (Chlorophyceae) to salt stress considering nutrient enrichment and intracellular proline accumulation. Int. J. Environ. Res. Public Health 2020, 17, 3624. [Google Scholar] [CrossRef] [PubMed]
- Simmons, J.A. Toxicity of major cations and anions (Na+, K+, Ca2+, Cl−, and SO42−) to a macrophyte and an alga. Environ. Toxicol. Chem. 2012, 31, 1370–1374. [Google Scholar] [CrossRef] [PubMed]
- Astorg, L.; Gagnon, J.-C.; Lazar, C.S.; Derry, A. Effects of freshwater salinization on a salt-naïve planktonic eukaryote community. Limnol. Oceanogr. Lett. 2022, in press. [Google Scholar] [CrossRef]
- Bray, J.P.; Reich, J.; Nichols, S.J.; Kon Kam King, G.; Mac Nally, R.; Thompson, R.; O’Reilly-Nugent, A.; Kefford, B.J. Biological interactions mediate context and species-specific sensitivities to salinity. Philos. Trans. R. Soc. B 2019, 374, 20180020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hintz, W.D.; Relyea, R.A. A salty landscape of fear: Responses of fish and zooplankton to freshwater salinization and predatory stress. Oecologia 2017, 185, 147–156. [Google Scholar] [CrossRef]
- Denis, C.H.S.; Pinheiro, M.D.O.; Power, M.E.; Bols, N.C. Effect of salt and urban water samples on bacterivory by the ciliate, Tetrahymena Thermophila. Environ. Pollut. 2010, 158, 502–507. [Google Scholar] [CrossRef]
- Merrick, A.M.; Searle, C.L. Combined effects of salinity and infectious disease on Daphnia dentifera at multiple scales. Freshw. Biol. 2019, 64, 601–607. [Google Scholar] [CrossRef]
- Venâncio, C.; Anselmo, E.; Soares, A.; Lopes, I. Does increased salinity influence the competitive outcome of two producer species? Environ. Sci. Pollut. Res. 2017, 24, 5888–5897. [Google Scholar] [CrossRef]
- Coveley, S.; Elshahed, M.S.; Youssef, N.H. Response of the rare biosphere to environmental stressors in a highly diverse ecosystem (Zodletone Spring, OK, USA). PeerJ 2015, 3, e1182. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Gao, G.; Tang, X.; Shao, K. Can the freshwater bacterial communities shift to the “marine-like” taxa? J. Basic Microbiol. 2014, 54, 1264–1272. [Google Scholar] [CrossRef]
- Brown, B.L.; Downing, A.L.; Leibold, M.A. Compensatory dynamics stabilize aggregate community properties in response to multiple types of perturbations. Ecology 2016, 97, 2021–2033. [Google Scholar] [CrossRef]
- Da Costa, N.B.; Fugère, V.; Hébert, M.P.; Xu, C.C.; Barrett, R.D.; Beisner, B.; Bell, G.; Yargeau, V.; Fussmann, G.F.; Gonzalez, A.; et al. Resistance, resilience, and functional redundancy of freshwater microbial communities facing multiple agricultural stressors in a mesocosm experiment. Mol. Ecol. 2021, 30, 4771–4788. [Google Scholar] [CrossRef]
- Spivak, A.C.; Vanni, M.J.; Mette, E.M. Moving on up: Can results from simple aquatic mesocosm experiments be applied across broad spatial scales? Freshw. Biol. 2011, 56, 279–291. [Google Scholar] [CrossRef]
- Reinosdotter, K.; Viklander, M. Road salt influence on pollutant releases from melting urban snow. Water Qual. Res. J. Can. 2007, 42, 153–161. [Google Scholar] [CrossRef]
