Fungal Diversity in Two Wastewater Treatment Plants in North Italy
Abstract
:1. Introduction
2. Materials and Methods
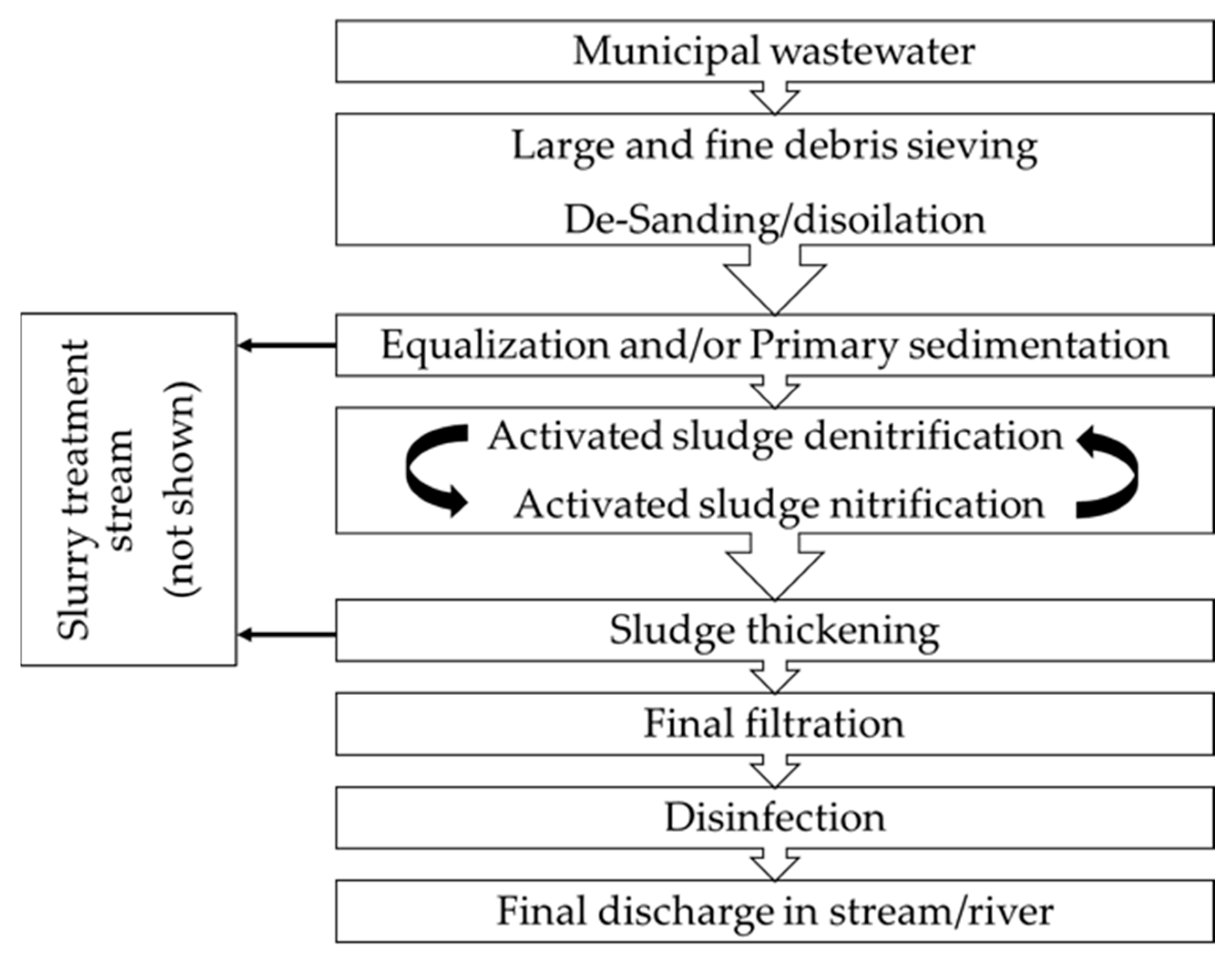
2.1. Structure of Wastewater Treatment Plants
- -
- Plant 1, managed by CAP Holding; this plant is located in the South-West sector of the Metropolitan City of Milan; it caters for a population equivalent of 320,000 people and treats an average wastewater volume of 100,000 m3 day−1;
- -
- Plant 2, managed by A2A Ciclo Idrico; this is located in Eastern Lombardia; it caters for a population equivalent of 296,000 people and treats an average wastewater volume of 70,000 m3 day−1.
