A Strategy for Gene Knockdown in Dinoflagellates
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culturing
2.2. Morpholino Customization and Delivery
2.3. Cell Counts and Fluorescence Quantification
2.4. Cell Imaging
2.5. Quantification of Protein Expression
2.6. Statistical Considerations
3. Results
3.1. Cell Viability
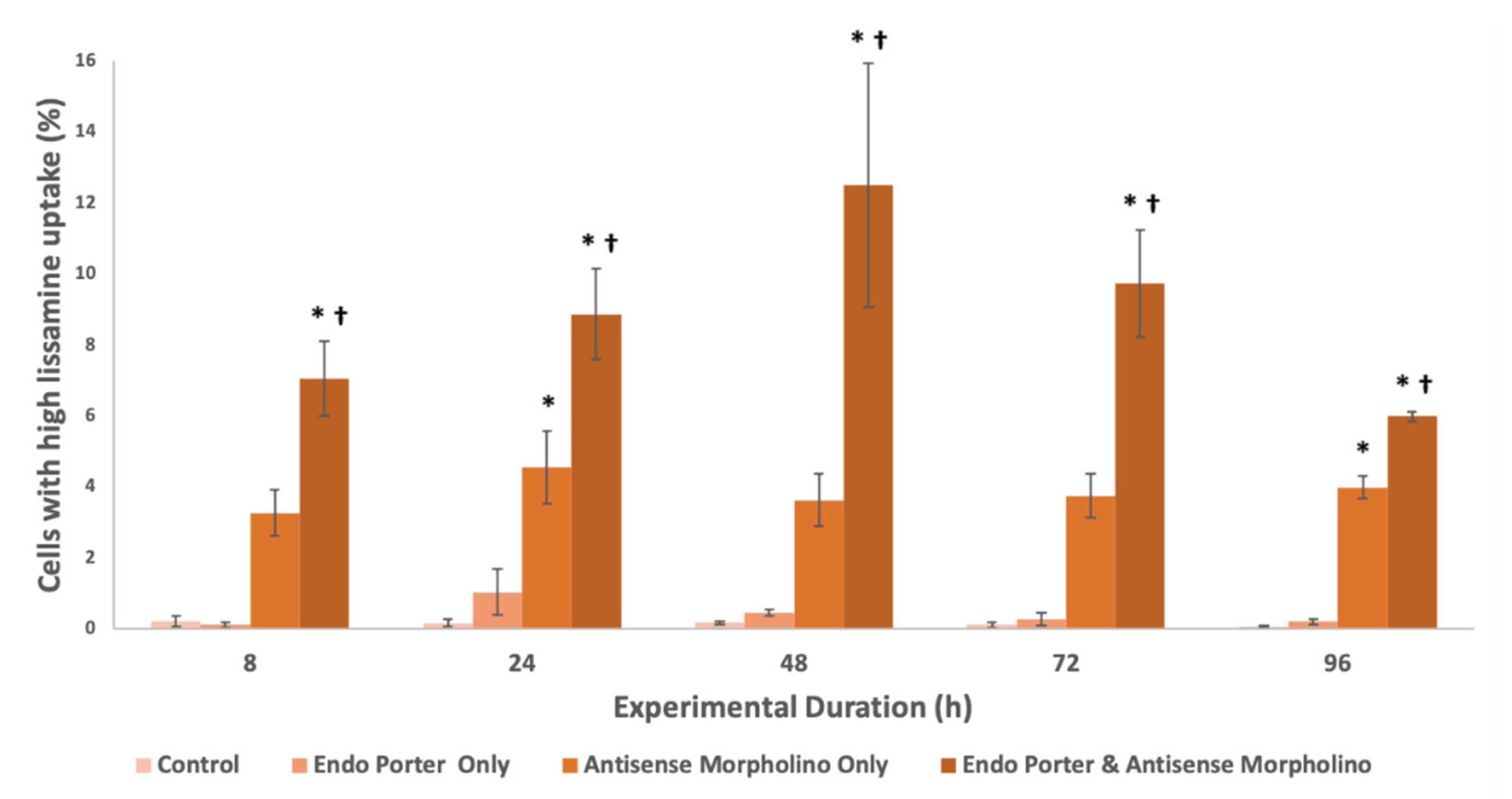
3.2. Uptake of the Morpholino
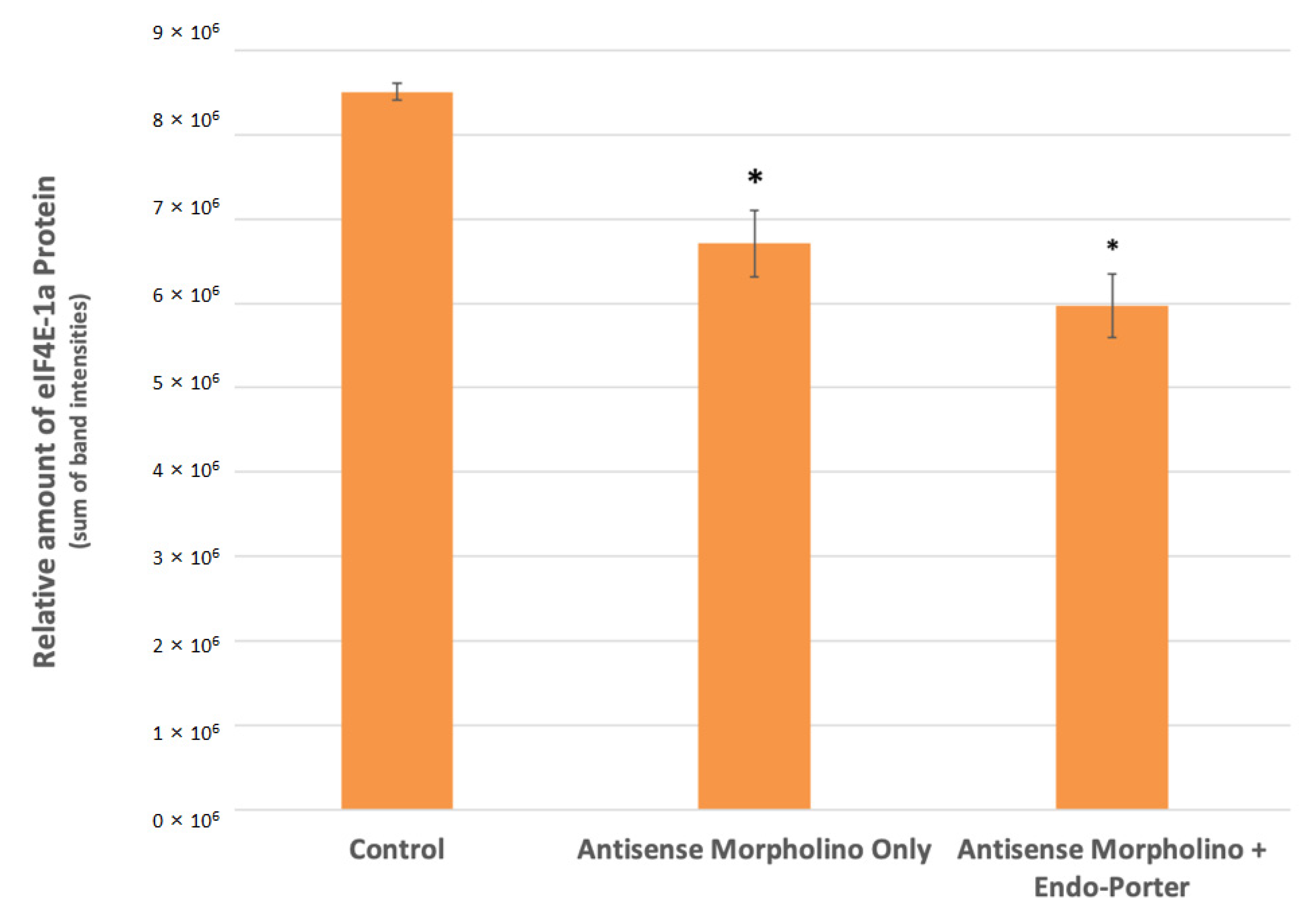
3.3. Initial Western Blot Analysis
3.4. Increase in MO Concentration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Not, F.; Siano, R.; Kooistra, W.H.C.F.; Simon, N.; Vaulot, D.; Probert, I. Diversity and Ecology of Eukaryotic Marine Phyto-plankton. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Adolf, J.E.; Stoecker, D.K.; Harding, L.W. The balance of autotrophy and heterotrophy during mixotrophic growth of Kar-lodinium micrum (Dinophyceae). J. Plankton Res. 2006, 28, 737–751. [Google Scholar] [CrossRef]
- Place, A.R.; Bowers, H.A.; Bachvaroff, T.R.; Adolf, J.E.; Deeds, J.R.; Sheng, J. Karlodinium veneficum-The little dinoflagellate with a big bite. Harmful Algae 2012, 14, 179–195. [Google Scholar] [CrossRef]
- Botana, L.M. Seafood and Freshwater Toxins: Pharmacology, Physiology, and Detection, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2008; pp. 1–943. [Google Scholar]
- Cusick, K.D.; Widder, E.A. Intensity differences in bioluminescent dinoflagellates impact foraging efficiency in a nocturnal predator. Bull. Mar. Sci. 2014, 90, 797–811. [Google Scholar] [CrossRef]
- Decelle, J.; Carradec, Q.; Pochon, X.; Henry, N.; Romac, S.; Mahé, F.; Dunthorn, M.; Kourlaiev, A.; Voolstra, C.R.; Wincker, P.; et al. Worldwide Occurrence and Activity of the Reef-Building Coral Symbiont Symbiodinium in the Open Ocean. Curr. Biol. 2018, 28, 3625–3633.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aquino-Cruz, A.; Okolodkov, Y.B. Impact of increasing water temperature on growth, photosynthetic efficiency, nutrient consumption, and potential toxicity of Amphidinium cf. carterae and Coolia monotis (Dinoflagellata). Rev. Biol. Mar. Oceanogr. 2016, 51, 565–580. [Google Scholar] [CrossRef] [Green Version]
