Physiological and Metabolic Response of Arthrospira maxima to Organophosphates
Abstract
:1. Introduction
2. Materials and Methods
2.1. Arthrospira maxima Culture Preparation
2.2. A. maxima Treated with Glyphosate (Gly)
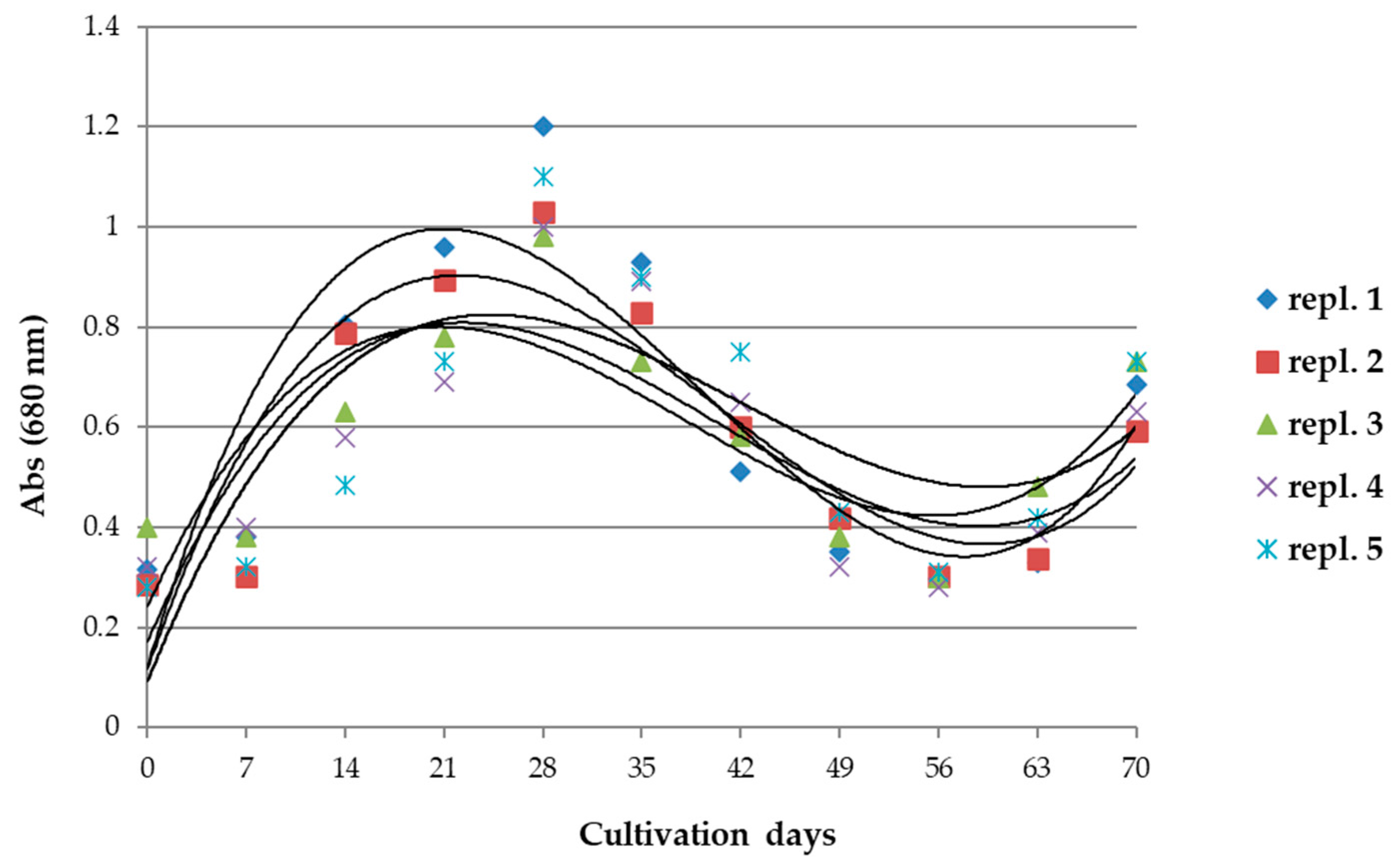
2.3. Evaluation of the Biomass of Cultivations
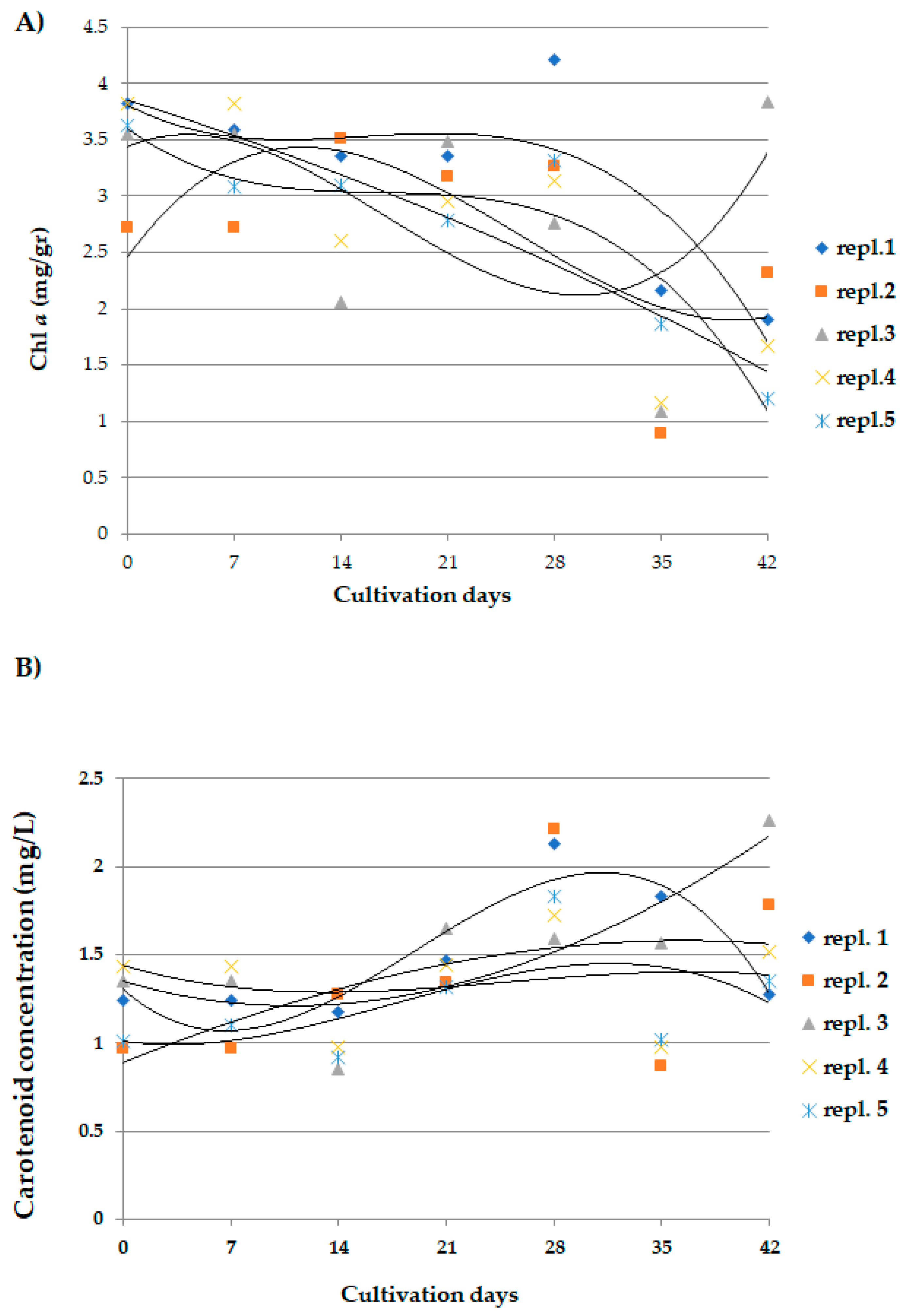
2.4. Analysis of the Concentration of Chlorophyll a and Carotenoids
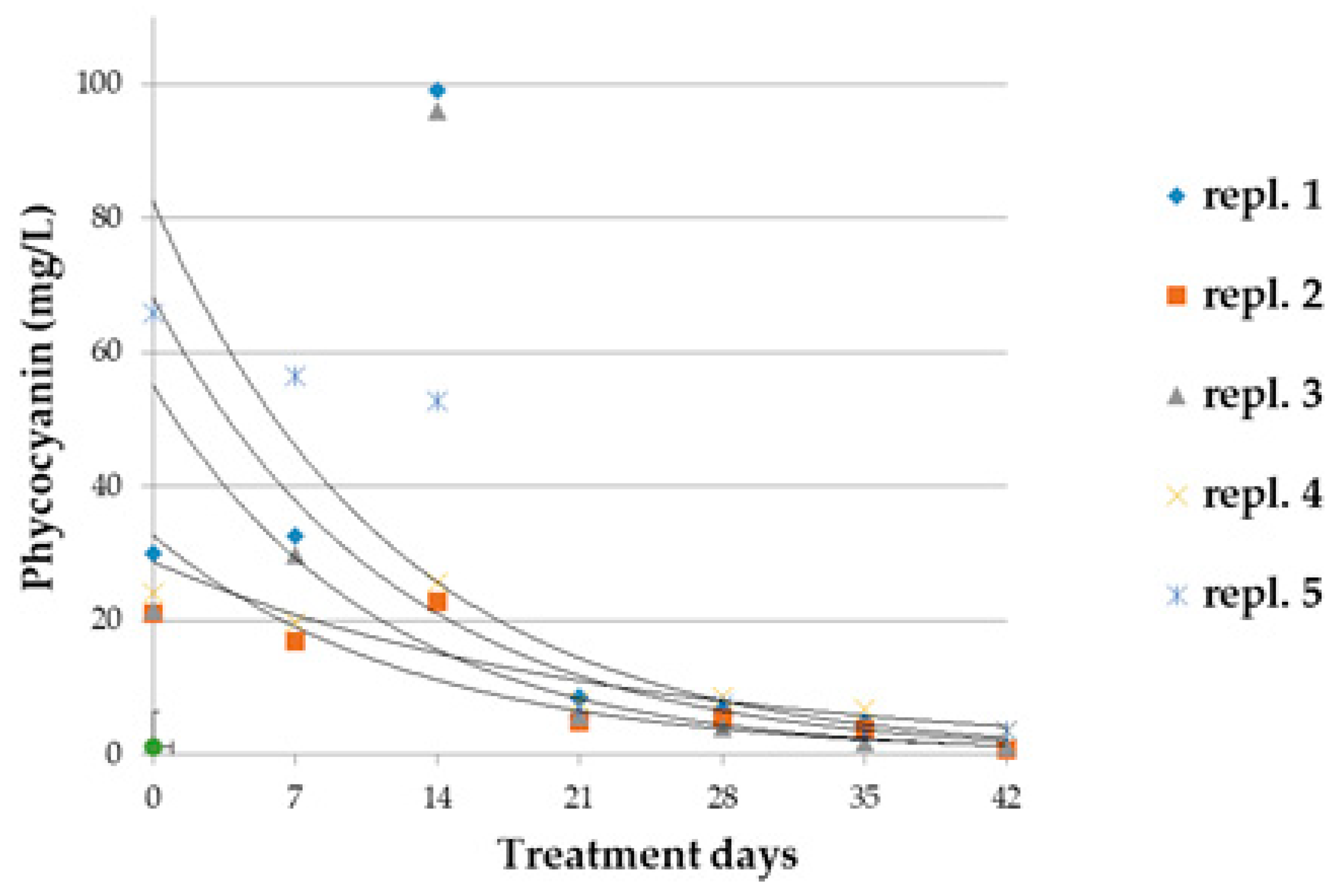
2.5. Extraction of Phycocyanins from A. maxima Cultivation
2.6. Extraction of Proteins from A. maxima Cultivation
2.7. Electrophoretic Separation of A. maxima Proteins by SDS-PAGE
2.8. Sample Preparation for Mass Analysis:Reduction, Alkylation, and in-Gel Digestion of Proteins
2.9. Mass Spectrometry Analysis
2.10. Bioinformatic Analysis and Proteins Identification of A. maxima
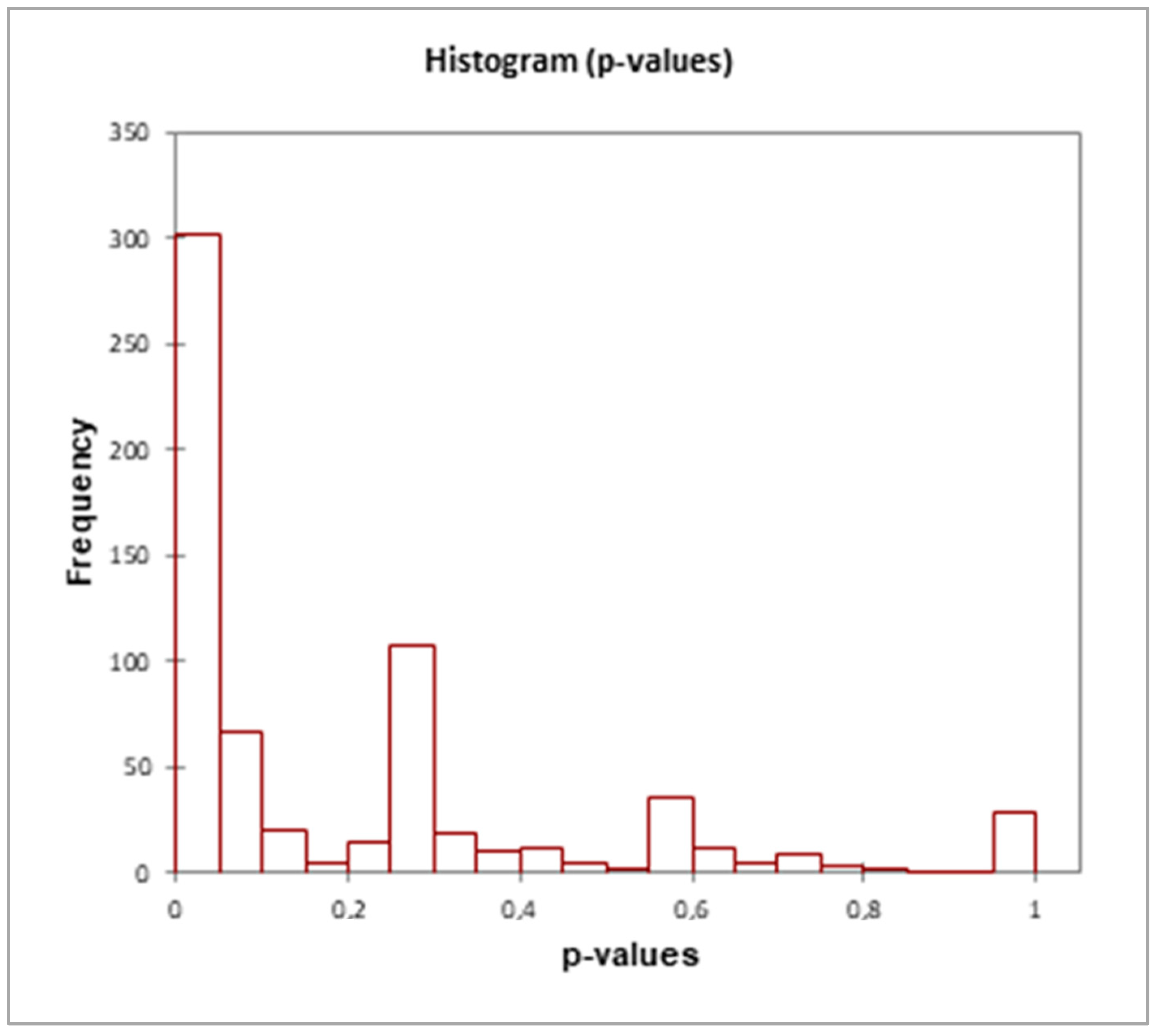
2.11. Statistics
3. Results
3.1. Dynamics of Arthrospira maxima Cultivation and Biochemical Parameters
3.2. A. maxima Cultivation Dynamics and Biochemical Parameters during Glyphosate Treatment
3.3. Proteins Differentially Expressed in Cultivations of Arthrospira maxima following Treatment with Glyphosate
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Casida, J.E.; Durkin, K.A. Pesticide Chemical Research in Toxicology: Lessons from Nature. Chem. Res. Toxicol. 2017, 30, 94–104. [Google Scholar] [CrossRef] [PubMed]
- ISPRA. Edizione 2020: Rapporto Nazionale pesticidi nelle acque—Dati 2017–2018. Edizione; ISPRA: Varese, Italy, 2020. [Google Scholar]
- van Bruggen, A.H.C.; Finckh, M.R.; He, M.; Ritsema, C.J.; Harkes, P.; Knuth, D.; Geissen, V. Indirect Effects of the Herbicide Glyphosate on Plant, Animal and Human Health Through its Effects on Microbial Communities. Front. Environ. Sci. 2021, 9, 464. [Google Scholar] [CrossRef]