2.2. Sampling and Isolation in Pure Culture
2.3. Molecular Identification of Selected Strains
2.4. Estimation of Ecological Parameters
3. Results and Discussion
3.1. Sampling, Isolation in Pure Culture and Identification
3.2. Diversity Patterns at Fungal Order Scale
3.3. Seasonal Variation
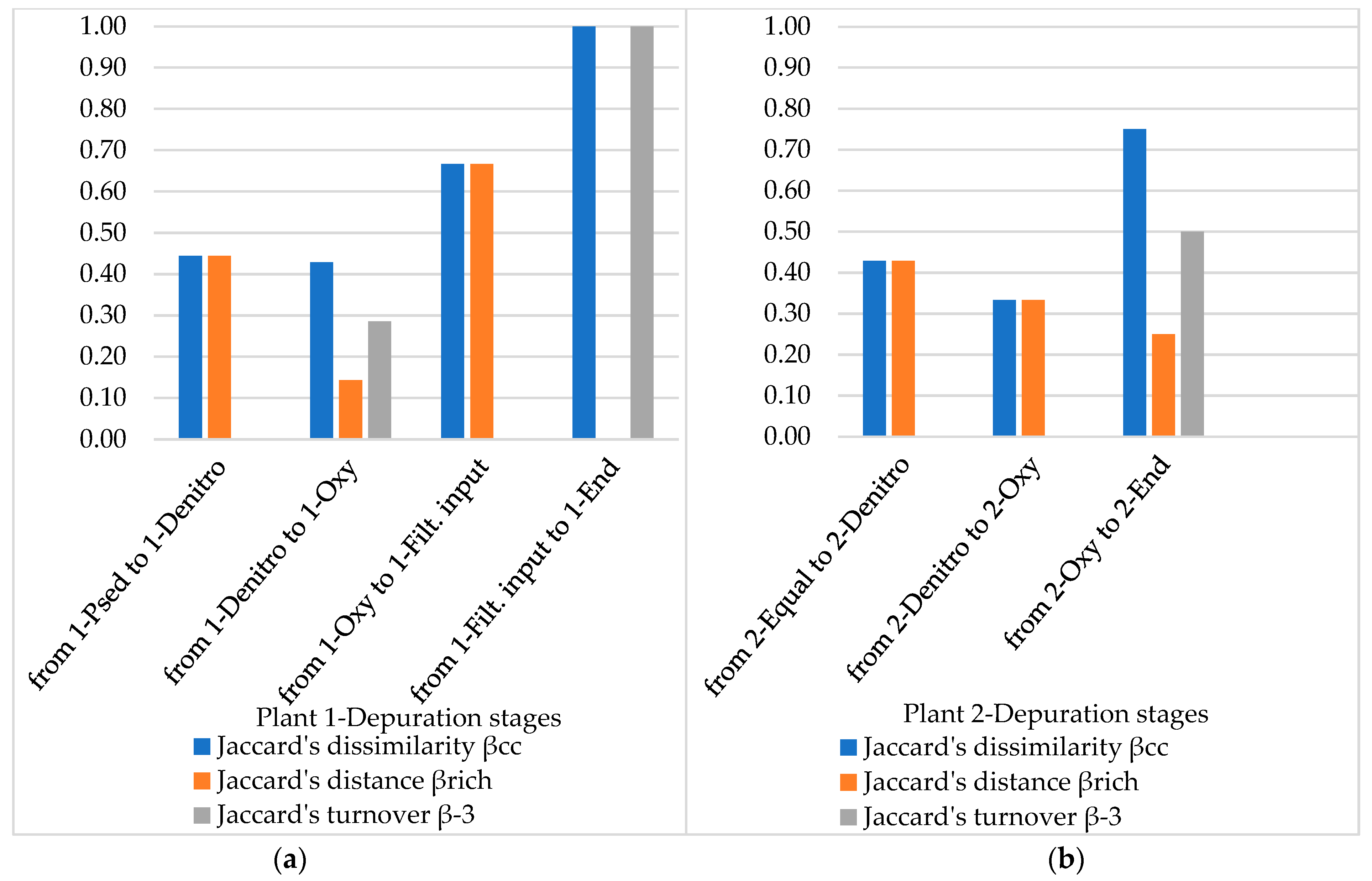
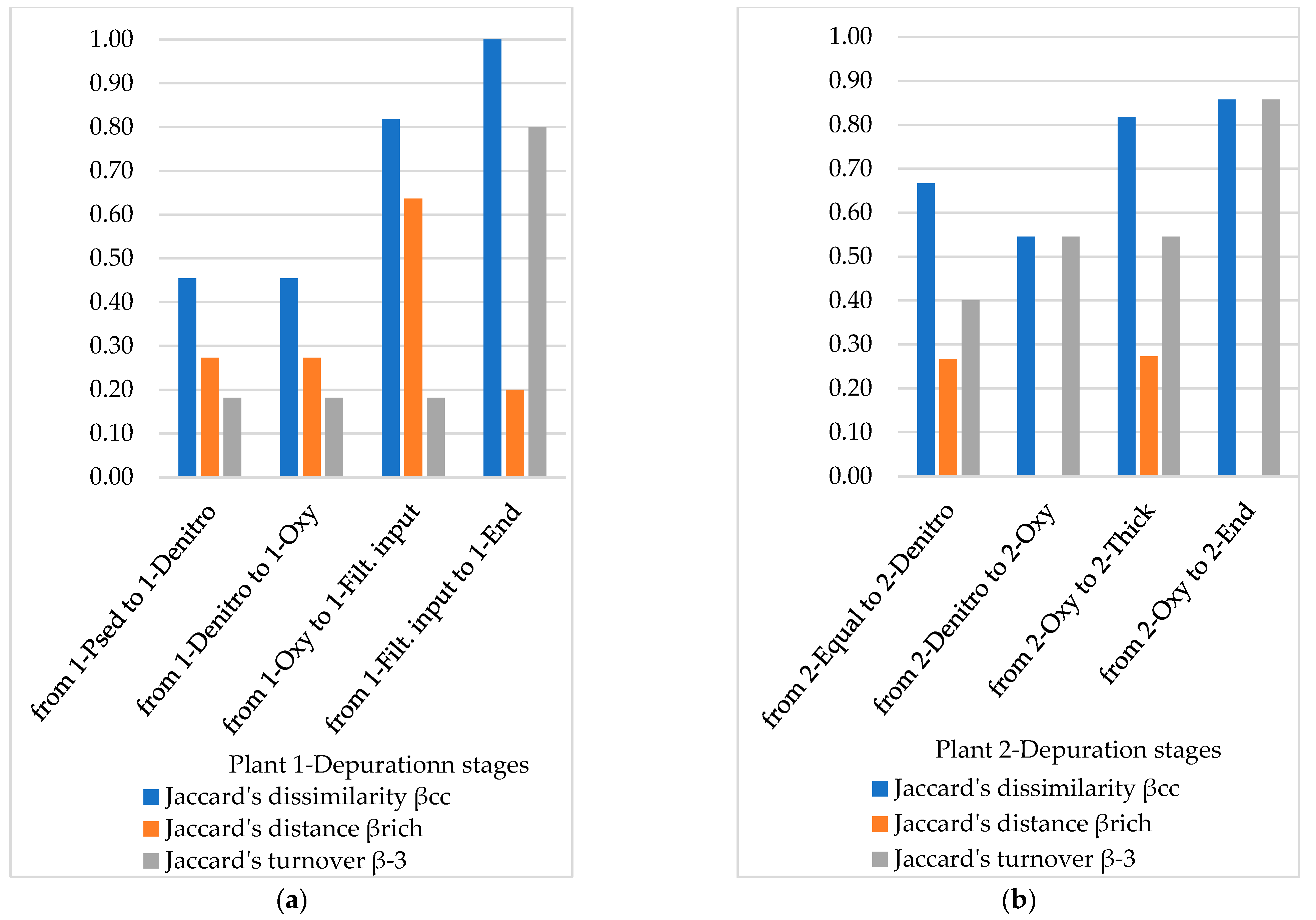
3.4. Diversity Indices
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alleman, J.E.; Prakasam, T.B.S. Reflections on seven decades of activated sludge history. J. Water Pollut. Control. Fed. 1983, 55, 436–443. [Google Scholar]
- Decreto Legislativo 3 Aprile 2006, n. 152 Norme in Materia Ambientale. Gazzetta Ufficiale della Repubblica Italiana n.88 del 14-4-2006—Suppl. Ordinario n. 96. Available online: https://www.gazzettaufficiale.it/dettaglio/codici/materiaAmbientale (accessed on 28 April 2022).
- Istituto Superiore per la Protezione e la Ricerca Ambientale. Available online: https://www.isprambiente.gov.it/ (accessed on 9 December 2021).
- Behnamia, A.; Benisb, K.Z.; Shakerkhatibic, M.; Derafshid, S.; Sabere, A.B.; Akbarif, N.A.R.; Yousefia, R. Comparative study on fungal communities of full scale municipal and industrial wastewater treatment plants. Desalination Water Treat. 2018, 131, 123–131. [Google Scholar] [CrossRef]
- Niu, L.; Li, Y.; Xu, L.; Wang, P.; Zhang, W.; Wang, C.; Cai, W.; Wang, L. Ignored fungal community in activated sludge wastewater treatment plants: Diversity and altitudinal characteristics. Environ. Sci. Pollut. Res. 2017, 24, 4185–4193. [Google Scholar] [CrossRef]
- ANPA—Dipartimento Prevenzione e Risanamento Ambientali. Guida alla Progettazione dei Sistemi di Collettamento e Depurazione delle Acque Reflue Urbane; ANPA e Ministero Ambiente: Roma, Italy, 2001; pp. 1–142.