- Gobler, C.J. Climate Change and Harmful Algal Blooms: Insights and perspective. Harmful Algae 2020, 91, 101731. [Google Scholar] [CrossRef] [PubMed]
- Griffith, A.W.; Gobler, C.J. Harmful algal blooms: A climate change co-stressor in marine and freshwater ecosystems. Harmful Algae 2020, 91, 101590. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Barua, A.; Ruvindy, R.; Savela, H.; Ajani, P.A.; Murray, S.A. The genetic basis of toxin biosynthesis in dinoflagel-lates. Microorganisms 2019, 7, 222. [Google Scholar] [CrossRef] [Green Version]
- Wells, M.L.; Trainer, V.L.; Smayda, T.J.; Karlson, B.S.; Trick, C.G.; Kudela, R.M.; Ishikawa, A.; Bernard, S.; Wulff, A.; Ander-son, D.M.; et al. Harmful algal blooms and climate change: Learning from the past and present to forecast the future. Harmful Algae 2015, 49, 68–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pagliara, P.; Caroppo, C. Toxicity assessment of Amphidinium carterae, Coolia cfr. monotis and Ostreopsis cfr. ovata (Di-nophyta) isolated from the northern Ionian Sea (Mediterranean Sea). Toxicon 2012, 60, 1203–1214. [Google Scholar] [CrossRef] [PubMed]
- Brownlee, E.F.; Sellner, S.G.; Sellner, K.G.; Nonogaki, H.; Adolf, J.E.; Bachvaroff, T.R.; Place, A.R. Responses of Crassostrea virginica (Gmelin) and C. ariakensis (Fujita) To Bloom-Forming Phytoplankton Including Ichthyotoxic Karlodinium veneficum (Ballantine). J. Shellfish. Res. 2008, 27, 581–591. [Google Scholar] [CrossRef]
- Galimany, E.; Place, A.R.; Ramón, M.; Jutson, M.; Pipe, R.K. The effects of feeding Karlodinium veneficum (PLY # 103; Gymnodinium veneficum Ballantine) to the blue mussel Mytilus edulis. Harmful Algae 2008, 7, 91–98. [Google Scholar]
- Baden, D.G. Brevetoxins: Unique polyether dinoflagellate toxins. FASEB J. 1989, 3, 1807–1817. [Google Scholar] [CrossRef] [PubMed]