- Barriuso, J.; Marin, S.; Mellado, R.P. Potential accumulative effect of the herbicide glyphosate on glyphosate-tolerant maize rhizobacterial communities over a three-year cultivation period. PLoS ONE 2011, 6, e27558. [Google Scholar] [CrossRef] [PubMed]
- Motta, E.V.S.; Raymann, K.; Moran, N.A. Glyphosate perturbs the gut microbiota of honey bees. Proc. Natl. Acad. Sci. USA 2018, 115, 10305–10310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, T.; Xu, N.; Zhang, Q.; Zhang, Z.; Debognies, A.; Zhou, Z.; Qian, H. Understanding the influence of glyphosate on the structure and function of freshwater microbial community in a microcosm. Environ. Pollut. 2020, 260, 114012. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, J.S. Cyanoremediation: A Green-Clean Tool for Decontamination of Synthetic Pesticides from Agro- and Aquatic Ecosystems. Agro-Environ. Sustain. 2017, 2, 59–83. [Google Scholar] [CrossRef]
- Lee, S.; Kim, J.; Kennedy, I.R.; Park, J.; Kwon, G.; Koh, S.; Kim, J.E. Biotransformation of an organochlorine insecticide, endosulfan, by Anabaena species. J. Agric. Food Chem. 2003, 51, 1336–1340. [Google Scholar] [CrossRef]
- Benbrook, C.M. Trends in glyphosate herbicide use in the United States and globally. Environ. Sci. Eur. 2016, 28, 3. [Google Scholar] [CrossRef] [Green Version]
- Forlani, G.; Pavan, M.; Gramek, M.; Kafarski, P.; Lipok, J. Biochemical Bases for a Widespread Tolerance of Cyanobacteria to the Phosphonate Herbicide Glyphosate. Plant Cell Physiol. 2008, 49, 443–456. [Google Scholar] [CrossRef]
- Nisticò, D.M.; Piro, A.; Oliva, D.; Osso, V.; Mazzuca, S.; Fagà, F.A.; Morelli, R.; Conidi, C.; Figoli, A.; Cassano, A. A Combination of Aqueous Extraction and Ultrafiltration for the Purification of Phycocyanin from Arthrospira maxima. Microorganisms 2022, 28, 308. [Google Scholar] [CrossRef]
