A Study and Modeling of Bifidobacterium and Bacillus Coculture Continuous Fermentation under Distal Intestine Simulated Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Inoculate Preparation
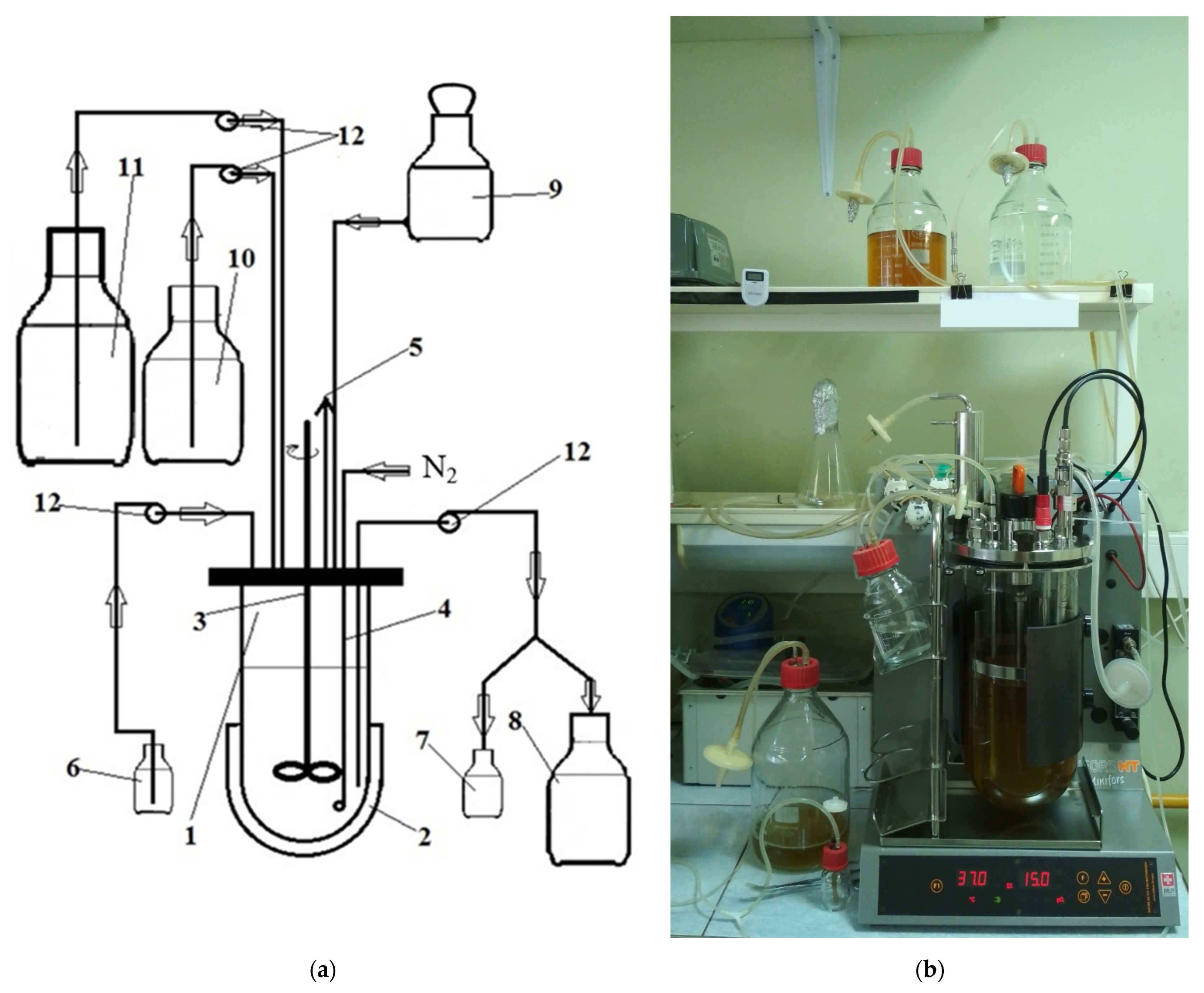
2.2. Continuous Fermentation
2.3. Enumeration of Bacteria
2.4. Quantification of Organic Acid
2.5. Carbohydrate Assay
2.6. Models and Calculations
2.7. Statistical Analysis
3. Results
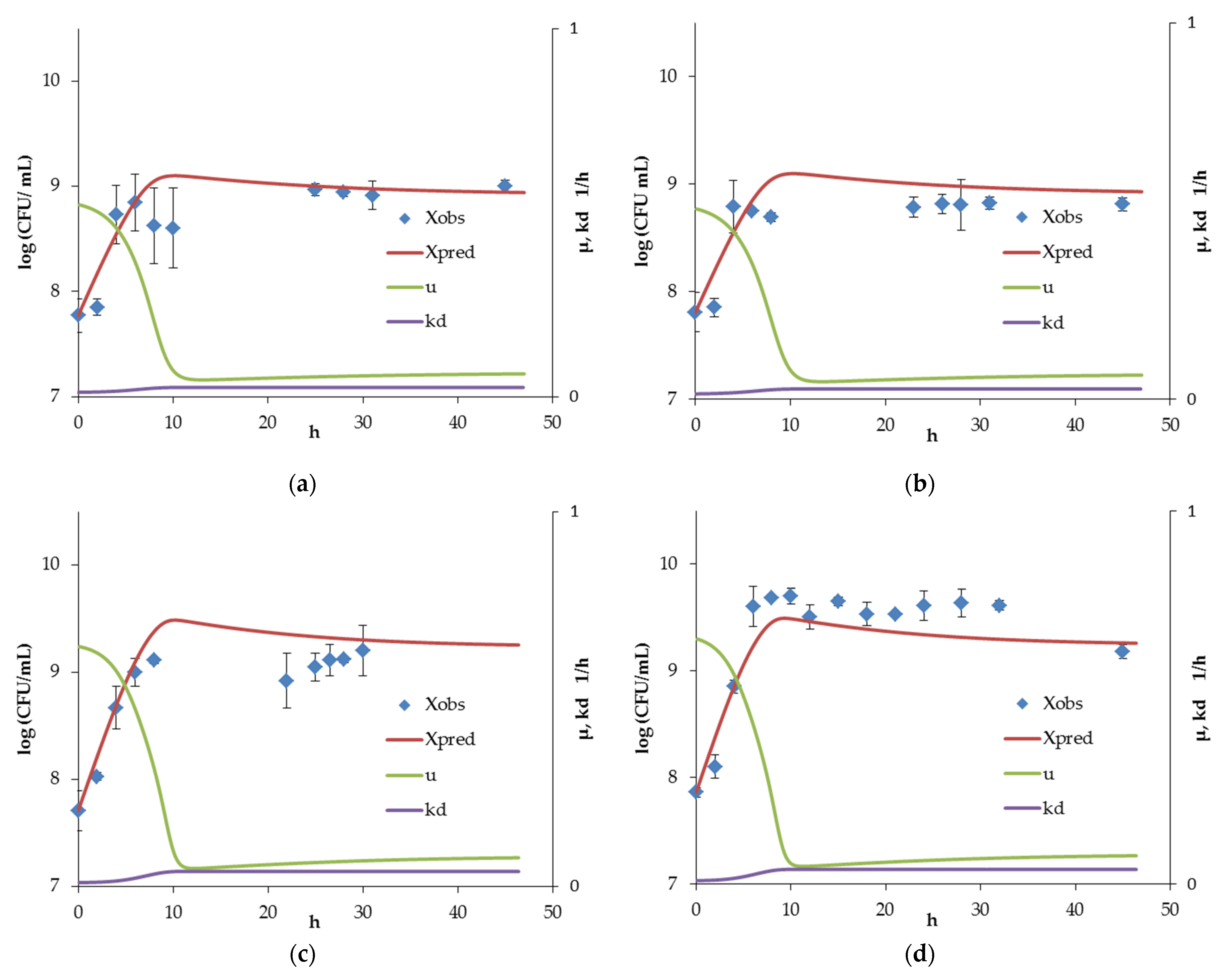
3.1. Bifidobacterium Monoculture Experiments and Model
3.2. Bacillus Monoculture and Coculture Experiments and Inhibition Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swanson, K.S.; Gibson, G.R.; Hutkins, R.; Reimer, R.A.; Reid, G.; Verbeke, K.; Scott, K.P.; Holscher, H.D.; Azad, M.B.; Delzenne, N.M.; et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 687–701. [Google Scholar] [CrossRef] [PubMed]
- Dronkers, T.M.G.; Ouwehand, A.C.; Rijkers, G.T. Global analysis of clinical trials with probiotics. Heliyon 2020, 6, e04467. [Google Scholar] [CrossRef]
- Williams, C.F.; Walton, G.E.; Jiang, L.; Plummer, S.; Garaiova, I.; Gibson, G.R. Comparative analysis of intestinal tract models. Annu. Rev. Food Sci. Technol. 2015, 6, 329–350. [Google Scholar] [CrossRef]
- Von Martels, J.; Sadaghian Sadabad, M.; Bourgonje, A.R.; Blokzijl, T.; Dijkstra, G.; Faber, K.N.; Harmsen, H. The role of gut microbiota in health and disease: In vitro modeling of host-microbe interactions at the aerobe-anaerobe interphase of the human gut. Anaerobe 2017, 44, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Gibson, G.R.; Cummings, J.H.; Macfarlane, G.T. Use of a three-stage continuous culture system to study the effect of mucin on dissimilatory sulfate reduction and methanogenesis by mixed populations of human gut bacteria. Appl. Environ. Microbiol. 1988, 54, 2750–2755. [Google Scholar] [CrossRef] [Green Version]