- Westerlund, C.; Viklander, M. Pollutant release from a disturbed urban snowpack in northern Sweden. Water Qual. Res. J. Can. 2011, 46, 98–109. [Google Scholar] [CrossRef] [Green Version]
- Tian, Z.; Gonzalez, M.; Rideout, C.A.; Zhao, H.N.; Hu, X.; Wetzel, J.; Mudrock, E.; James, C.A.; McIntyre, J.K.; Kolodziej, E.P. 6PPD-quinone: Revised toxicity assessment and quantification with a commercial standard. Environ. Sci. Technol. Lett. 2022, 9, 140–146. [Google Scholar] [CrossRef]
- Brinkmann, M.; Montgomery, D.; Selinger, S.; Miller, J.G.; Stock, E.; Alcaraz, A.J.; Challis, J.K.; Weber, L.; Janz, D.; Hecker, M.; et al. Acute toxicity of the tire rubber-derived chemical 6ppd-quinone to four fishes of commercial, cultural, and ecological importance. Environ. Sci. Technol. Lett. 2022, in press. [Google Scholar] [CrossRef]


| 515F/806R | 1389F/1510R | |||||
|---|---|---|---|---|---|---|
| Steps | Temperature | Time | Cycles | Temperature | Time | Cycles |
| (°C) | (s) | (°C) | (s) | |||
| Initial denaturation | None | NA | 98 | 30 | 1 | |
| Denaturation | 94 | 45 | 36 | 98 | 10 | 30 |
| Annealing | 50 | 60 | 52 | 30 | ||
| Extension | 72 | 90 | 72 | 30 | ||
| Final extension | 72 | 60 | 1 | 72 | 270 | 1 |
| Variable | Natural Medium | Treatments | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lake (t0) | Snow | Control a | P-Only a | Snow a | Salt a | |||||||
| Alkalinity | 15.6 | (2) | 40.9 | (13) | 14.7 | (2) | 14.7 | (2) | 18.0 | (1) | 14.7 | (2) |
| Calcium | 6.4 | (1) | 14.8 | (23) | 6.0 | (1) | 6.0 | (1) | 7.3 | (1) | 6.0 | (1) |
| Chloride | 20.0 | (9) | 17.8 | (68) | 18.9 | (9) | 18.9 | (9) | 49.6 | (3) | 48.9 | (3) |
| Nitrogen b | 0.8 | (6) | 0.4 | (46) | 0.7 | (6) | 0.7 | (6) | 0.7 | (6) | 0.7 | (6) |
| Phosphorus b | 11.4 | (22) | 84.3 | (46) | 10.8 | (22) | 14.8 | (16) | 16.5 | (15) | 14.8 | (16) |
| Potassium | 0.7 | (2) | 0.3 | (27) | 0.7 | (2) | 0.7 | (2) | 0.7 | (2) | 0.7 | (2) |
| Sodium | 11.1 | (7) | 10.6 | (63) | 10.4 | (7) | 10.4 | (7) | 30.4 | (2) | 30.1 | (2) |
| Sulfate | 4.6 | (3) | 5.3 | (72) | 4.3 | (3) | 4.3 | (3) | 4.9 | (3) | 4.3 | (3) |
| Taxon | Control | Taxon | P-Only | Taxon | Snow | Taxon | Salt | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Comamonadaceae_unclass. | 22 | (52) | Roseateles sp. | 12 | (31) | Comamonadaceae_unclass. | 14 | (23) | Comamonadaceae_unclass. | 18 | (10) |
| Roseateles sp. | 15 | (34) | Comamonadaceae_unclass. | 12 | (19) | ML635J-21_unclass. | 11 | (87) | FamilyI_unclass. | 8 | (57) |
| Polynucleobacter sp. | 8 | (85) | Polynucleobacter sp. | 10 | (21) | Polynucleobacter sp. | 10 | (30) | Polynucleobacter sp. | 6 | (17) |
| Albidiferax sp. | 5 | (13) | Flavobacterium sp. | 10 | (87) | Roseateles sp. | 9 | (16) | NS11-12_marine_group_unclass. | 5 | (17) |
| Rhodobacter sp. | 4 | (28) | Albidiferax sp. | 5 | (51) | Albidiferax sp. | 6 | (32) | Roseateles sp. | 5 | (46) |
| Flavobacterium sp. | 3 | (66) | Caulobacter sp. | 5 | (43) | Flavobacterium sp. | 4 | (38) | Caulobacter sp. | 4 | (36) |
| Flectobacillus sp. | 3 | (171) | ML635J-21_unclass. | 4 | (155) | NS11-12_marine_group_unclass. | 3 | (116) | Albidiferax sp. | 4 | (44) |