- Herrera, A.; Boussiba, S.; Napoleone, V.; Hohlberg, A. Recovery of c-phycocyanin from the cyanobacterium Spirulina maxima. J. Appl. Phycol. 1989, 1, 325–331. [Google Scholar] [CrossRef]
- Norling, B.; Zak, E.; Andersson, B.; Pakrasi, H. 2D-isolation of pure plasma and thylakoid membranes from the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 1998, 436, 189–192. [Google Scholar] [CrossRef] [Green Version]
- Hongsthong, A.; Sirijuntarut, M.; Yutthanasirikul, R.; Senachak, J.; Kurdrid, P.; Cheevadhanarak, S.; Tanticharoen, M. Subcellular proteomic characterization of the high-temperature stress response of the cyanobacterium Spirulina platensis. Proteome Sci. 2009, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Matallana-Surget, S.; Derock, J.; Leroy, B.; Badri, H.; Deschoenmaeker, F.; Wattiez, R. Proteome-Wide Analysis and Diel Proteomic Profiling of the Cyanobacterium Arthrospira platensis PCC 8005. PLoS ONE 2014, 9, e99076. [Google Scholar] [CrossRef] [PubMed]
- Janssen, P.J.; Morin, N.; Mergeay, M.; Leroy, B.; Wattiez, R.; Vallaeys, T.; Waleron, K.; Waleron, M.; Wilmotte, A.; Quillardet, P.; et al. Genome sequence of the edible cyanobacterium Arthrospira sp. PCC 8005. J. Bacteriol. 2010, 192, 2465–2466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- United International Bureaux for the Protection of Intellectual Property (BIRPI). Paris Convention for the Protection of Industrial Property of March 20, 1883, as Revised at Brussels on December 14, 1900, at Washington on June 2, 1911, at The Hague on November 6, 1925, at London on June 2, 1934, at Lisbon on October 31, 1958, and at Stockholm on July 14, 1967; United International Bureaux for the Protection of Intellectual Property (BIRPI): Geneva, Switzerland, 1883. [Google Scholar]
