Land-Use Driven Changes in Soil Microbial Community Composition and Soil Fertility in the Dry-Hot Valley Region of Southwestern China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Soil Sampling
2.3. Analysis of Soil Physicochemical Properties
2.4. Total DNA Extraction, PCR Amplification, and Purification
2.5. Library Preparation and Sequencing
2.6. Data Preprocessing and Species Annotation
2.7. Data Analysis
3. Results
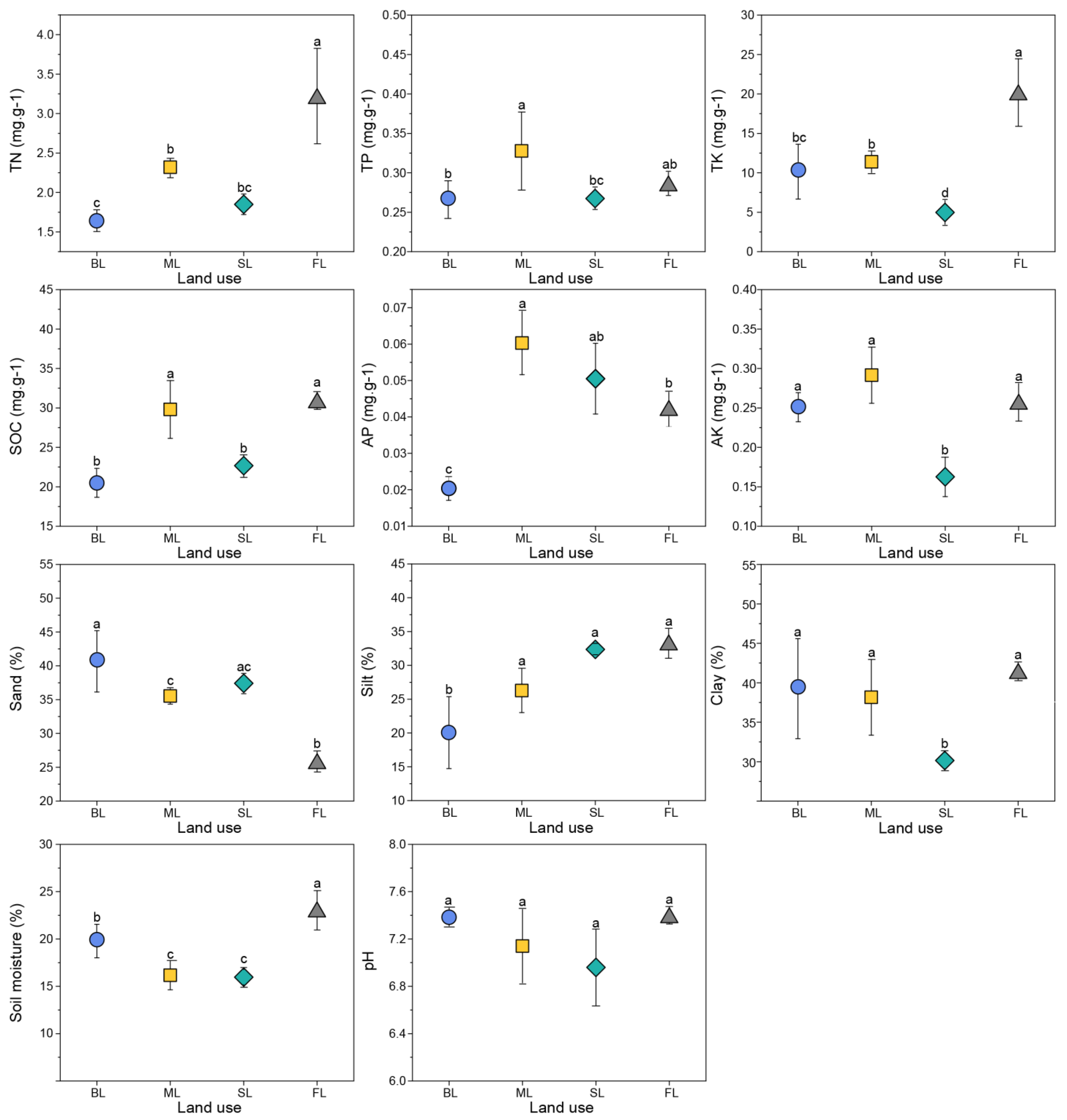
3.1. Soil Physicochemical Properties
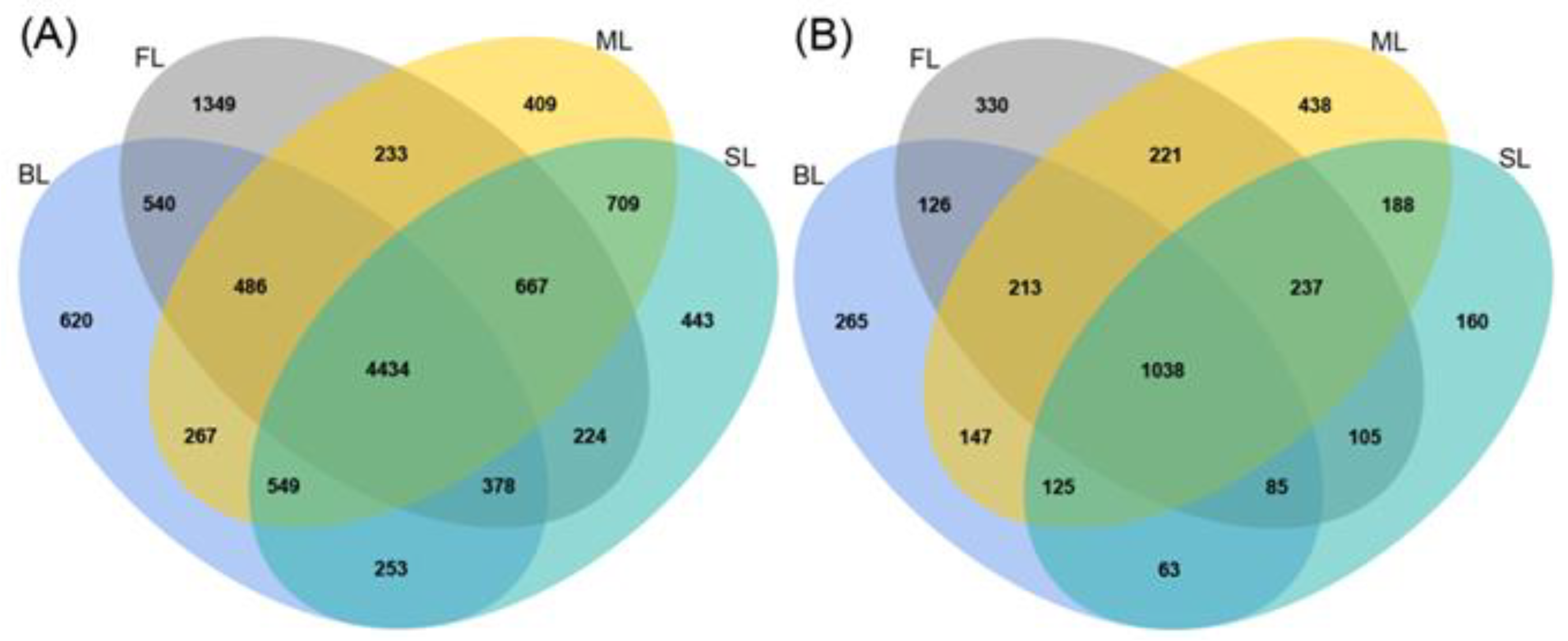
3.2. Analysis of Soil Microbial Sequences and OTUs
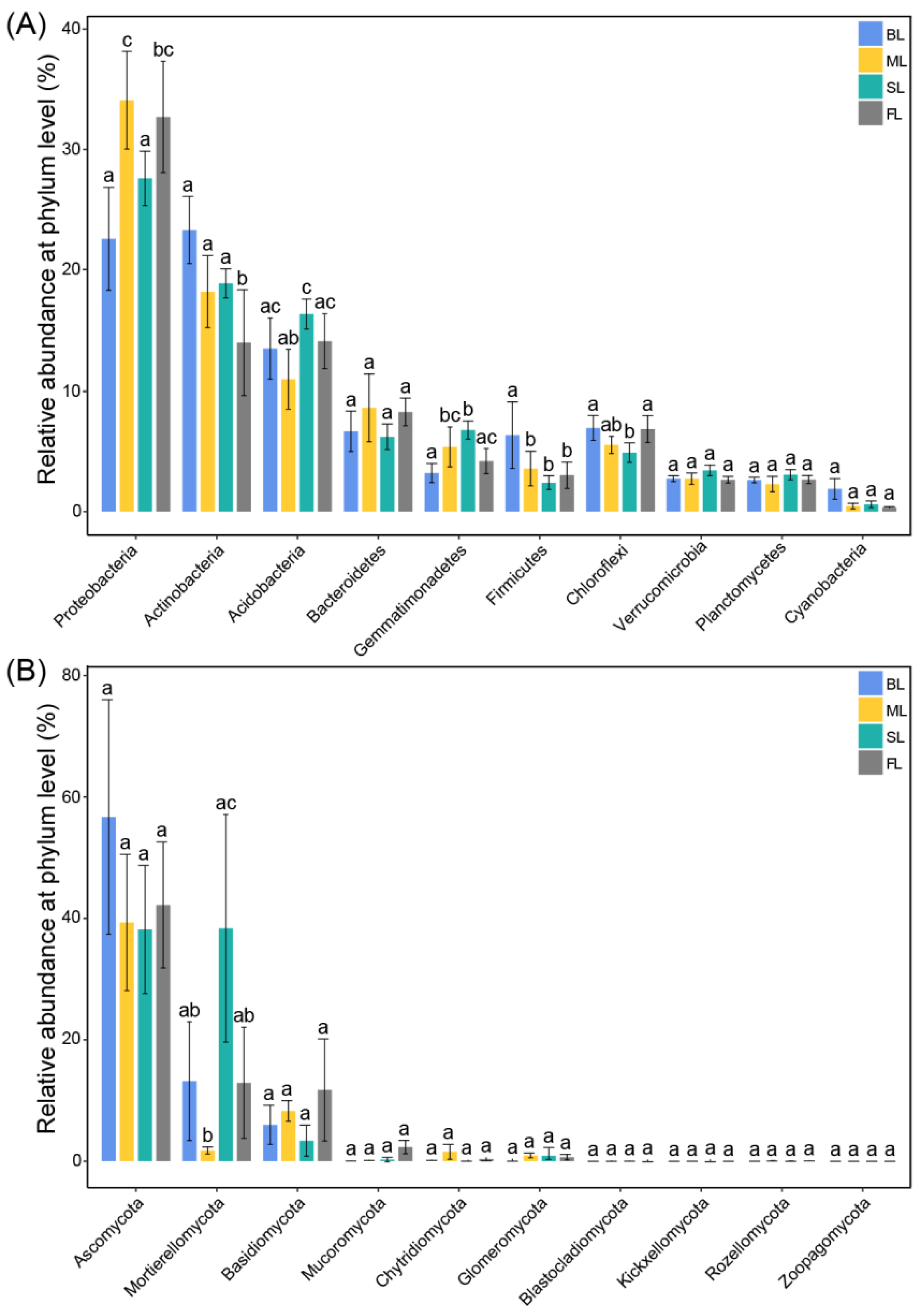
3.3. Comparison of Soil Microbial Community Composition and Relative Abundance
3.4. Soil Microbial Community Composition and Diversity
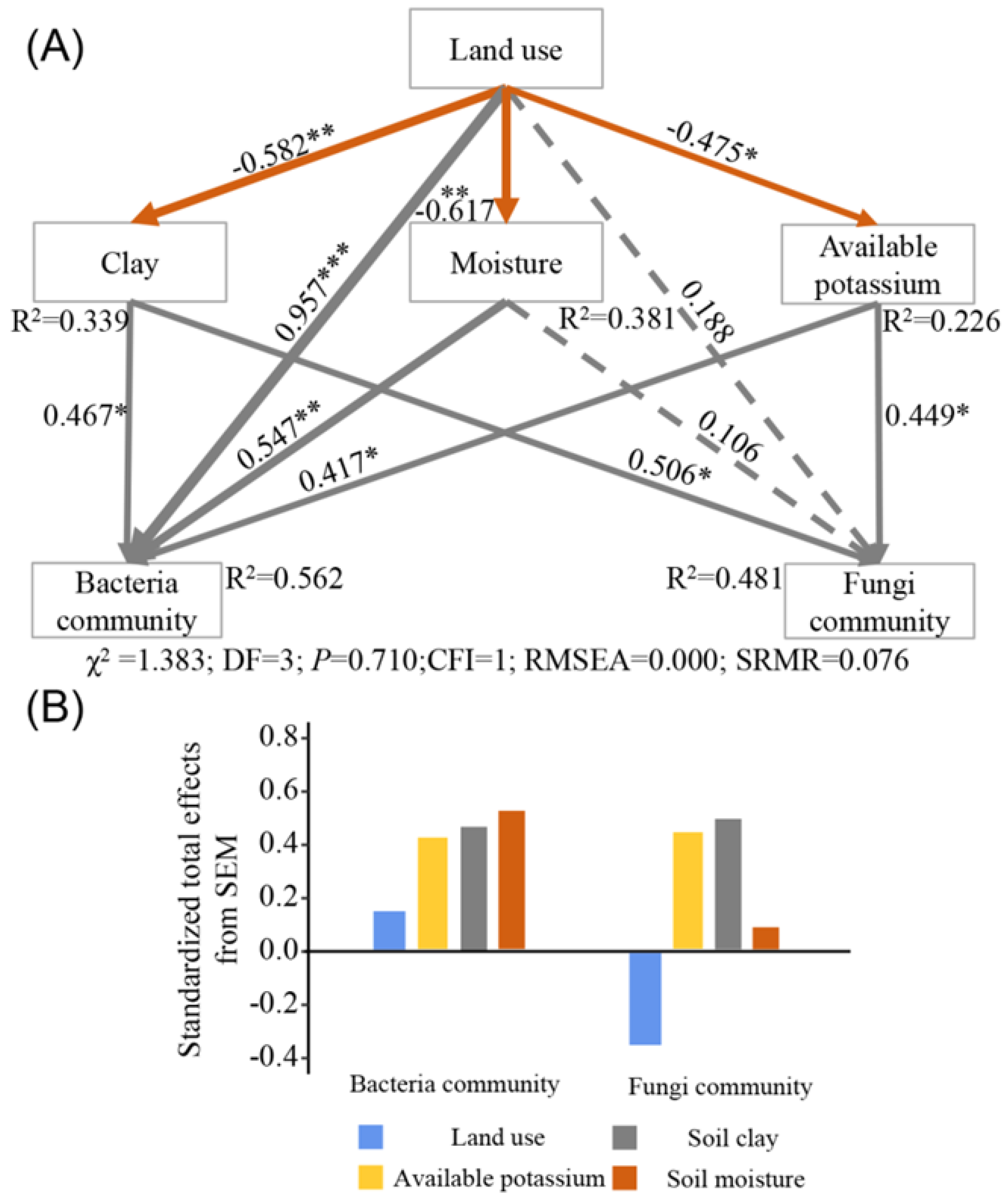
3.5. Relationships among Soil Microbial Community Abundance and Soil Physicochemical Properties
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fedele, G.; Locatelli, B.; Djoudi, H.; Colloff, M.J. Reducing risks by transforming landscapes: Cross-scale effects of land-use changes on ecosystem services. PLoS ONE 2018, 13, e195895. [Google Scholar] [CrossRef] [PubMed]
- Schjønning, P.; Jensen, J.L.; Bruun, S.; Jensen, L.S.; Christensen, B.T.; Munkholm, L.J. The Role of Soil Organic Matter for Maintaining Crop Yields: Evidence for a Renewed Conceptual Basis. Adv. Agron. 2018, 150, 35–79. [Google Scholar] [CrossRef]
- Song, X.; Hansen, M.C.; Stehman, S.V.; Potapov, P.V.; Tyukavina, A.; Vermote, E.F.; Townshend, J.R. Global land change from 1982 to 2016. Nature 2018, 560, 639–643. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.B.; Gifford, R.M. Soil carbon stocks and land use change: A meta-analysis. Glob. Chang. Biol. 2002, 8, 345–360. [Google Scholar] [CrossRef]
- Dong, Y.; Xiong, D.; Su, Z.; Li, J.; Yang, D.; Shi, L.; Liu, G. The distribution of and factors influencing the vegetation in a gully in the Dry-hot Valley of southwest China. Catena 2014, 116, 60–67. [Google Scholar] [CrossRef]
- Wang, G.; Huang, W.; Zhou, G.; Mayes, M.A.; Zhou, J. Modeling the processes of soil moisture in regulating microbial and carbon-nitrogen cycling. J. Hydrol. 2020, 585, 124777. [Google Scholar] [CrossRef]
- Borrelli, P.; Robinson, D.A.; Panagos, P.; Lugatod, E.; Yangb, J.Y.; Alewell, C.; Wuepper, D.; Montanarella, L.; Ballabio, C. Land use and climate change impacts on global soil erosion by water (2015—2070). Proc. Natl. Acad. Sci. USA 2020, 117, 21994–22001. [Google Scholar] [CrossRef]