- Deeds, J.R.; Hoesch, R.E.; Place, A.R.; Kao, J.P. The cytotoxic mechanism of karlotoxin 2 (KmT × 2) from Karlodinium veneficum (Dinophyceae). Aquat. Toxicol. 2015, 159, 148–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, J.; Place, A.R.; Yoshida, W.; Anklin, C.; Hamann, M.T. Structure and absolute configuration of karlotoxin-2, an ichthy-otoxin from the marine dinoflagellate karlodinium veneficum. J. Am. Chem. Soc. 2010, 132, 3277–3279. [Google Scholar] [CrossRef] [Green Version]
- Sheng, J.; Malkiel, E.; Katz, J.; Adolf, J.E.; Place, A.R. A dinoflagellate exploits toxins to immobilize prey prior to ingestion. Proc. Natl. Acad. Sci. USA 2010, 107, 2082–2087. [Google Scholar] [CrossRef] [Green Version]
- Van Wagoner, R.M.; Deeds, J.R.; Satake, M.; Ribeiro, A.A.; Place, A.R.; Wright, J.L. Isolation and characterization of karlo-toxin 1, a new amphipathic toxin from Karlodinium veneficum. Tetrahedron Lett. 2008, 49, 6457–6461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, P.J. Oceans and Human Health: Risks and Remedies from the Seas; Academic Press: Cambridge, MA, USA, 2008. [Google Scholar]
- Hieda, M.; Sorada, A.; Kinoshita, M.; Matsumori, N. Amphidinol 3 preferentially binds to cholesterol in disordered do-mains and disrupts membrane phase separation. Biochem. Biophys. Rep. 2021, 26, 100941. [Google Scholar] [PubMed]
- Jones, G.D.; Williams, E.P.; Place, A.R.; Jagus, R.; Bachvaroff, T.R. The alveolate translation initiation factor 4E family reveals a custom toolkit for translational control in core dinoflagellates. BMC Evol. Biol. 2015, 15, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brayton, K.A.; Lau, A.O.; Herndon, D.R.; Hannick, L.; Kappmeyer, L.S.; Berens, S.J.; Bidwell, S.L.; Brown, W.C.; Crabtree, J.; Fadrosh, D.; et al. Genome sequence of Babesia bovis and comparative analysis of apicomplexan hemoprotozoa. PLoS Pathog. 2007, 3, 1401–1413. [Google Scholar] [CrossRef] [PubMed]