- Zarrouk, C. Contribution à L’étude D’une Cyanophycée. Influence de Divers Facteurs Physiques et Chimiques Sur la Croissance et la Photosynthèse de Spirulina maxima. Ph.D. Thesis, Université De Paris, Paris, France, 1966. [Google Scholar]
- El-Kassas, H.Y.; Heneash, A.M.M.; Hussein, N.R. Cultivation of Arthrospira (Spirulina) platensis using confectionary wastes for aquaculture feeding. J. Genet. Eng. Biotechnol. 2015, 13, 145–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Cheng, J.; Kubar, A.A.; Guo, W.; Song, Y.; Liu, S.; Chen, S.; Tian, J. Orange light spectra filtered through transparent colored polyvinyl chloride sheet enhanced pigment content and growth of Arthrospira cells. Bioresour. Technol. 2021, 319, 124179. [Google Scholar] [CrossRef]
- Porra, R.J.; Thompson, W.; Kriedemann, P.E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 1989, 975, 384–394. [Google Scholar] [CrossRef]
- Bennett, A.; Bogorad, L. Complementary chromatic adaption in a filamentous bluegreen alga. J. Cell Biol. 1973, 58, 419–435. [Google Scholar] [CrossRef]
- Mostafa, M.S.I.; Piercey-Normorea, M.D.; Rampitsch, C. Proteomic analyses of the cyanobacterium Arthrospira (Spirulina) platensis under iron and salinity stress. Environ. Exp. Bot. 2018, 147, 63–74. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Bradford, M.M. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Wilm, M.; Shevchenko, A.; Houthaeve, T.; Breit, S.; Schweigerer, L.; Fotsis, T.; Mann, M. Fentomole sequencing of proteins from polyacrylamide gels by nanoelectrospray mass spettrometry. Nature 1996, 379, 466–469. [Google Scholar] [CrossRef] [PubMed]
- Searle, B.C. Scaffold: A bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics 2010, 10, 1265–1269. [Google Scholar] [CrossRef] [PubMed]