- Mcdougall, R.; Rader, R.; Kristiansen, P. Urban agriculture could provide 15% of food supply to Sydney, Australia, under expanded land use scenarios. Land Use Policy 2020, 94, 104554. [Google Scholar] [CrossRef]
- Li, B.; Liu, Y.; Zhang, H.; Liu, Y.; Liu, Y.; Xie, P. Research progress in the functionalization of microcystin-LR based on interdisciplinary technologies. Coord. Chem. Rev. 2021, 443, 214041. [Google Scholar] [CrossRef]
- Roy, K.; Ghosh, D.; DeBruyn, J.M.; Dasgupta, T.; Wommack, K.E.; Liang, X.; Wagner, R.E.; Radosevich, M. Temporal Dynamics of Soil Virus and Bacterial Populations in Agricultural and Early Plant Successional Soils. Front. Microbiol. 2020, 11, 1494. [Google Scholar] [CrossRef]
- Six, J.; Paustian, K. Aggregate-associated soil organic matter as an ecosystem property and a measurement tool. Soil Biol. Biochem. 2014, 68, A4–A9. [Google Scholar] [CrossRef]
- Yang, X.; Leng, Y.; Zhou, Z.; Shang, H.; Ni, K.; Ma, L.; Yi, X.; Cai, Y.; Ji, L.; Ruan, J.; et al. Ecological management model for the improvement of soil fertility through the regulation of rare microbial taxa in tea (Camellia sinensis L.) plantation soils. J. Environ. Manag. 2022, 308, 114595. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, P.B.; Fuentes-Alburquenque, S.; Díez, B.; Vargas, I.; Bonilla, C.A. Soil microbial community responses to labile organic carbon fractions in relation to soil type and land use along a climate gradient. Soil Biol. Biochem. 2020, 141, 107692. [Google Scholar] [CrossRef]
- Ji, L.; Yang, Y.; Yang, L.; Zhang, D. Effect of land uses on soil microbial community structures among different soil depths in northeastern China. Eur. J. Soil Biol. 2020, 99, 103205. [Google Scholar] [CrossRef]
- Li, Y.; Xie, X.; Zhu, Z.; Liu, K.; Liu, W.; Wang, J. Land use driven change in soil organic carbon affects soil microbial community assembly in the riparian of Three Gorges Reservoir Region. Appl. Soil Ecol. 2022, 176, 104467. [Google Scholar] [CrossRef]
- Emmert, E.A.B.; Geleta, S.B.; Rose, C.M.; Seho-Ahiable, G.E.; Hawkins, A.E.; Baker, K.T.; Evans, A.S.; Harris, M.E.; Mrozinski, A.C.; Folkoff, M.E.; et al. Effect of land use changes on soil microbial enzymatic activity and soil microbial community composition on Maryland’s Eastern Shore. Appl. Soil Ecol. 2021, 161, 103824. [Google Scholar] [CrossRef]