- Molina-Muñoz, M.; Poyatos, J.; Sánchez-Peinado, M.; Hontoria, E.; González-López, J.; Rodelas, B. Microbial community structure and dynamics in a pilot-scale submerged membrane bioreactor aerobically treating domestic wastewater under real operation conditions. Sci. Total Environ. 2009, 407, 3994–4003. [Google Scholar] [CrossRef]
- Wan, C.-Y.; De Wever, H.; Diels, L.; Thoeye, C.; Liang, J.-B.; Huang, L.-N. Biodiversity and population dynamics of microorganisms in a full-scale membrane bioreactor for municipal wastewater treatment. Water Res. 2011, 45, 1129–1138. [Google Scholar] [CrossRef]
- Matsunaga, K.; Kubota, K.; Harada, H. Molecular Diversity of Eukaryotes in Municipal Wastewater Treatment Processes as Revealed by 18S rRNA Gene Analysis. Microbes Environ. 2014, 29, 401–407. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Wang, L.; Xiang, F.; Zhao, L.; Qiao, Z. Activated Sludge Microbial Community and Treatment Performance of Wastewater Treatment Plants in Industrial and Municipal Zones. Int. J. Environ. Res. Public Health 2020, 17, 436. [Google Scholar] [CrossRef] [Green Version]
- Urban Waste Water Treatment Directive. Directive 91/271/EEC of 21 May 1991 concerning urban waste-water treatment. J. Eur. Commun. 1991, 34, 40. [Google Scholar]
- Philippot, L.; Hallin, S. Finding the missing link between diversity and activity using denitrifying bacteria as a model functional community. Curr. Opin. Microbiol. 2005, 8, 234–239. [Google Scholar] [CrossRef]
- Ji, B.; Yang, K.; Zhu, L.; Jiang, Y.; Wang, H.; Zhou, J.; Zhang, H. Aerobic denitrification: A review of important advances of the last 30 years. Biotechnol. Bioprocess Eng. 2015, 20, 643–651. [Google Scholar] [CrossRef]
- Mohsenpour, S.F.; Hennige, S.; Willoughby, N.; Adeloye, A.; Gutierrez, T. Integrating micro-algae into wastewater treatment: A review. Sci. Total Environ. 2021, 752, 142168. [Google Scholar] [CrossRef]
- Hyde, K.D.; Xu, J.; Rapior, S.; Jeewon, R.; Lumyong, S.; Niego, A.G.T.; Abeywickrama, P.D.; Aluthmuhandiram, J.V.S.; Brahamanage, R.S.; Brooks, S.; et al. The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers. 2019, 97, 1–136. [Google Scholar] [CrossRef] [Green Version]
- Selbmann, L.; Egidi, E.; Isola, D.; Onofri, S.; Zucconi, L.; de Hoog, G.S.; Chinaglia, S.; Testa, L.; Tosi, S.; Balestrazzi, A.; et al. Biodiversity, evolution and adaptation of fungi in extreme environments. Plant Biosyst. Int. J. Deal. All Asp. Plant Biol. 2013, 147, 237–246. [Google Scholar] [CrossRef] [Green Version]
- Hillmann, F.; Shekhova, E.; Kniemeyer, O. Insights into the cellular responses to hypoxia in filamentous fungi. Curr. Genet. 2015, 61, 441–455. [Google Scholar] [CrossRef]
- Harms, H.; Schlosser, D.; Wick, L.Y. Untapped potential: Exploiting fungi in bioremediation of hazardous chemicals. Nat. Rev. Microbiol. 2011, 9, 177–192. [Google Scholar] [CrossRef]
- Asif, M.B.; Hai, F.I.; Singh, L.; Price, W.E.; Nghiem, L.D. Degradation of Pharmaceuticals and Personal Care Products by White-Rot Fungi—A Critical Review. Curr. Pollut. Rep. 2017, 3, 88–103. [Google Scholar] [CrossRef] [Green Version]
- Zahmatkesh, M.; Spanjers, H.; van Lier, J.B. A novel approach for application of white rot fungi in wastewater treatment under non-sterile conditions: Immobilization of fungi on sorghum. Environ. Technol. 2018, 39, 2030–2040. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, C. Fungal potential for the degradation of petroleum-based polymers: An overview of macro- and microplastics biodegradation. Biotechnol. Adv. 2020, 40, 107501. [Google Scholar] [CrossRef]
- Singh, R.K.; Tripathi, R.; Ranjan, A.; Srivastava, A.K. Fungi as potential candidates for bioremediation. In Abatement of Environmental Pollutants; Elsevier: Amsterdam, The Netherlands, 2019; pp. 177–191. [Google Scholar]
- Benguenab, A.; Chibani, A. Biodegradation of petroleum hydrocarbons by filamentous fungi (Aspergillus ustus and Purpureocillium lilacinum) isolated from used engine oil contaminated soil. Acta Ecol. Sin. 2021, 41, 416–423. [Google Scholar] [CrossRef]
- Gadd, G.M. Interactions of Fungi with Toxic Metals; Springer: Boston, MA, USA, 1994; pp. 361–374. [Google Scholar] [CrossRef]
- Abbas, S.H.; Ismail, I.M.; Mostafa, T.M.; Sulaymon, A.H. Biosorption of heavy metals: A review. J. Chem. Sci. Technol. 2014, 3, 74–102. [Google Scholar]