- Vidal, N.P.; Manful, C.F.; Pham, T.H.; Stewart, P.; Keough, D.; Thomas, R. The use of XLSTAT in conducting principal component analysis (PCA) when evaluating the relationships between sensory and quality attributes in grilled foods. MethodsX 2020, 7, 100835. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Gutteridge, S.; Cardona, T.; Shao, S.; Nixon, P.J. Enhancing photosynthesis in plants: The light reactions. Essays Biochem. 2018, 62, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Deniz, F.; Saygideger, S.D.; Karaman, S. Response to Copper and Sodium Chloride Excess in Spirulina sp. (Cyanobacteria). Bull. Environ. Contam. Toxicol. 2011, 87, 11–15. [Google Scholar] [CrossRef]
- Maslowska, K.H.; Makiela-Dzbenska, K.; Fijalkowska, I.J. The SOS system: A complex and tightly regulated response to DNA damage. Environ. Mol. Mutagenesis 2019, 60, 368–384. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.M.; Ross, C.T.; Zeng, B.B.; Singleton, S.F. A molecular target for suppression of the evolution of antibiotic resistance: Inhibition of the Escherichia coli RecA Protein by N6-(1-Naphthyl)-ADP. J. Med. Chem. 2005, 48, 5408–5411. [Google Scholar] [CrossRef]
- Dörr, T.; Vulić, M.; Lewis, K. Ciprofloxacin Causes Persister Formation by Inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010, 8, e1000317. [Google Scholar] [CrossRef] [Green Version]
- Nagib, A.; Dong-Gi, L.; Ki-Won, L.; Iftekhar, A.; Sang-Hoon, L.; Bahk, J.D.; Byung-Hyun, L. Glyphosate-induced oxidative stress in rice leaves revealed by proteomic approach. Plant Physiol. Biochem. 2008, 46, 1062–1070. [Google Scholar] [CrossRef]
- Ravishankar, A.; Pupo, A.; Gallagher, J.E.G. Resistance Mechanisms of Saccharomyces cerevisiae to Commercial Formulations of Glyphosate Involve DNA Damage Repair, the Cell Cycle, and the Cell Wall Structure. G3 Genes Genomes Genet. 2020, 10, 2043–2056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinrücken, H.C.; Amrhein, N. The herbicide glyphosate is a potent inhibitor of 5-enolpyruvyl-shikimic acid-3-phosphate synthase. Biochem. Biophys. Res. Commun. 1980, 94, 1207–1212. [Google Scholar] [CrossRef]