- Xu, A.; Liu, J.; Guo, Z.; Wang, C.; Pan, K.; Zhang, F.; Pan, X. Soil microbial community composition but not diversity is affected by land-use types in the agro-pastoral ecotone undergoing frequent conversions between cropland and grassland. Geoderma 2021, 401, 115165. [Google Scholar] [CrossRef]
- Mhete, M.; Eze, P.N.; Rahube, T.O.; Akinyemi, F.O. Soil properties influence bacterial abundance and diversity under different land-use regimes in semi-arid environments. Sci. Am. 2020, 7, e246. [Google Scholar] [CrossRef]
- Gömöryová, E.; Barančíková, G.; Tobiašová, E.; Halás, J.; Skalský, R.; Koco, Š.; Gömöry, D. Responses of soil microorganisms to land use in different soil types along the soil profiles. Soil Water Res. 2020, 15, 125–134. [Google Scholar] [CrossRef]
- Naldini, M.B.; Pérez Harguindeguy, N.; Kowaljow, E. Soil carbon release enhanced by increased litter input in a degraded semi-arid forest soil. J. Arid. Environ. 2021, 186, 104400. [Google Scholar] [CrossRef]
- Chernov, T.I.; Zhelezova, A.D.; Tkhakakhova, A.K.; Ksenofontova, N.A.; Zverev, A.O.; Tiunov, A.V. Soil microbiome, organic matter content and microbial abundance in forest and forest-derived land cover in Cat Tien National Park (Vietnam). Appl. Soil Ecol. 2021, 165, 103957. [Google Scholar] [CrossRef]
- Medorio-García, H.P.; Alarcón, E.; Flores-Esteves, N.; Montaño, N.M.; Perroni, Y. Soil carbon, nitrogen and phosphorus dynamics in sugarcane plantations converted from tropical dry forest. Appl. Soil Ecol. 2020, 154, 103600. [Google Scholar] [CrossRef]
- de Vries, F.T.; Hoffland, E.; van Eekeren, N.; Brussaard, L.; Bloem, J. Fungal/bacterial ratios in grasslands with contrasting nitrogen management. Soil Biol. Biochem. 2006, 38, 2092–2103. [Google Scholar] [CrossRef] [Green Version]
- Rousk, J.; Brookes, P.C.; Bååth, E. Investigating the mechanisms for the opposing pH relationships of fungal and bacterial growth in soil. Soil Biol. Biochem. 2010, 42, 926–934. [Google Scholar] [CrossRef]
- Fanin, N.; Bertrand, I. Aboveground litter quality is a better predictor than belowground microbial communities when estimating carbon mineralization along a land-use gradient. Soil Biol. Biochem. 2016, 94, 48–60. [Google Scholar] [CrossRef]
- Veldkamp, E.; Schmidt, M.; Powers, J.S.; Corre, M.D. Deforestation and reforestation impacts on soils in the tropics. Nat. Rev. Earth Environ. 2020, 1, 590–605. [Google Scholar] [CrossRef]
- Zhang, R.Z. Dry Valley in Hengduan Mountainous Region; Science Press: Beijing, China, 1992; pp. 120–162. [Google Scholar]