| Caulobacter sp. | 3 | (45) | FamilyI_unclass | 3 | (124) | Proteobacteria_unclass. | 3 | (132) | Candidatus Odyssella | 3 | (169) |
| PRD01a011B | 3 | (159) | Proteobacteria_unclass | 3 | (122) | Sphingomonadaceae_unclass. | 3 | (49) | Rhizobiales_unclass. | 3 | (57) |
| Pseudarcicella sp. | 3 | (52) | Myxococcales_unclass. | 3 | (45) | GKS98_freshwater_group | 2 | (35) | Sediminibacterium sp. | 3 | (39) |
| Ferruginibacter sp. | 2 | (39) | Rhodobacter sp. | 3 | (76) | Caulobacter sp. | 2 | (28) | ML635J-21_unclass. | 3 | (118) |
| OM27_clade | 2 | (168) | Pseudarcicella sp. | 2 | (52) | Planctomycetaceae_unclass. | 2 | (31) | Myxococcales_unclass. | 2 | (81) |
| Taxon | Control | Taxon | P-Only | Taxon | Snow | Taxon | Salt | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cyclidium sp. | 30 | (86) | Cyclidium | 27 | (38) | Cyclidium sp. | 23 | (87) | Cyclidium sp. | 21 | (55) |
| Conthreep_unclass. | 16 | (77) | Conthreep_unclass. | 26 | (113) | Cercozoa_unclass. | 13 | (82) | SA1-3C06_unclass. | 18 | (72) |
| Chrysophyceae_unclass. | 9 | (18) | Choreotrichia_unclass | 10 | (116) | Conthreep_unclass. | 10 | (60) | A31_unclass. | 13 | (77) |
| Choreotrichia_unclass. | 8 | (170) | Haptoria_unclass. | 7 | (173) | Rimostrombidium sp. | 10 | (173) | Cercozoa_unclass. | 10 | (73) |
| Cercozoa_unclass. | 6 | (92) | Spirotrichea_unclass | 7 | (132) | Choanoflagellida_unclass. | 6 | (131) | Chrysophyceae_unclass. | 9 | (54) |
| Choanoflagellida_unclass. | 6 | (173) | Chrysophyceae_unclass | 4 | (71) | Chrysophyceae_unclass. | 4 | (97) | Conthreep_unclass. | 9 | (79) |
| Glissomonadida_unclass. | 5 | (173) | Pedinellales_unclass. | 3 | (127) | Choreotrichia_unclass. | 4 | (123) | Choreotrichia_unclass | 6 | (133) |
| Leucocryptos sp. | 2 | (83) | Epipyxis sp. | 2 | (173) | A31_unclass. | 4 | (91) | Paraphysomonas sp. | 3 | (169) |
| A31_unclass. | 2 | (138) | Leucocryptos sp. | 1 | (158) | Pedinellales_unclass. | 2 | (73) | Epipyxis sp. | 3 | (42) |
| SA1-3C06_unclass. | 1 | (157) | Cercozoa_unclass. | 1 | (35) | AMT-15-27-30 | 2 | (173) | Synura sp. | 1 | (72) |
| Pedinellales_unclass. | 1 | (39) | A31_unclass. | 1 | (81) | SA1-3C06_unclass. | 2 | (170) | Haptoria_unclass. | 1 | (171) |
| Chrysamoeba sp. | 1 | (119) | Bolidomonas sp. | 1 | (141) | Spirotrichea_unclass. | 1 | (121) | Bolidomonas sp. | 1 | (60) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fournier, I.B.; Lovejoy, C.; Vincent, W.F. Road Salt versus Urban Snow Effects on Lake Microbial Communities. Microorganisms 2022, 10, 803. https://doi.org/10.3390/microorganisms10040803
Fournier IB, Lovejoy C, Vincent WF. Road Salt versus Urban Snow Effects on Lake Microbial Communities. Microorganisms. 2022; 10(4):803. https://doi.org/10.3390/microorganisms10040803
Chicago/Turabian StyleFournier, Isabelle B., Connie Lovejoy, and Warwick F. Vincent. 2022. "Road Salt versus Urban Snow Effects on Lake Microbial Communities" Microorganisms 10, no. 4: 803. https://doi.org/10.3390/microorganisms10040803