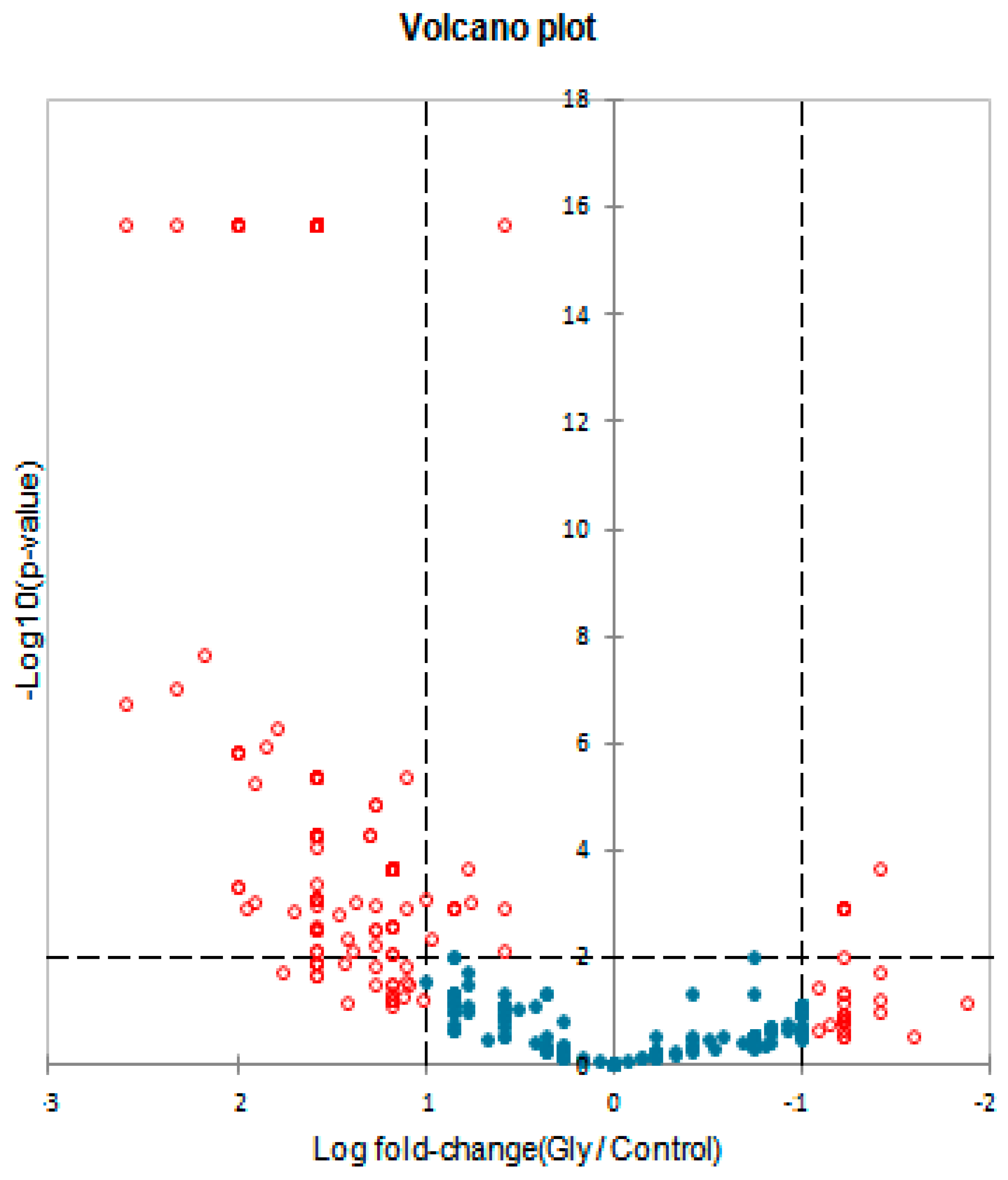
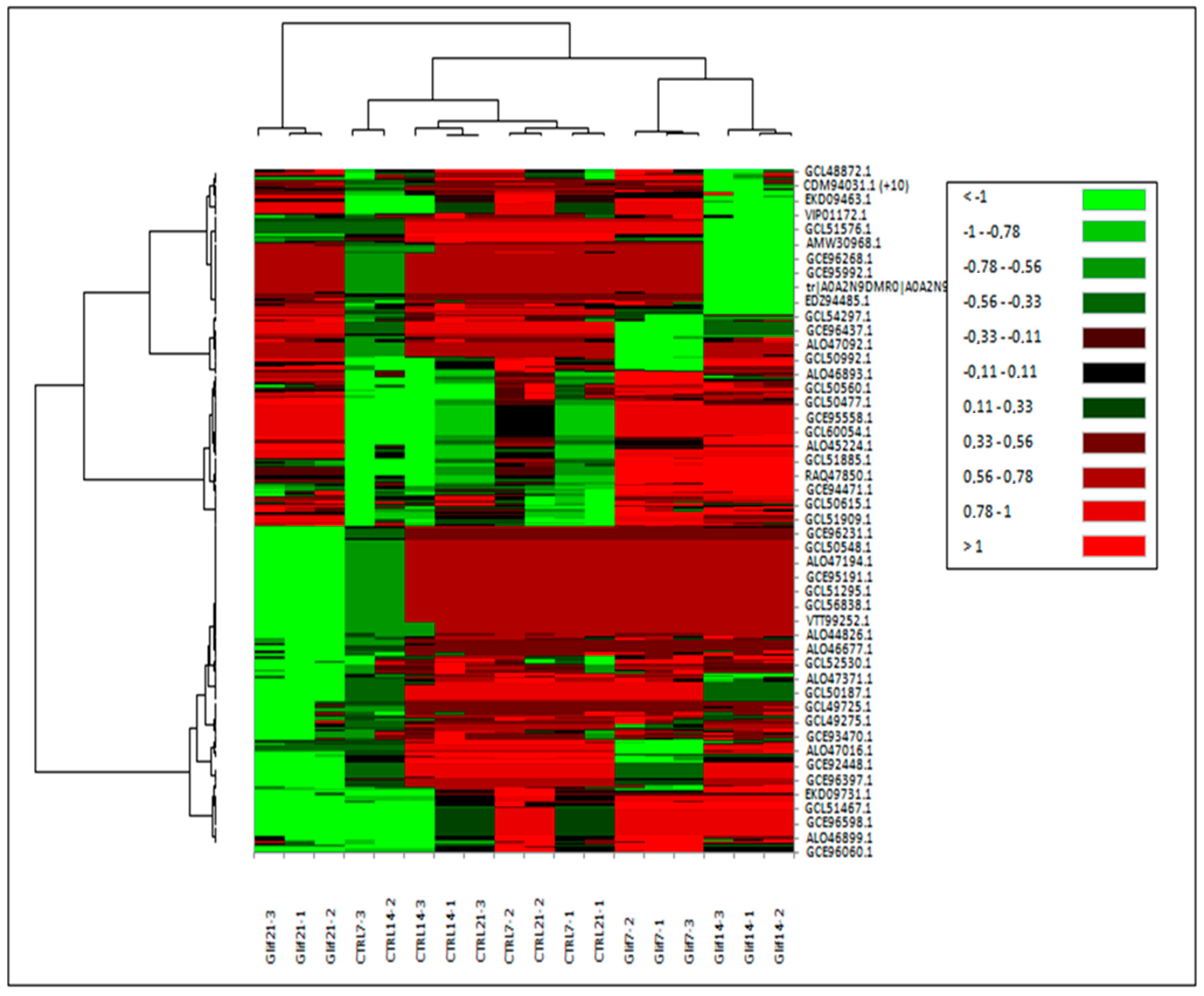
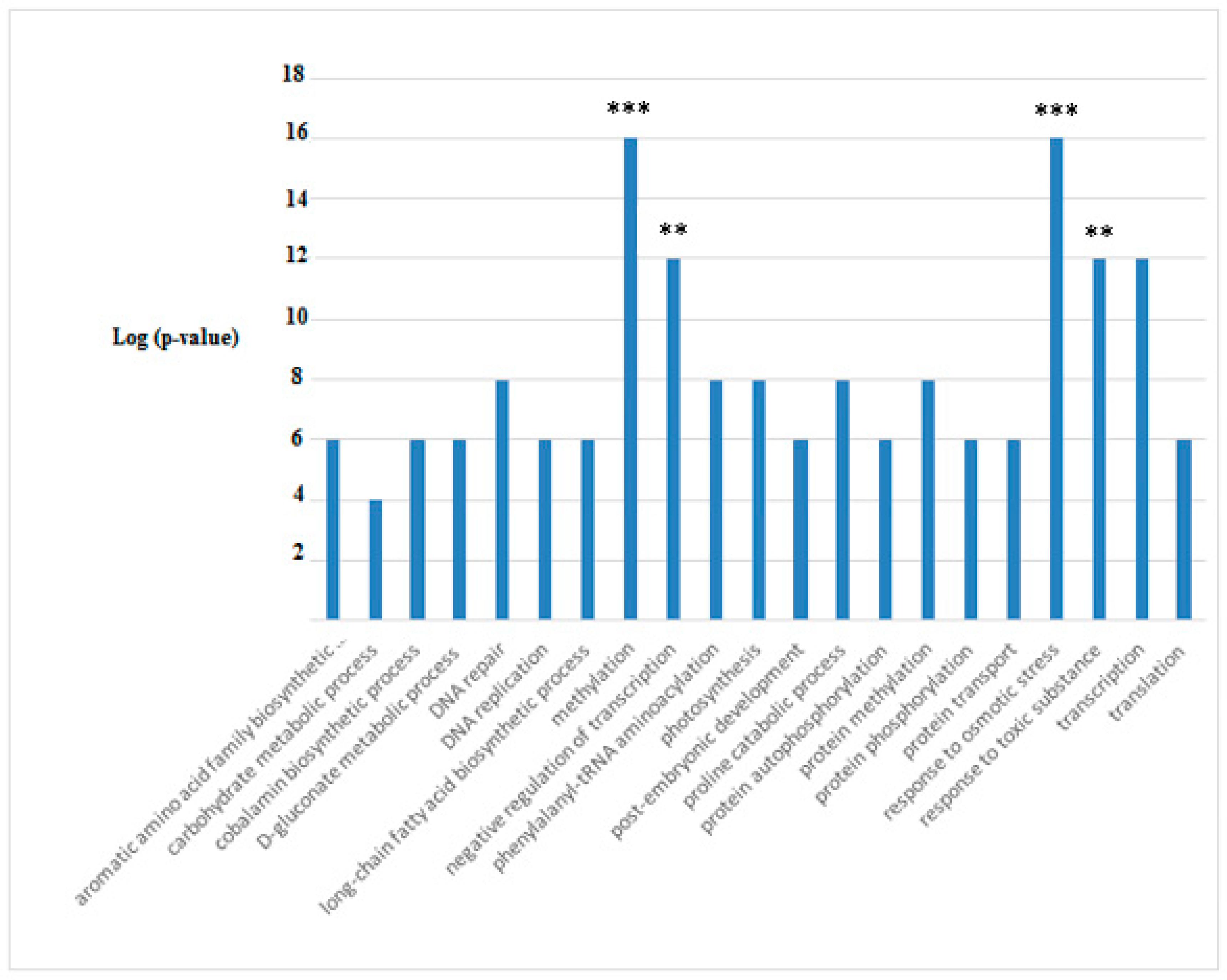
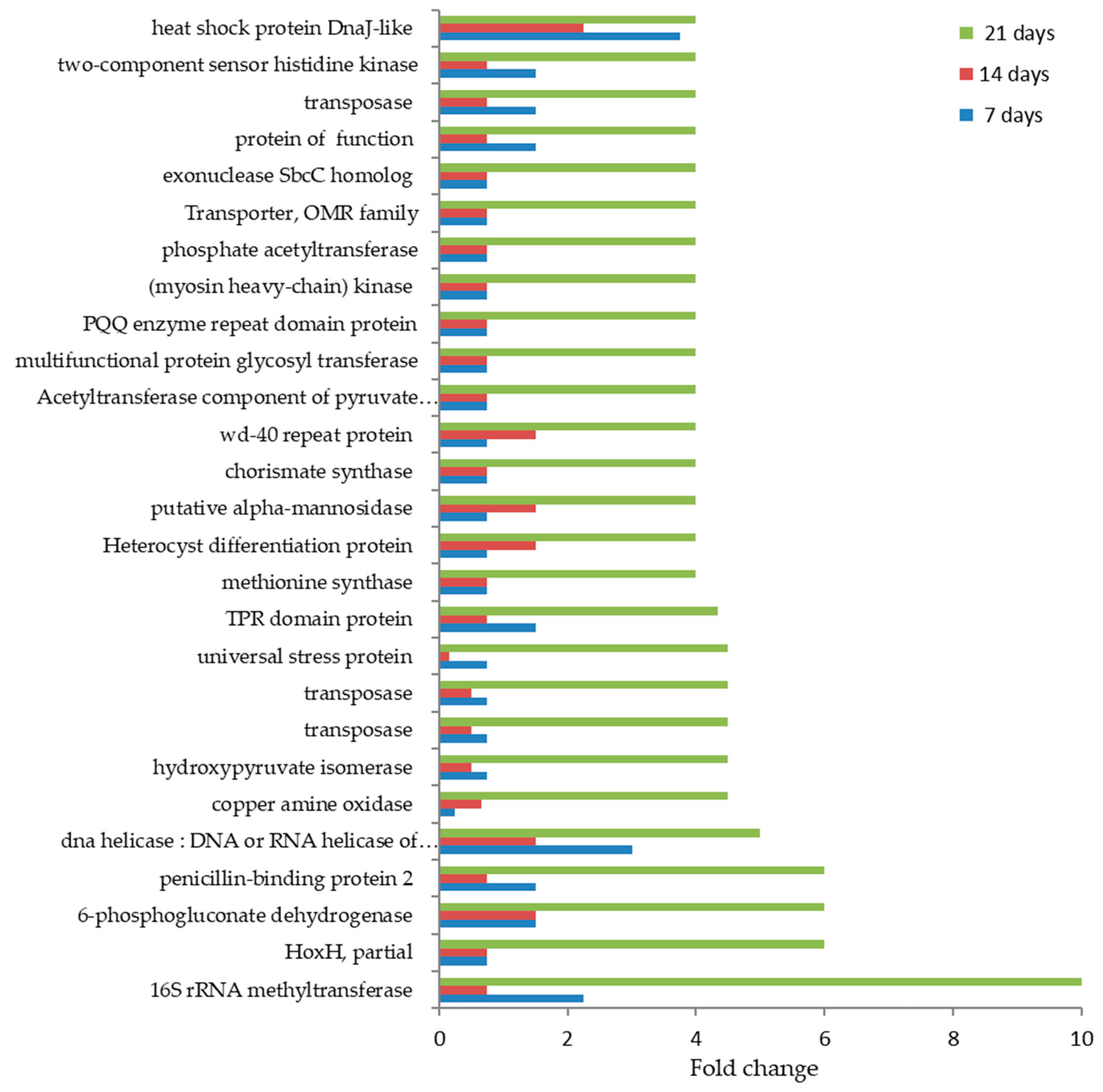
| Protein name and Taxonomy | IDs | p-Value | Quantitative Profile ** (Fold Change) | Qualitative Profile | GO Terms | |
|---|---|---|---|---|---|---|
| Reference | Gly | |||||
| 16S rRNA methyltransferase [Pseudohongiella spirulinae] | GCL59050.1 | <0.0001 | 1667 (b) | 10 (a) | H.A. | SOS response |
| HoxH, partial [Arthrospira platensis FACHB-440] | EDZ95376.1 | <0.0001 | 1 (b) | 6 (a) | H.A. | ADP binding; transferase activity |
| Penicillin-binding protein 2 [Arthrospira sp. PLM2.Bin9] | GCE96397.1 | <0.0001 | 1333 (b) | 6 (a) | H.A. | molybdopterin cofactor binding |
| 6-phosphogluconate dehydrogenase [Arthrospira platensis NIES-46] | GCE96312.1 | <0.0001 | 1667 (b) | 6 (a) | H.A. | glycolytic process |
| ATP-dependent DNA helicase [Pseudohongiella spirulinae] | ALO44826.1 | <0.0001 | 1444 (b) | 5 (a) | H.A. | cell division |
| hypothetical protein NIES46_38410 [Arthrospira platensis NIES-46] | ALO47614.1 | <0.0001 | 1 (b) | 5 (a) | H.A. | DNA restriction-modification system |
| Transporter, OMR family [Pseudohongiella spirulinae] | GCL52209.1 | <0.0001 | 1 (b) | 4 (a) | H.A. | aromatic amino acid family biosynthetic process |
| Chorismate synthase [Microcystis aeruginosa NIES-3804] | GCE92370.1 | <0.0001 | 1 (b) | 4 (a) | H.A. | aromatic amino acid family biosynthetic process; chorismate biosynthetic process |
| hypothetical protein NIES46_48480 [Arthrospira platensis NIES-46] | GCL50795.1 | <0.0001 | 1333 (b) | 4 (a) | H.A. | ATPase activity; ATP binding; DNA binding |