- Cecchi, G.; Marescotti, P.; Di Piazza, S.; Zotti, M. Native fungi as metal remediators: Silver myco-accumulation from metal contaminated waste-rock dumps (Libiola Mine, Italy). J. Environ. Sci. Health Part B 2017, 52, 191–195. [Google Scholar] [CrossRef]
- Cecchi, G.; Vagge, G.; Cutroneo, L.; Greco, G.; Di Piazza, S.; Faga, M.; Zotti, M.; Capello, M. Fungi as potential tool for polluted port sediment remediation. Environ. Sci. Pollut. Res. 2019, 26, 35602–35609. [Google Scholar] [CrossRef]
- More, T.; Yan, S.; Tyagi, R.; Surampalli, R. Potential use of filamentous fungi for wastewater sludge treatment. Bioresour. Technol. 2010, 101, 7691–7700. [Google Scholar] [CrossRef]
- Kacprzak, M.; Neczaj, E.; Okoniewska, E. The comparative mycological analysis of wastewater and sewage sludges from selected wastewater treatment plants. Desalination 2005, 185, 363–370. [Google Scholar] [CrossRef]
- Awad, M.F.; Kraume, M. Fungal diversity in activated sludge from membrane bioreactors in Berlin. Can. J. Microbiol. 2011, 57, 693–698. [Google Scholar] [CrossRef]
- Zhang, H.; Feng, J.; Chen, S.; Li, B.; Sekar, R.; Zhao, Z.; Jia, J.; Wang, Y.; Kang, P. Disentangling the Drivers of Diversity and Distribution of Fungal Community Composition in Wastewater Treatment Plants Across Spatial Scales. Front. Microbiol. 2018, 9, 1291. [Google Scholar] [CrossRef]
- Wei, Z.; Liu, Y.; Feng, K.; Li, S.; Wang, S.; Jin, D.; Zhang, Y.; Chen, H.; Yin, H.; Xu, M.; et al. The divergence between fungal and bacterial communities in seasonal and spatial variations of wastewater treatment plants. Sci. Total Environ. 2018, 628, 969–978. [Google Scholar] [CrossRef]
- Zhang, S.; Fan, F.; Meng, F. Seasonality and Community Separation of Fungi in a Municipal Wastewater Treatment Plant. Appl. Environ. Microbiol. 2020, 86, e00991-20. [Google Scholar] [CrossRef]
- Istituto Regionale di Ricerca della Lombardia (IRER). Depurazione delle Acque Reflue Urbane: Tecnologie Innovative Idonee Acontesti Molto Urbanizzati. Rapporto Finale; Istituto Regionale di Ricerca della Lombardia (IRER): Milano, Italy, 2010; pp. 1–342. [Google Scholar]
- Lacap, D.C.; Hyde, K.D.; Liew, E.C.Y. An evaluation of the fungal ‘morphotype’ concept based on ribosomal DNA sequences. Fungal Divers. 2003, 12, 53–66. [Google Scholar]
- Sutherland, W.J. Ecological Census Techniques: A Handbook; Cambridge University Press: Cambridge, UK, 2006. [Google Scholar]
- Nilsson, R.H.; Ryberg, M.; Abarenkov, K.; Sjã¶kvist, E.; Kristiansson, E. The ITS region as a target for characterization of fungal communities using emerging sequencing technologies. FEMS Microbiol. Lett. 2009, 296, 97–101. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Barcoding Consortium; Fungal Barcoding Consortium Author List; Bolchacova, E.; et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [Green Version]
- Lücking, R.; Aime, M.C.; Robbertse, B.; Miller, A.N.; Ariyawansa, H.A.; Aoki, T.; Cardinali, G.; Crous, P.W.; Druzhinina, I.S.; Geiser, D.M.; et al. Unambiguous identification of fungi: Where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus 2020, 11, 14. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Girometta, C.E.; Bernicchia, A.; Baiguera, R.M.; Bracco, F.; Buratti, S.; Cartabia, M.; Picco, A.M.; Savino, E. An Italian Research Culture Collection of Wood Decay Fungi. Diversity 2020, 12, 58. [Google Scholar] [CrossRef] [Green Version]
- McGeoch, M.A.; Latombe, G.; Andrew, N.R.; Nakagawa, S.; Nipperess, D.A.; Roigé, M.; Marzinelli, E.M.; Campbell, A.H.; Vergés, A.; Thomas, T.; et al. Measuring continuous compositional change using decline and decay in zeta diversity. Ecology 2019, 100, e02832. [Google Scholar] [CrossRef]
- Mouillot, D.; Lepretre, A. A comparison of species diversity estimators. Res. Popul. Ecol. 1999, 41, 203–215. [Google Scholar]
- Bullini, L.; Pignatti, S.; De Santo, A.V. Ecologia Generale; Utet: Torino, Italy, 1998; pp. 234–237.
- Baselga, A. Partitioning the turnover and nestedness components of beta diversity. Glob. Ecol. Biogeogr. 2010, 19, 134–143. [Google Scholar] [CrossRef]
- Baselga, A.; Leprieur, F. Comparing methods to separate components of beta diversity. Methods Ecol. Evol. 2015, 6, 1069–1079. [Google Scholar] [CrossRef]
- Hui, C.; McGeoch, M.A. Zeta Diversity as a Concept and Metric That Unifies Incidence-Based Biodiversity Patterns. Am. Nat. 2014, 184, 684–694. [Google Scholar] [CrossRef] [Green Version]
- Mycobank. Available online: www.mycobank.org (accessed on 9 December 2021).