- Macfarlane, S.; Quigley, M.E.; Hopkins, M.J.; Newton, D.F.; Macfarlane, G.T. Polysaccharide degradation by human intestinal bacteria during growth under multi-substrate limiting conditions in a three-stage continuous culture system. FEMS Microbiol. Ecol 1998, 26, 231–243. [Google Scholar] [CrossRef]
- Probert, H.M.; Apajalahti, J.H.; Rautonen, N.; Stowell, J.; Gibson, G.R. Polydextrose, lactitol, and fructo-oligosaccharide fermentation by colonic bacteria in a three-stage continuous culture system. Appl. Environ. Microbiol. 2004, 70, 4505–4511. [Google Scholar] [CrossRef] [Green Version]
- Costabile, A.; Walton, G.E.; Tzortzis, G.; Vulevic, J.; Charalampopoulos, D.; Gibson, G.R. Effects of orange juice formulation on prebiotic functionality using an in vitro colonic model system. PLoS ONE 2015, 10, e0121955. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Gibson, G.R.; Sailer, M.; Theis, S.; Rastall, R.A. Prebiotics Inhibit Proteolysis by Gut Bacteria in a Host Diet-Dependent Manner: A Three-Stage Continuous In Vitro Gut Model Experiment. Appl. Environ. Microbiol. 2020, 86, e02730-19. [Google Scholar] [CrossRef] [PubMed]
- Astó, E.; Méndez, I.; Rodríguez-Prado, M.; Cuñé, J.; Espadaler, J.; Farran-Codina, A. Effect of the Degree of Polymerization of Fructans on Ex Vivo Fermented Human Gut Microbiome. Nutrients 2019, 11, 1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sannasiddappa, T.H.; Costabile, A.; Gibson, G.R.; Clarke, S.R. The influence of Staphylococcus aureus on gut microbial ecology in an in vitro continuous culture human colonic model system. PLoS ONE 2011, 6, e23227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vesely, O.; Marsik, P.; Jarosova, V.; Doskocil, I.; Smejkal, K.; Kloucek, P.; Havlik, J. Metabolism of Selected 2-Arylbenzofurans in a Colon In Vitro Model System. Foods 2021, 10, 2754. [Google Scholar] [CrossRef] [PubMed]
- Feria-Gervasio, D.; Denis, S.; Alric, M.; Brugère, J.F. In vitro maintenance of a human proximal colon microbiota using the continuous fermentation system P-ECSIM. Appl. Microbiol. Biotechnol. 2011, 91, 1425–1433. [Google Scholar] [CrossRef] [PubMed]