| Phosphate acetyltransferase [Arthrospira platensis NIES-46] | GCL50871.1 | <0.0001 | 1 (b) | 4 (a) | H.A. | cell wall organization; peptidoglycan biosynthetic process; regulation of cell shape |
| putative DNA helicase [Microcystis aeruginosa NIES-3804] | GCL52800.1 | <0.0001 | 1333 (b) | 4 (a) | H.A. | DNA metabolism |
| (myosin heavy-chain) kinase [Gemmata massiliana] | GCL49992.1 | <0.0001 | 1 (b) | 4 (a) | H.A. | DNA methylation |
| PBS lyase heat-like repeat protein [Microcystis aeruginosa NIES-3804] | GCE96060.1 | <0.0001 | 1667 (b) | 4 (a) | H.A. | DNA repair |
| wd-40 repeat protein: [Gemmata massiliana] | GCE92418.1 | <0.0001 | 1333 (b) | 4 (a) | H.A. | DNA synthesis involved in DNA repair; double-strand break repair |
| Acetyltransferase component of pyruvate dehydrogenase complex [Pseudohongiella spirulinae] | GCE92614.1 | <0.0001 | 1 (b) | 4 (a) | H.A. | glucose metabolic process |
| Exonuclease SbcC homolog [Microcystis aeruginosa NIES-3804] | VTU01960.1 | <0.0001 | 1 (b) | 4 (a) | H.A. | integral component of membrane |
| Putative multifunctional protein glycosyl transferase, tetratricopeptide domains and SAM methyltransferase domains [Limnospira indica PCC 8005] | GCE93275.1 | <0.0001 | 1 (b) | 4 (a) | H.A. | methylation |
| PQQ enzyme repeat domain protein [Gemmataceae bacterium] | GCE96231.1 | <0.0001 | 1 (b) | 4 (a) | H.A. | methylation |
| Lytic transglycosylase catalytic precursor [Microcystis aeruginosa NIES-3804] | GCL46959.1 | <0.0001 | 1333 (b) | 4 (a) | H.A. | nitrogen compound metabolic process |
| Aldolase/epimerase [Microcystis aeruginosa NIES-3804] | ALO45161.1 | <0.0001 | 1333 (b) | 4 (a) | H.A. | organic acid metabolic process |
| Transcription-repair coupling factor [Pseudohongiella spirulinae] | ALO46521.1 | <0.0001 | 1333 (b) | 4 (a) | H.A. | regulation of transcription, DNA-templated |