- Xiong, D.; Zhou, H.; Yang, Z.; Zhang, X. Slope lithologic property, soil moisture condition and revegetation in dry-hot valley of jinsha river. Chin. Geogr. Sci. 2005, 15, 186–192. [Google Scholar] [CrossRef]
- Long, H.; Zhang, D.; He, G. The effects of planted and plowed Stylosanthes guianensis on degrading soil fertility in hilly countries of dry-hot valley. Acta Ecol. Sin. 2017, 37, 279–283. [Google Scholar] [CrossRef]
- Rong, L.; Duan, X.; Zhang, G.; Gu, Z.; Feng, D. Impacts of tillage practices on ephemeral gully erosion in a dry-hot valley region in southwestern China. Soil Tillage Res. 2019, 187, 72–84. [Google Scholar] [CrossRef]
- Li, Z.; Sun, X.; Huang, Z.; Zhang, X.; Wang, Z.; Li, S.; Zheng, W.; Zhai, B. Changes in nutrient balance, environmental effects, and green development after returning farmland to forests: A case study in Ningxia, China. Sci. Total Environ. 2020, 735, 139370. [Google Scholar] [CrossRef]
- Li, S.; Huang, X.; Lang, X.; Shen, J.; Xu, F.; Su, J. Cumulative effects of multiple biodiversity attributes and abiotic factors on ecosystem multifunctionality in the Jinsha River valley of southwestern China. For. Ecol. Manag. 2020, 472, 118281. [Google Scholar] [CrossRef]
- Li, Y.; Duan, X.; Li, Y.; Li, Y.; Zhang, L. Interactive effects of land use and soil erosion on soil organic carbon in the dry-hot valley region of southern China. Catena 2021, 201, 105187. [Google Scholar] [CrossRef]
- Yang, D.; Xiong, D.; Zhang, B.; Guo, M.; Su, Z.; Dong, Y.; Zhang, S.; Xiao, L.; Lu, X. Effect of grass basal diameter on hydraulic properties and sediment yield processes in gully beds in the dry-hot valley region of Southwest China. Catena 2017, 152, 299–310. [Google Scholar] [CrossRef]
- Yuan, Y.; Xiong, D.; Wu, H.; Zhang, S.; Zhang, B.; Dahal, N.M.; Liu, L.; Li, W.; Zhang, W.; Shi, L. Spatial variation of soil physical properties and its relationship with plant biomass in degraded slopes in dry-hot valley region of Southwest China. J. Soil Sediments 2020, 20, 2354–2366. [Google Scholar] [CrossRef]
- FAO. World Reference Base for Soil Resources; FAO-ISRIC-ISSS: Rome, Italy, 2006. [Google Scholar]
- Duka, I.; Shallari, S.; Maçi, A.; Shehu, J. Heavy Metal Concentration and Physico-Chemical Parameters in Koder Kamza Soils. J. Agric. Sci. 2015, 14, 157. [Google Scholar]