- Ebenezer, T.E.; Zoltner, M.; Burrell, A.; Nenarokova, A.; Vanclová, A.M.G.N.; Prasad, B.; Soukal, P.; Santana-Molina, C.; O’Neill, E.; Nankissoor, N.N.; et al. Transcriptome, proteome and draft genome of Euglena gracilis. BMC Biol. 2019, 17, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Lin, S. Initial evidence of functional siRNA machinery in dinoflagellates. Harmful Algae 2019, 81, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Amorim, I.S.; Lach, G.; Gkogkas, C.G. The Role of the Eukaryotic Translation Initiation Factor 4E (eIF4E) in Neuropsychiat-ric Disorders. Front. Genet. 2018, 9, 561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batool, A.; Aashaq, S.; Andrabi, K.I. Eukaryotic initiation factor 4E (eIF4E): A recap of the cap-binding protein. J. Cell. Biochem. 2019, 120, 14201–14212. [Google Scholar] [CrossRef] [PubMed]
- Altmann, M.; Müller, P.P.; Pelletier, J.; Sonenberg, N.; Trachsel, H. A Mammalian Translation Initiation Factor Can Substi-tute for Its Yeast Homologue in Vivo. J. Biol. Chem. 1989, 264, 12145–12147. [Google Scholar] [CrossRef]
- Feoktistova, K.; Tuvshintogs, E.; Do, A.; Fraser, C.S. Human eIF4E promotes mRNA restructuring by stimulating eIF4A hel-icase activity. Proc. Natl. Acad. Sci. USA 2013, 110, 13339–13344. [Google Scholar] [CrossRef] [Green Version]
- Ibrahimo, S.; Holmes, L.E.; Ashe, M.P. Regulation of translation initiation by the yeast eIF4E binding proteins is required for the pseudohyphal response. Yeast 2006, 23, 1075–1088. [Google Scholar] [CrossRef]
- Joshi, B.; Lee, K.; Maeder, D.L.; Jagus, R. Phylogenetic analysis of eIF4E-family members. BMC Evol. Biol. 2005, 5, 48. [Google Scholar] [CrossRef] [Green Version]
- Piserà, A.; Campo, A.; Campo, S. Structure and functions of the translation initiation factor eIF4E and its role in cancer de-velopment and treatment. J. Genet. Genom. 2018, 45, 13–24. [Google Scholar] [CrossRef]
- Grifo, J.A.; Tahara, S.M.; Leis, J.P.; Morgan, M.A.; Shatkin, A.J.; Merrick, W.C. Characterization of eukaryotic initiation factor 4A, a protein involved in ATP-dependent binding of globin mRNA. J. Biol. Chem. 1982, 257, 5246–5252. [Google Scholar] [CrossRef]
- Hernández, G.; Vazquez-Pianzola, P. Functional diversity of the eukaryotic translation initiation factors belonging to eIF4 families. Mech. Dev. 2005, 122, 865–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, B.; Cameron, A.; Jagus, R. Characterization of mammalian eIF4E-family members. Eur. J. Biochem. 2004, 271, 2189–2203. [Google Scholar] [CrossRef] [PubMed]