- Domsch, K.H.; Gams, W.; Anderson, T.H. Compendium of Soil Fungi; Academic Press: London, UK, 1980; Volume 1. [Google Scholar]
- Ziaee, A.; Zia, M.; Goli, M. Identification of saprophytic and allergenic fungi in indoor and outdoor environments. Environ. Monit. Assess. 2018, 190, 574. [Google Scholar] [CrossRef]
- Liu, X.Z.; Wang, G.M.; Göker, M.; Groenewald, M.; Kachalkin, A.V.; Lumbsch, H.T.; Millanes, A.M.; Wedin, M.; Yurkov, A.M.; Bai, F.Y.; et al. Towards an integrated phylogenetic classification of the Tremellomycetes. Stud. Mycol. 2015, 81, 85–147. [Google Scholar] [CrossRef] [Green Version]
- de Hoog, G.S.; Guarro, J.; Gené, J.; Figueras, M.J. Atlas of Clinical Fungi; Centraalbureau voor Schimmelcultures: Utrecht, The Netherlands, 2005. [Google Scholar]
- Montoya Mendoza, A.M.; González González, G.M. Trichosporon spp.: An emerging fungal pathogen. Med. Univ. 2014, 16, 37–43. [Google Scholar]
- Singh, A.; Shukla, N.; Kabadwal, B.; Tewari, A.; Kumar, J. Review on Plant-Trichoderma-Pathogen Interaction. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 2382–2397. [Google Scholar] [CrossRef]
- Kubicek, C.P.; Steindorff, A.S.; Chenthamara, K.; Manganiello, G.; Henrissat, B.; Zhang, J.; Cai, F.; Kopchinskiy, A.G.; Kubicek, E.M.; Kuo, A.; et al. Evolution and comparative genomics of the most common Trichoderma species. BMC Genom. 2019, 20, 485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerin, D.; Pollastro, S.; Raguseo, C.; De Miccolis Angelini, R.M.; Faretra, F. A ready-to-use single-and duplex-TaqMan-qPCR assay to detect and quantify the biocontrol agents Trichoderma asperellum and Trichoderma gamsii. Front. Microbiol. 2018, 9, 2073. [Google Scholar] [CrossRef] [PubMed]
- Win, T.T.; Bo, B.; Malec, P.; Khan, S.; Fu, P. Newly isolated strain of Trichoderma asperellum from disease suppressive soil is a potential bio-control agent to suppress Fusarium soil borne fungal phytopathogens. J. Plant Pathol. 2021, 103, 549–561. [Google Scholar] [CrossRef]
- Gräfenhan, T.; Schroers, H.-J.; Nirenberg, H.; Seifert, K. An overview of the taxonomy, phylogeny, and typification of nectriaceous fungi in Cosmospora, Acremonium, Fusarium, Stilbella, and Volutella. Stud. Mycol. 2011, 68, 79–113. [Google Scholar] [CrossRef] [Green Version]
- Seifert, K.A.; Gams, W. The Genera of Hyphomycetes—2011 update. Pers. Mol. Phylogeny Evol. Fungi 2011, 27, 119. [Google Scholar] [CrossRef] [Green Version]
- Dragičević, T.; Hren, M.; Gmajnić, M.; Pelko, S.; Kungulovski, D.; Kungulovski, I.; Čvek, D.; Frece, J.; Markov, K.; Delaš, F. Biodegradation of Olive Mill Wastewater by Trichosporon Cutaneum and Geotrichum Candidum. Arch. Ind. Hyg. Toxicol. 2010, 61, 399–405. [Google Scholar] [CrossRef]
- Kurtzman, C.; Fell, J.W.; Boekhout, T. The Yeasts: A Taxonomic Study; Elsevier: London, UK, 2011. [Google Scholar]
- Samuels, G.J.; Hebbar, P.K. Developing Trichoderma-based products for application in agriculture. In Trichoderma Identification and Agricultural Applications; The American Phytopatholog: St. Paul, MN, USA, 2015; pp. 7–34. [Google Scholar]
- ARPA-Lombardia. Available online: www.arpalombardia.it (accessed on 14 May 2022).
- Simpson, E.H. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Pielou, E.C. The measurement of diversity in different types of biological collections. J. Theor. Biol. 1966, 13, 131–144. [Google Scholar] [CrossRef]
- Hasan, M.T.; Sneddon, G.; Ma, R. Modeling binomial amphibian roadkill data in distance sampling while accounting for zero-inflation, serial correlation and varying cluster sizes simultaneously. Environ. Ecol. Stat. 2017, 24, 201–217. [Google Scholar] [CrossRef]
- Bonadonna, L.; Musmeci, L. Metodi Analitici di Riferimento per la Valutazione Microbiologica dei Fanghi di Depurazione e di Matrici ad Essi Assimilabili; Rapporti ISTISAN 14/18; Istituto Superiore di Sanità: Roma, Italy, 2014.