- Vamanu, E.; Pelinescu, D.; Avram, I.; Niță, S.; Vamanu, A. Study of PROBAC product influence on infant microbiota in a single-chamber colonic fermentation model GIS1. Ann. Microbiol. 2013, 63, 1029–1038. [Google Scholar] [CrossRef]
- Vamanu, E.; Pelinescu, D.; Avram, I.; Nita, S. An in vitro evaluation of antioxidant and colonic microbial profile levels following mushroom consumption. Biomed Res. Int. 2013, 2013, 289821. [Google Scholar] [CrossRef] [Green Version]
- Konopka, A. What is microbial community ecology? ISME J. 2009, 3, 1223–1230. [Google Scholar] [CrossRef] [Green Version]
- Qian, Y.; Lan, F.; Venturelli, O.S. Towards a deeper understanding of microbial communities: Integrating experimental data with dynamic models. Curr. Opin. Microbiol. 2021, 62, 84–92. [Google Scholar] [CrossRef]
- Giménez, B.; Dalgaard, P. Modelling and predicting the simultaneous growth of Listeria monocytogenes and spoilage micro-organisms in cold-smoked salmon. J. Appl. Microbiol. 2004, 96, 96–109. [Google Scholar] [CrossRef]
- Ye, K.; Wang, H.; Jiang, Y.; Xu, X.; Cao, J.; Zhou, G. Development of interspecific competition models for the growth of Listeria monocytogenes and Lactobacillus on vacuum-packaged chilled pork by quantitative real-time PCR. Food Res. Int. 2014, 64, 626–633. [Google Scholar] [CrossRef] [PubMed]
- Ačai, P.; Valík, L.; Medved’ová, A.; Rosskopf, F. Modelling and predicting the simultaneous growth of Escherichia coli and lactic acid bacteria in milk. Food. Sci. Technol. Int. 2016, 22, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Panebianco, F.; Giarratana, F.; Caridi, A.; Sidari, R.; De Bruno, A.; Giuffrida, A. Lactic acid bacteria isolated from traditional Italian dairy products: Activity against Listeria monocytogenes and modelling of microbial competition in soft cheese. LWT 2021, 137, 110446. [Google Scholar] [CrossRef]