- Song, J.; Duan, X.; Han, X.; Li, Y.; Li, Y.; He, D. The accumulation and redistribution of heavy metals in the water-level fluctuation zone of the Nuozhadu Reservoir, Upper Mekong. Catena 2019, 172, 335–344. [Google Scholar] [CrossRef]
- Nelson, D.W.; Sommers, L.E. Total carbon, organic carbon and organic matter. In Methods of Soil Analysis Part 2, Chemical and Microbial Properties; Soil Science Society of America and the American Society of Agronomy: Madison, WI, USA, 1982; pp. 961–1010. [Google Scholar]
- Liu, G.S. Soil Physics and Chemistry Analysis and Description of Soil Profiles; Science Press: Beijing, China, 1996; pp. 320–346. [Google Scholar]
- Bao, S.D. Soil Agri-Chemistrical Analysis; Science Press: Beijing, China, 2000; pp. 120–195. [Google Scholar]
- Mori, H.; Maruyama, F.; Kato, H.; Toyoda, A.; Dozono, A.; Ohtsubo, Y.; Nagata, Y.; Fujiyama, A.; Tsuda, M.; Kurokawa, K. Design and Experimental Application of a Novel Non-Degenerate Universal Primer Set that Amplifies Prokaryotic 16S rRNA Genes with a Low Possibility to Amplify Eukaryotic rRNA Genes. DNA Res. 2014, 21, 217–227. [Google Scholar] [CrossRef] [PubMed]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics; Gelfand, D., Sninsky, J., White, T., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Ward, T.; Larson, J.; Meulemans, J.; Hillmann, B.; Lynch, J.; Sidiropoulos, D.; Spear, J.R.; Gaporaso, G.; Blekhman, R.; Knight, R.; et al. BugBase predicts organism level microbiome phenotypes. BioRxiv 2017, 2, 133462. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Jeffries, T.C.; Gaitan, J.J.; Encinar, D.; Berdugo, M.; Campbell, C.D.; Singh, B.K. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 2016, 7, 10541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Liu, W.; Chen, C. Can intercropping with the world’s three major beverage plants help improve the water use of rubber trees? J. Appl. Ecol. 2016, 53, 1787–1799. [Google Scholar] [CrossRef]
- Watanabe, K.; Yamamoto, T.; Yamada, T.; Sakuratani, T.; Nawata, E.; Noichana, C.; Sributta, A.; Higuchi, H. Changes in seasonal evapotranspiration, soil water content, and crop coefficients in sugarcane, cassava, and maize fields in Northeast Thailand. Agr. Water Manag. 2004, 67, 133–143. [Google Scholar] [CrossRef]
- Estrada-Bonilla, G.A.; Durrer, A.; Cardoso, E.J.B.N. Use of compost and phosphate-solubilizing bacteria affect sugarcane mineral nutrition, phosphorus availability, and the soil bacterial community. Appl. Soil Ecol. 2021, 157, 103760. [Google Scholar] [CrossRef]