- Rousk, J.; Bååth, E. Growth of saprotrophic fungi and bacteria in soil. FEMS Microbiol. Ecol. 2011, 78, 17–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konieczny, K. Effectiveness of wastewater treatment with the use of the biological membrane reactors. Annu. Set—Environ. Prot. 2015, 17, 1034. [Google Scholar]




| Scheme | Plant | Code in This Study |
|---|---|---|
| Primary sedimentation of input wastewater | 1 | 1-PSed |
| Activated sludge–denitrification | 1 | 1-Denitro |
| Activated sludge–oxidation | 1 | 1-Oxy |
| Filtration input–discharge post-secondary sedimentation | 1 | 1-Filt. input |
| Post ozonation | 1 | 1-End |
| Equalization of input wastewater | 2 | 2-Equal |
| Activated sludge–denitrification | 2 | 2-Denitro |
| Activated sludge–oxidation | 2 | 2-Oxy |
| Filtration input | 2 | 2-End |
| Stage | Bulk | 1:10 | 1:100 | 1:1000 | 1:10,000 |
|---|---|---|---|---|---|
| 1-PSed | x | x | x | x | |
| 1-Denitro | x | x | |||
| 1-Oxy | x | x | x | ||
| 1-Filt. input | x | x | x | ||
| 1-End | x | x | x | ||
| 2-Equal | x | x | x | x | |
| 2-Denitro | x | x | |||
| 2-Oxy | x | x | x | ||
| 2-End | x | x | x |
| Fungal Taxa | Author | Depuration Stages of Provenance |
|---|---|---|
| Acremonium spp. | Link | 1-Psed, 1-Denitro, 1-End; 2-Equal, 2-Denitro, 2-Oxy |
| Alternaria spp. | Nees | 1-Denitro, 2- Equal |
| Apiotrichum domesticum | (Sugita, A. Nishikawa & Shinoda) Yurkov & Boekhout | 2-Oxy |
| Apiotrichum montevideense | (L.A. Queiroz) Yurkov & Boekhout | 2-Equal |
| Apiotrichum laibachii | (Windisch) Yurkov & Boekhout | 1-Psed, 1-Denitro, 1-Oxy |
| Aspergillus flavus | Link | 1-Psed, 1-Denitro, 1-Oxy, 1-Filt. Input; 2-Equal, 2-Denitro, 2-Oxy |
| Aspergillus fumigatus | Fresen. | 1-Psed, 1-Denitro, 1-Oxy, 1-Filt. Input, 1-End; 2-Equal, 2-Denitro, 2-Oxy, 2-End |
| Aspergillus niger | Tiegh. | 1-Psed, 1-Oxy, 1-Filt. Input, 1-End; 2-Equal, 2-Denitro, 2-Oxy |
| Aspergillus tubingensis | Mosseray | 1-Oxy |
| Aspergillus spp. | P. Micheli ex Haller | 1-Psed, 1-Denitro, 1-Oxy, 1-Filt. Input; 2-Oxy, 2-End |
| Candida pseudolambica | M.T. Sm. & Poot | 1-Denitro |
| Cladosporium spp. | Link | 1-Psed, 1-Denitro, 1-Oxy, 1-Filt. Input, 1-End; 2-Equal, 2-Denitro, 2-Oxy, 2-End |
| Chaetomium sp. | Kunze | 2-Oxy |
| Chrysosporium tropicum | J.W. Carmich. | 1-Denitro, 1-Oxy, 1-Filt. Input |
| Cosmospora butyri | (J.F.H. Beyma) Gräfenhan | 2-Equal |
| Cutaneotrichosporon cutaneum | (Beurm., Gougerot & Vaucher bis) Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout | 1-Psed, 1-Denitro, 1-Oxy, 1-Filt. Input; 2-Equal, 2-Oxy |
| Cutaneotrichosporon jirovecii | (Frágner) Xin Zhan Liu, F.Y. Bai | 1-Oxy; 2-Equal, 2-Oxy |
| Cutaneotrichosporon mucoides | (E. Guého & M.T. Sm.) Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout | 1-Psed, 1-Denitro, 1-Oxy |
| Debaryomyces hansenii | (Zopf) Lodder & Kreger | 1-Psed |
| Dipodascus fermentans | (Diddens & Lodder) P.M. Kirk | 1-Denitro |
| Diutina neorugosa | (Paredes, Deanna A. Sutton, Cano & Guarro) Khunnamw., Jindam., Limtong & Lachance | 1-Psed |
| Engyodontium sp. | de Hoog | 2-Equal |
| Exophiala lecanii-corni | (Benedek & G. Specht) Haase & de Hoog | 2-Equal, 2-End |
| Fusarium fujikuroi | Nirenberg | 1-Oxy |
| Fusarium oxysporum | Schltdl. | 2-Denitro |
| Fusarium spp. | Link | 1-Psed, 1-Denitro, 1-Oxy, 1-Filt. Input; 2-Equal, 2-Denitro, 2-Oxy, 2-End |
| Fusicladium sp. | Bonord. | 2-Equal |
| Geotrichum candidum | Link | 1-Psed, 1-Denitro, 1-Oxy, 1-Filt. Input, 1-End; 2-Equal, 2-Denitro, 2-Oxy, 2-End |
| Geotrichum fragrans | Morenz | 2-Oxy |
| Geotrichum spp. | Link | 1-Psed, 1-Denitro, 1-Oxy 2-Equal, 2-Denitro, 2-Oxy, 2-End |