- Ačai, P.; Valík, Ľ.; Medveďová, A. One-and Two-Step Kinetic Data Analysis Applied for Single and Co-Culture Growth of Staphylococcus aureus, Escherichia coli, and Lactic Acid Bacteria in Milk. Appl. Sci. 2021, 11, 8673. [Google Scholar] [CrossRef]
- Venturelli, O.S.; Carr, A.C.; Fisher, G.; Hsu, R.H.; Lau, R.; Bowen, B.P.; Hromada, S.; Northen, T.; Arkin, A.P. Deciphering microbial interactions in synthetic human gut microbiome communities. Mol. Syst. Biol. 2018, 14, e8157. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Tamayo, R.; Laroche, B.; Walter, E.; Doré, J.; Leclerc, M. Mathematical modelling of carbohydrate degradation by human colonic microbiota. J. Theor. Biol. 2010, 266, 189–201. [Google Scholar] [CrossRef]
- Muñoz-Tamayo, R.; Laroche, B.; Walter, E.; Doré, J.; Duncan, S.H.; Flint, H.J.; Leclerc, M. Kinetic modelling of lactate utilization and butyrate production by key human colonic bacterial species. FEMS Microbial. Ecol. 2011, 76, 615–624. [Google Scholar] [CrossRef] [Green Version]
- Pinto, F.; Medina, D.A.; Pérez-Correa, J.R.; Garrido, D. Modeling Metabolic Interactions in a Consortium of the Infant Gut Microbiome. Front. Microbial. 2017, 8, 2507. [Google Scholar] [CrossRef]
- Andrews, J.F. A mathematical model for the continuous culture of microorganisms utilizing inhibitory substrates. Biotechnol. Bioeng. 1968, 10, 707–723. [Google Scholar] [CrossRef]
- Karetkin, B.A.; Guseva, E.V.; Evdokimova, S.A.; Mishchenko, A.S.; Khabibulina, N.V.; Grosheva, V.D.; Menshutina, N.V.; Panfilov, V.I. A quantitative model of Bacillus cereus ATCC 9634 growth inhibition by bifidobacteria for synbiotic effect evaluation. World J. Microbiol. Biotechnol. 2019, 35, 89. [Google Scholar] [CrossRef]