- Cherubin, M.R.; Franco, A.L.C.; Guimarães, R.M.L.; Tormena, C.A.; Cerri, C.E.P.; Karlen, D.L.; Cerri, C.C. Assessing soil structural quality under Brazilian sugarcane expansion areas using Visual Evaluation of Soil Structure (VESS). Soil Tillage Res. 2017, 173, 64–74. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Liu, J.; Chen, H.; Zheng, L.; Wang, K. Soil microbial community responses to forage grass cultivation in degraded karst soils, Southwest China. Land Degrad. Dev. 2018, 29, 4262–4270. [Google Scholar] [CrossRef]
- Lewin, G.R.; Carlos, C.; Chevrette, M.G.; Horn, H.A.; McDonald, B.R.; Stankey, R.J.; Fox, B.G.; Currie, C.R. Evolution and Ecology of Actinobacteria and Their Bioenergy Applications. Annu. Rev. Microbiol. 2016, 70, 235–254. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, A.A.; Araujo, M.V.F.; Soares, M.D.S.; Oliveira, B.F.R.D.; Sibov, S.T.; Vieira, J.D.G. Isolation and Screening for Multi-trait Plant Growth Promoting Actinobacteria From Organic Sugarcane Rhizosphere. Int. J. Microbiol. Res. 2018, 10, 1193–1198. [Google Scholar] [CrossRef]
- Praeg, N.; Seeber, J.; Leitinger, G.; Tasser, E.; Newesely, C.; Tappeiner, U.; Illmer, P. The role of land management and elevation in shaping soil microbial communities: Insights from the Central European Alps. Soil Biol. Biochem. 2020, 150, 107951. [Google Scholar] [CrossRef]
- Ward, N.L.; Challacombe, J.F.; Janssen, P.H.; Henrissat, B.; Coutinho, P.M.; Wu, M.; Xie, G.; Haft, D.H.; Sait, M.; Badger, J.; et al. Three Genomes from the PhylumAcidobacteria Provide Insight into the Lifestyles of These Microorganisms in Soils. Appl. Environ. Microbiol. 2009, 75, 2046–2056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manpoong, C.; De Mandal, S.; Bangaruswamy, D.K.; Perumal, R.C.; Benny, J.; Beena, P.S.; Ghosh, A.; Kumar, N.S.; Tripathi, S.K. Linking rhizosphere soil biochemical and microbial community characteristics across different land use systems in mountainous region in Northeast India. Meta Gene 2020, 23, 100625. [Google Scholar] [CrossRef]
- Xun, W.; Huang, T.; Zhao, J.; Ran, W.; Wang, B.; Shen, Q.; Zhang, R. Environmental conditions rather than microbial inoculum composition determine the bacterial composition, microbial biomass and enzymatic activity of reconstructed soil microbial communities. Soil Biol. Biochem. 2015, 90, 10–18. [Google Scholar] [CrossRef]