| Graphium sp. | Corda | 1-Psed |
| Mucor spp. | Fresen. | 1-Psed, 1-Denitro, 1-Oxy; 2-Equal, 2-Denitro, 2-Oxy, 2-End |
| Oxyporus latemarginatus | (Durieu & Mont.) Donk | 1-Oxy |
| Phialophora spp. | Medlar | 1-Psed, 1-Denitro, 1-Oxy, 1-Filt. Input |
| Phycomyces sp. | Kunze | 1-Psed, 1-Denitro, 1-Oxy, 1-Filt. Input |
| Phoma spp. | Sacc. | 1-Denitro, 1-Filt. Input; 2-Equal |
| Penicillium albocoremium | (Frisvad) Frisvad | 1-Oxy |
| Penicillium crustosum | Thom | 2-Equal |
| Penicillium expansum | Link | 2-Oxy |
| Penicillium griseofulvum | Dierckx | 1-Oxy, 1-Equal |
| Penicillium olsonii | Bainier & Sartory | 1-Psed |
| Penicillium verrucosum | Dierckx | 1-Oxy |
| Penicillium spp. | Link | 1-Psed, 1-Denitro, 1-Oxy, 1-Filt. Input, 1-End; 2-Equal, 2-Denitro, 2-Oxy |
| Purpureocillium lilacinum | (Thom) Luangsa-ard, Houbraken, Hywel-Jones & Samson | 1-Oxy; 2- Equal, 2-Denitro, 2-Oxy, 2-End |
| Rhizopus oryzae | Went & Prins. Geerl. | 1-Psed, 1-Denitro, 1-Oxy; 2-Equal, 2-Denitro, 2-Oxy |
| Rhodotorula glutinis | (Fresen.) F.C. Harrison | 1-Psed, 1-Denitro, 1-Oxy, 1-End; 2-Equal, 2-End |
| Rhodotorula mucilaginosa | (A. Jörg.) F.C. Harrison | 1-Oxy |
| Sampaiozyma ingeniosa | (Di Menna) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout | 1-Psed |
| Scedosporium dehoogi | Gilgado, Cano, Gené & Guarro | 2-Oxy |
| Scheffersomyces spartinae | (Ahearn, Yarrow & Meyers) Kurtzman & M. Suzuki | 1-Denitro |
| Scopulariopsis brevicaulis | (Sacc.) Bainier | 1-Psed, 1-Denitro; 2-Equal, 2-Denitro, 2-Oxy, 2-End |
| Sporobolomyces spp. | Kluyver & C.B. Niel | 2-Equal 2-Denitro |
| Talaromyces flavus | (Klöcker) Stolk & Samson | 2-Equal |
| Talaromyces spp. | C.R. Benj. | 2-Equal |
| Trichoderma asperellum | Samuels, Lieckf. & Nirenberg | 1-Psed, 1-Denitro, 1-Oxy; 2-Equal, 2-Oxy |
| Trichoderma citrinoviride | Bissett | 2-Oxy |
| Trichoderma harzianum | Rifai | 1-Psed |
| Trichoderma saturnisporum | Hammill | 2-Denitro |
| Trichoderma virens | (J.H. Mill., Giddens & A.A. Foster) Arx | 1-Denitro |
| Trichoderma spp. | Pers. | 1-Psed, 1-Denitro, 1-Oxy, 1-End; 2-Equal, 2-Denitro, 2-Oxy, 2-End |
| Trichosporon asahii | Akagi ex Sugita, A. Nishikawa & Shinoda | 1-Psed, 1-Denitro, 1-Oxy, 1-Filt. Input |
| Verticillium sp. | Nees | 2-Oxy |
| Yarrowia lipolytica | (Wick., Kurtzman & Herman) Van der Walt & Arx | 2-Denitro |
| Zygoascus polysorbophila | (Kurtzman) Nagats., Kiyuna & Sugiy. | 2-Denitro |
| Other yeasts | 1-Psed, 1-Denitro, 1-Oxy, 1-Filt. Input, 1-End; 2-Equal, 2-Denitro, 2-End | |
| Sporigenous fungi | 1-Psed, 1-Denitro, 1-Oxy, 1-Filt. Input | |
| Mycelia sterilia | 1-Psed, 1-Oxy, 1-Filt. Input; 2-Denitro |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buratti, S.; Girometta, C.E.; Baiguera, R.M.; Barucco, B.; Bernardi, M.; De Girolamo, G.; Malgaretti, M.; Oliva, D.; Picco, A.M.; Savino, E. Fungal Diversity in Two Wastewater Treatment Plants in North Italy. Microorganisms 2022, 10, 1096. https://doi.org/10.3390/microorganisms10061096
Buratti S, Girometta CE, Baiguera RM, Barucco B, Bernardi M, De Girolamo G, Malgaretti M, Oliva D, Picco AM, Savino E. Fungal Diversity in Two Wastewater Treatment Plants in North Italy. Microorganisms. 2022; 10(6):1096. https://doi.org/10.3390/microorganisms10061096
Chicago/Turabian StyleBuratti, Simone, Carolina Elena Girometta, Rebecca Michela Baiguera, Barbara Barucco, Marco Bernardi, Giuseppe De Girolamo, Maura Malgaretti, Desdemona Oliva, Anna Maria Picco, and Elena Savino. 2022. "Fungal Diversity in Two Wastewater Treatment Plants in North Italy" Microorganisms 10, no. 6: 1096. https://doi.org/10.3390/microorganisms10061096