- Evdokimova, S.A.; Nokhaeva, V.S.; Karetkin, B.A.; Guseva, E.V.; Khabibulina, N.V.; Kornienko, M.A.; Grosheva, V.D.; Menshutina, N.V.; Shakir, I.V.; Panfilov, V.I. A Study on the Synbiotic Composition of Bifidobacterium bifidum and Fructans from Arctium lappa Roots and Helianthus tuberosus Tubers against Staphylococcus aureus. Microorganisms 2021, 9, 930. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Corradini, C.; Amaretti, A.; Nicolini, M.; Pompei, A.; Zanoni, S.; Matteuzzi, D. Fermentation of fructooligosaccharides and inulin by bifidobacteria: A comparative study of pure and faecal cultures. Appl. Environ. Microbiolog. 2005, 71, 6150–6158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Man, J.C.; Rogosa, D.; Sharpe, M.E. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 1960, 23, 130–135. [Google Scholar] [CrossRef]
- Nebra, Y.; Blanch, A.R. A new selective medium for Bifidobacterium spp. Appl. Environ. Microbiol. 1999, 65, 5173–5176. [Google Scholar] [CrossRef] [Green Version]
- Scherer, R.; Rybka, A.C.P.; Ballus, C.A.; Meinhart, A.D.; Filho, J.T.; Godoy, H.T. Validation of a HPLC method for simultaneous determination of main organic acids in fruits and juices. Food Chem. 2012, 135, 150–154. [Google Scholar] [CrossRef] [Green Version]
- Andersen, K.E.; Bjergegaard, C.; Møller, P.; Sørensen, J.C.; Sørensen, H. High-Performance Capillary Electrophoresis with Indirect UV Detection for Determination of α-Galactosides in Leguminosae and Brassicaceae. J. Agric. Food Chem. 2003, 51, 6391–6397. [Google Scholar] [CrossRef]
- Arentoft, A.M.; Michaelsen, S.; Sørensen, H. Determination of oligosaccharides by capillary zone electrophoresis. J. Chromatogr. A 1993, 652, 517–524. [Google Scholar] [CrossRef]
- Pirt, S.J. Principles of Microbe and Cell Cultivation; Halsted Press: New York, NY, USA, 1975; p. 274. ISBN 9780470690383. [Google Scholar] [CrossRef]
- Monod, J. The growth of bacterial cultures. Annu. Rev. Microbiol. 1949, 3, 371–394. [Google Scholar] [CrossRef] [Green Version]
- Legner, M.; McMillen, D.R.; Cvitkovitch, D.G. Role of Dilution Rate and Nutrient Availability in the Formation of Microbial Biofilms. Front. Microbiol. 2019, 10, 916. [Google Scholar] [CrossRef] [Green Version]