- Knappová, J.; Pánková, H.; Münzbergová, Z. Roles of Arbuscular Mycorrhizal Fungi and Soil Abiotic Conditions in the Establishment of a Dry Grassland Community. PLoS ONE 2016, 11, e158925. [Google Scholar] [CrossRef]
- Pesaro, M.; Nicollier, G.; Zeyer, J.; Widmeret, F. Impact of soil drying-rewetting stress on microbial communities and activities and on degradation of two crop protection products. Appl. Environ. Microbiol. 2004, 70, 2577–2587. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Du, H.; Zeng, F.; Song, T.; Peng, W. Diminished rhizosphere and bulk soil microbial abundance and diversity across succession stages in Karst area, southwest China. Appl. Soil Ecol. 2021, 158, 103799. [Google Scholar] [CrossRef]
- Boer, W.D.; Folman, L.B.; Summerbell, R.C.; Boddy, L. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 2005, 29, 795–811. [Google Scholar] [CrossRef] [Green Version]
- Ewbank, G.; Edwards, D.; Abbott, G.D. Chemical characterization of Lower Devonian vascular plants. Org. Geochem. 1996, 25, 461–473. [Google Scholar] [CrossRef]
- Tardy, V.; Spor, A.; Mathieu, O.; Lévèque, J.; Terrat, S.; Plassart, P.; Regnier, T.; Bardgett, R.D.; van der Putten, W.H.; Roggero, P.P.; et al. Shifts in microbial diversity through land use intensity as drivers of carbon mineralization in soil. Soil Biol. Biochem. 2015, 90, 204–213. [Google Scholar] [CrossRef]
- Haghverdi, K.; Kooch, Y. Soil carbon and nitrogen fractions in response to land use/cover changes. Acta Oecol. 2020, 109, 103659. [Google Scholar] [CrossRef]
- Orwin, K.H.; Dickie, I.A.; Wood, J.R.; Bonner, K.I.; Holdaway, R.J. Soil microbial community structure explains the resistance of respiration to a dry–rewet cycle, but not soil functioning under static conditions. Funct. Ecol. 2015, 30, 1430–1439. [Google Scholar] [CrossRef] [Green Version]
- Preece, C.; Verbruggen, E.; Liu, L.; Weedon, J.T.; Peñuelas, J. Effects of past and current drought on the composition and diversity of soil microbial communities. Soil Biol. Biochem. 2019, 131, 28–39. [Google Scholar] [CrossRef]
- Wang, G.; Or, D. Hydration dynamics promote bacterial coexistence on rough surfaces. ISME J. 2013, 7, 395–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brady, N.C.; Weil, R.R. The Nature and Properties of Soils; Prentice Hall Press: London, UK, 2017; pp. 215–243. [Google Scholar]
- Pan, X.; Zhang, S.; Zhong, Q.; Gong, G.; Wang, G.; Guo, X.; Xu, X. Effects of soil chemical properties and fractions of Pb, Cd, and Zn on bacterial and fungal communities. Sci. Total Environ. 2020, 715, 136904. [Google Scholar] [CrossRef] [PubMed]