| Chromosome segregation protein [Microcystis aeruginosa NIES-3804] | GCE95530.1 | <0.0001 | 1333 (b) | 4 (a) | H.A. | reproductive process |
| 60 kDa molecular chaperonin 2 [Microcystis aeruginosa NIES-3804] | RAQ47071.1 | <0.0001 | 1333 (b) | 4 (a) | H.A. | response to stimulus |
| Transposase [Microcystis aeruginosa NIES-3804] | GCE94368.1 | <0.0001 | 1333 (b) | 4 (a) | H.A. | signal transduction |
| hypothetical protein NIES46_27100 [Arthrospira platensis NIES-46] | GCL55249.1 | <0.0001 | 1 (b) | 4 (a) | H.A. | transmembrane transport |
| Methionine synthase [Arthrospira sp. O9.13F] | ALO46677.1 | <0.0001 | 1 (b) | 4 (a) | H.A. | transmembrane transporter activity |
| hypothetical protein NIES46_44690 [Arthrospira platensis NIES-46] | ALO46533.1 | <0.0001 | 1 (b) | 4 (a) | H.A. | tricarboxylic acid cycle |
| hypothetical protein NIES46_24140 [Arthrospira platensis NIES-46] | EKD07239.1 | <0.0001 | 1 (b) | 4 (a) | H.A. | ubiquitin binding |
| Two-component sensor histidine kinase [Microcystis aeruginosa NIES-3804] | GCL51973.1 | <0.0001 | 1333 (b) | 4 (a) | H.A. | viral tail assembly |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piro, A.; Nisticò, D.M.; Oliva, D.; Fagà, F.A.; Mazzuca, S. Physiological and Metabolic Response of Arthrospira maxima to Organophosphates. Microorganisms 2022, 10, 1063. https://doi.org/10.3390/microorganisms10051063
Piro A, Nisticò DM, Oliva D, Fagà FA, Mazzuca S. Physiological and Metabolic Response of Arthrospira maxima to Organophosphates. Microorganisms. 2022; 10(5):1063. https://doi.org/10.3390/microorganisms10051063
Chicago/Turabian StylePiro, Amalia, Dante Matteo Nisticò, Daniela Oliva, Francesco Antonio Fagà, and Silvia Mazzuca. 2022. "Physiological and Metabolic Response of Arthrospira maxima to Organophosphates" Microorganisms 10, no. 5: 1063. https://doi.org/10.3390/microorganisms10051063