- Shan, H.; Yasuda, T.; Ohkura, K. A self adaptive hybrid enhanced artificial bee colony algorithm for continuous optimization problems. Biosystems 2015, 132–133, 43–53. [Google Scholar] [CrossRef]
- Roeva, O.; Zoteva, D.; Lyubenova, V. Escherichia coli Cultivation Process Modelling Using ABC-GA Hybrid Algorithm. Processes 2021, 9, 1418. [Google Scholar] [CrossRef]
- Lu, J.; Wu, Y.; Yuan, J.; Yuan, J.; Wang, Z.; Gao, J.; Chen, H. Characterization of Bacillus cereus AFA01 Capable of Degrading Gluten and Celiac-Immunotoxic Peptides. Foods 2021, 10, 1725. [Google Scholar] [CrossRef]
- Muñoz-Tamayo, R.; Giger-Reverdin, S.; Sauvant, D. Mechanistic modelling of in vitro fermentation and methane production by rumen microbiota. Anim. Feed Sci. Technol. 2016, 220, 1–21. [Google Scholar] [CrossRef]
- Cruz Ramos, H.; Hoffmann, T.; Marino, M.; Nedjari, H.; Presecan-Siedel, E.; Dreesen, O.; Glaser, P.; Jahn, D. Fermentative metabolism of Bacillus subtilis: Physiology and regulation of gene expression. J. Bacteriol. 2000, 182, 3072–3080. [Google Scholar] [CrossRef] [Green Version]
- Baranyi, J.; Roberts, T.A. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 1994, 23, 277–294. [Google Scholar] [CrossRef]
- Likotrafiti, E.; Tuohy, K.M.; Gibson, G.R.; Rastall, R.A. An in vitro study of the effect of probiotics, prebiotics and synbiotics on the elderly faecal microbiota. Anaerobe 2014, 27, 50–55. [Google Scholar] [CrossRef] [PubMed]


| Parameter Description | Unit | Value |
|---|---|---|
| Bifidobacterium monoculture | ||
| h−1 | 1.003 | |
| mg/mL | 0.727 | |
| mg/mL | 5.46 | |
| mg/mL | 5.33 | |
| h−1 | 0.117 | |
| mg/mL | 2.12 | |
| mg/mL | 2.05 | |
| mg/cell | 1.40 × 10−6 | |
| mg/mg | 0.313 | |
| mg/mg | 0.631 | |
| Bacillus coculture | ||
| h−1 | 1.214 | |
| mg/mL | 8.85 | |
| mg/mL | 10.15 | |
| mg/mL | 0.18 | |
| mg/mL | 0.71 | |
| - | 0.55 | |
| - | 2.23 | |