- Weed, S.B.; Davey, C.B.; Cook, M.G. Weathering of Mica by Fungi. Soil Sci. Soc. Am. J. 1969, 33, 702–706. [Google Scholar] [CrossRef]
- Miransari, M. Soil microbes and the availability of soil nutrients. Acta Physiol. Plant 2013, 35, 3075–3084. [Google Scholar] [CrossRef]
- Pereira, L.B.; Vicentini, R.; Ottoboni, L.M.M.; Moustafa, A. Changes in the bacterial community of soil from a neutral mine drainage channel. PLoS ONE 2014, 9, e96605. [Google Scholar] [CrossRef]

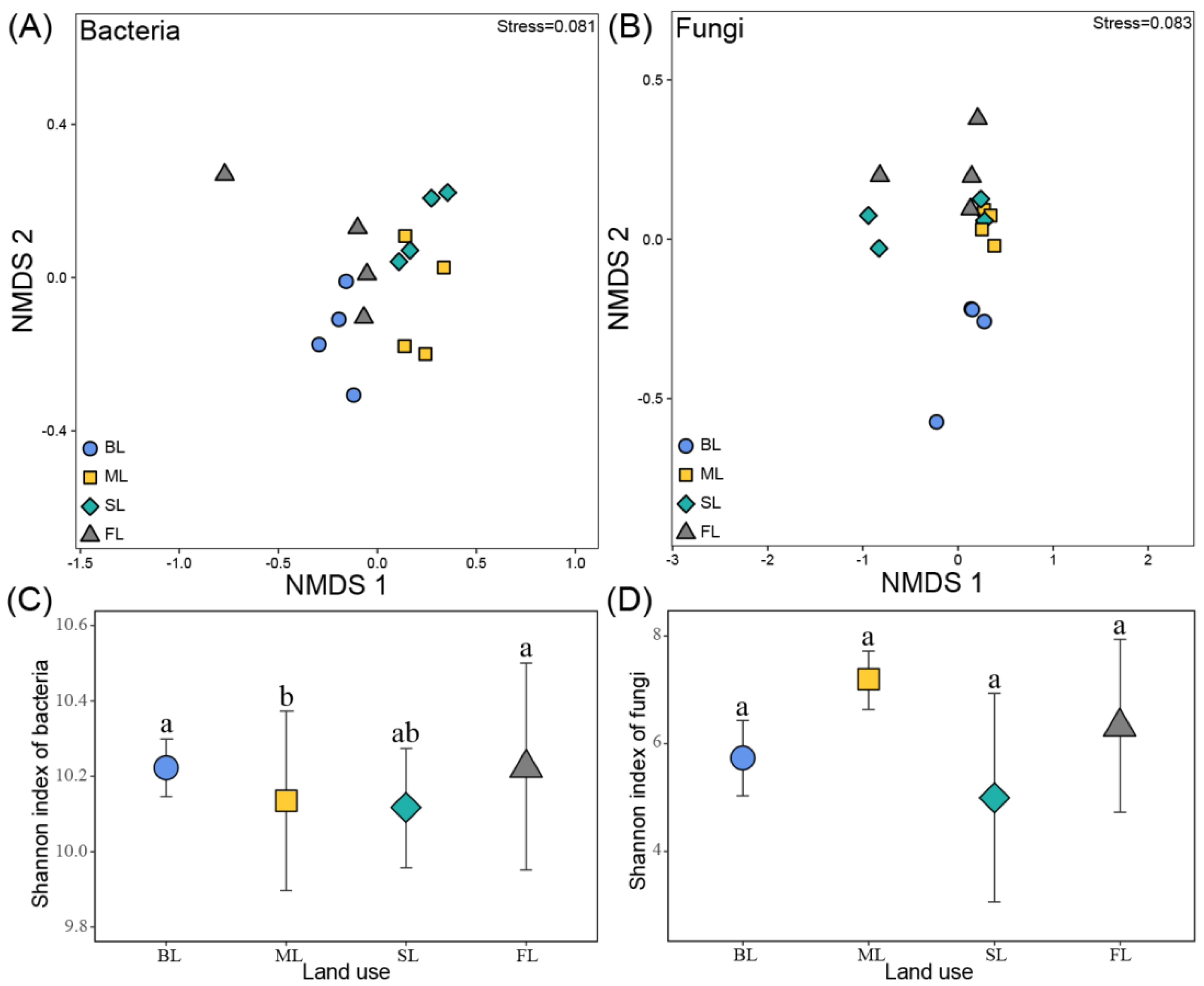
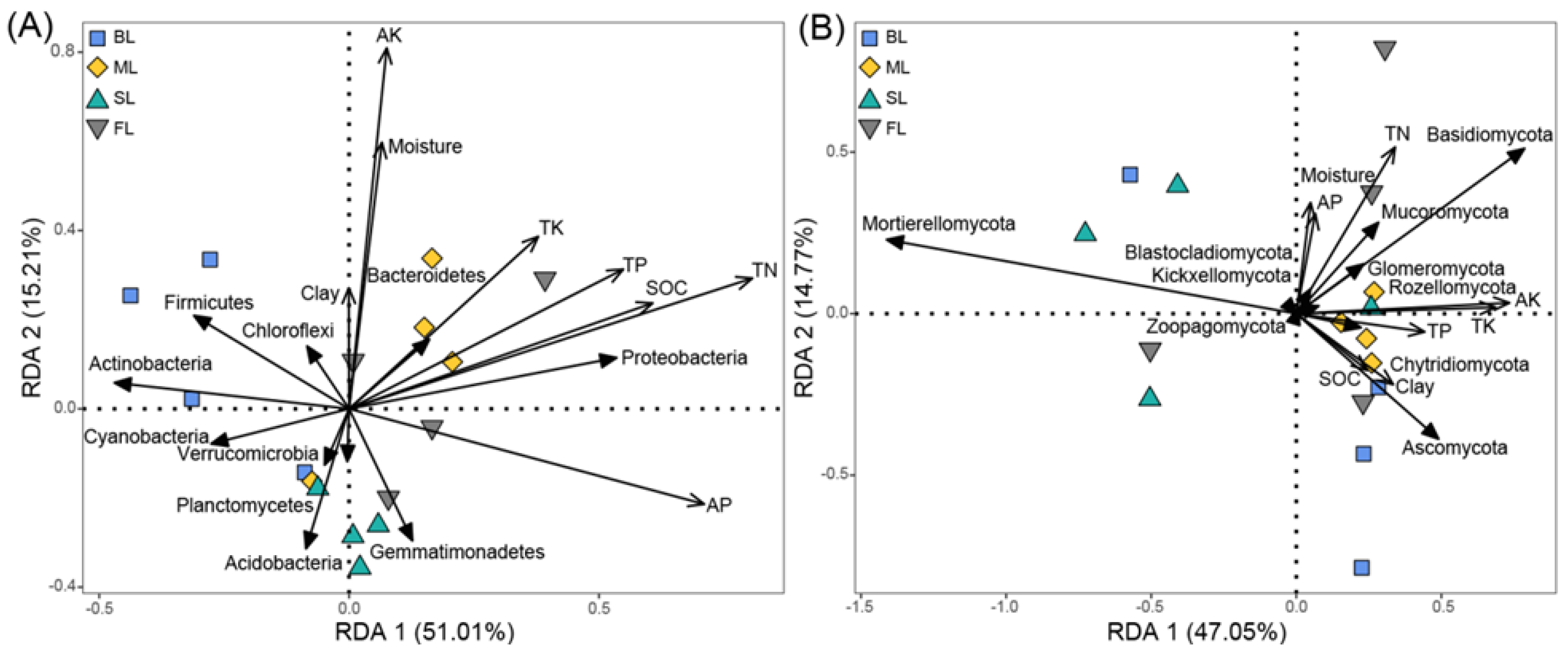
| Bacteria | Fungi | ||||
|---|---|---|---|---|---|
| Vs_Group | R | p | Vs_Group | R 1 | p |
| SL-ML | 0.17 | 0.17 | SL-ML | 0.36 | 0.03 |
| SL-BL | 0.86 | 0.04 | SL-BL | 0.39 | 0.18 |
| SL-FL | 0.49 | 0.03 | SL-FL | 0.15 | 0.15 |
| ML-BL | 0.85 | 0.02 | ML-BL | 0.53 | 0.03 |
| ML-FL | 0.42 | 0.03 | ML-FL | 0.40 | 0.03 |
| BL-FL | 0.47 | 0.02 | BL-FL | 0.40 | 0.06 |
| Bacteria | Fungi | ||||||
|---|---|---|---|---|---|---|---|
| Soil Property | Explains (%) | pseudo-F | p | Name | Explains (%) | pseudo-F | p |
| TN | 33.3 | 7.0 | 0.002 * | AK | 22.1 | 4.0 | 0.020 * |
| SOC | 15.1 | 3.8 | 0.012 * | SOC | 11.6 | 2.3 | 0.078 |
| AK | 10.2 | 3.0 | 0.030 * | TK | 6.6 | 1.4 | 0.244 |
| TK | 6.9 | 2.2 | 0.082 | Moisture | 5.8 | 1.2 | 0.364 |
| TP | 3.0 | 0.9 | 0.447 | AP | 5.7 | 1.0 | 0.400 |
| Moisture | 2.3 | 0.7 | 0.536 | Clay | 5.2 | 1.1 | 0.384 |
| Clay | 2.4 | 0.7 | 0.528 | TP | 4.8 | 0.9 | 0.458 |
| AP | 1.3 | 0.3 | 0.746 | TN | 4.4 | 0.9 | 0.430 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, T.; Chen, Z.; Rong, L.; Duan, X. Land-Use Driven Changes in Soil Microbial Community Composition and Soil Fertility in the Dry-Hot Valley Region of Southwestern China. Microorganisms 2022, 10, 956. https://doi.org/10.3390/microorganisms10050956
Liu T, Chen Z, Rong L, Duan X. Land-Use Driven Changes in Soil Microbial Community Composition and Soil Fertility in the Dry-Hot Valley Region of Southwestern China. Microorganisms. 2022; 10(5):956. https://doi.org/10.3390/microorganisms10050956
Chicago/Turabian StyleLiu, Taicong, Zhe Chen, Li Rong, and Xingwu Duan. 2022. "Land-Use Driven Changes in Soil Microbial Community Composition and Soil Fertility in the Dry-Hot Valley Region of Southwestern China" Microorganisms 10, no. 5: 956. https://doi.org/10.3390/microorganisms10050956