| Inlet OF Concentration | Bifidobacterium Monoculture | Bacillus Cell Count in Coculture | ||
|---|---|---|---|---|
| (mg/mL) | Viable Cell Count | LA | AA | |
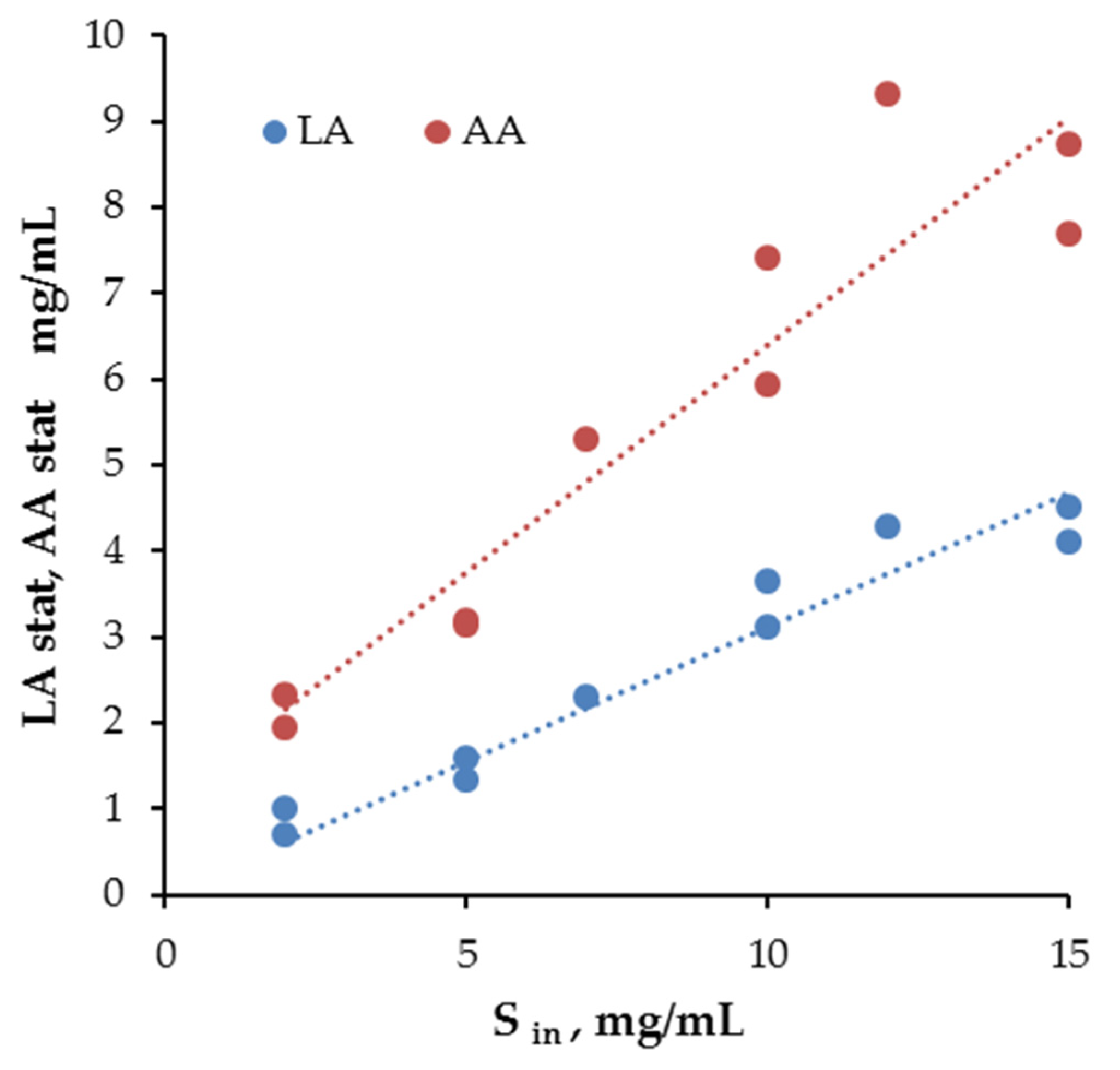
| 2 | 1.93 | 27.2 | 19.3 | - |
| 2 | 2.17 | 68.9 | 10.3 | - |
| 5 | 2.03 | 24.4 | 24.8 | - |
| 5 | 2.11 | 33.1 | 25.3 | 0.75 |
| 7 | 2.10 | 27.5 | 10.5 | 0.93 |
| 10 | 1.14 | 9.8 | 18.3 | - |
| 10 | 2.58 | 7.39 | 5.66 | 2.78 |
| 12 | 2.12 | 13.7 | 11.9 | 0.75 |
| 15 | 2.39 | 29.9 | 18.9 | - |
| 15 | 1.00 | 14.2 | 27.5 | 0.74 |
| Sin (mg/mL) | LA (mg/mL) | AA (mg/mL) | Total Acids (mg/mL) | at log Phase (h−1) | |
|---|---|---|---|---|---|
| Monoculture | 0.78 | 0 | |||
| 5 | 1.23 | 3.44 | 4.67 | 0.38 | 3.5 |
| 7 | 1.03 | 6.60 | 7.63 | 0.53 | 3.0 |
| 10 | 3.20 | 8.02 | 11.22 | 0.66 | 3.5 |
| 12 | 3.22 | 8.28 | 11.50 | 0.59 | 4.0 |
| 15 | 3.54 | 7.49 | 11.03 | 0.62 | 8.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Evdokimova, S.A.; Karetkin, B.A.; Guseva, E.V.; Gordienko, M.G.; Khabibulina, N.V.; Panfilov, V.I.; Menshutina, N.V.; Gradova, N.B. A Study and Modeling of Bifidobacterium and Bacillus Coculture Continuous Fermentation under Distal Intestine Simulated Conditions. Microorganisms 2022, 10, 929. https://doi.org/10.3390/microorganisms10050929
Evdokimova SA, Karetkin BA, Guseva EV, Gordienko MG, Khabibulina NV, Panfilov VI, Menshutina NV, Gradova NB. A Study and Modeling of Bifidobacterium and Bacillus Coculture Continuous Fermentation under Distal Intestine Simulated Conditions. Microorganisms. 2022; 10(5):929. https://doi.org/10.3390/microorganisms10050929
Chicago/Turabian StyleEvdokimova, Svetlana A., Boris A. Karetkin, Elena V. Guseva, Maria G. Gordienko, Natalia V. Khabibulina, Victor I. Panfilov, Natalia V. Menshutina, and Nina B. Gradova. 2022. "A Study and Modeling of Bifidobacterium and Bacillus Coculture Continuous Fermentation under Distal Intestine Simulated Conditions" Microorganisms 10, no. 5: 929. https://doi.org/10.3390/microorganisms10050929