- Lasko, P. The drosophila melanogaster genome: Translation factors and RNA binding proteins. J. Cell Biol. 2000, 150, F51–F56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachvaroff, T.R.; Place, A.R. From stop to start: Tandem gene arrangement, copy number and Trans-splicing sites in the di-noflagellate Amphidinium carterae. PLoS ONE 2008, 3, e2929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beauchemin, M.; Roy, S.; Daoust, P.; Dagenais-Bellefeuille, S.; Bertomeu, T.; Letourneau, L.; Lang, B.F.; Morse, D. Dinoflag-ellate tandem array gene transcripts are highly conserved and not polycistronic. Proc. Natl. Acad. Sci. USA 2012, 109, 15793–15798. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Wang, C.C. Identification in the ancient protist Giardia lamblia of two eukaryotic translation initiation factor 4E homologues with distinctive functions. Eukaryot. Cell 2005, 4, 948–959. [Google Scholar] [CrossRef] [Green Version]
- Jagus, R.; Bachvaroff, T.R.; Joshi, B.; Place, A.R. Diversity of eukaryotic translational initiation factor eIF4E in protists. Comp. Funct. Genom. 2012, 2012, 134839. [Google Scholar] [CrossRef] [Green Version]
- Morf, L.; Pearson, R.J.; Wang, A.S.; Singh, U. Robust gene silencing mediated by antisense small RNAs in the pathogenic protist Entamoeba histolytica. Nucleic Acids Res. 2013, 41, 9424–9437. [Google Scholar] [CrossRef] [Green Version]
- Kwok, A.C.M.; Mak, C.C.M.; Wong, F.T.W.; Wong, J.T.Y. Novel method for preparing spheroplasts from cells with an in-ternal cellulosic cell wall. Eukaryot. Cell 2007, 6, 563–567. [Google Scholar] [CrossRef] [Green Version]
- Yan, T.H.K.; Wu, Z.; Kwok, A.C.M.; Wong, J.T.Y. Knockdown of dinoflagellate condensin CcSMC4 subunit leads to s-phase impediment and decompaction of liquid crystalline chromosomes. Microorganisms 2020, 8, 565. [Google Scholar] [CrossRef] [Green Version]
- Chan, W.S.; Kwok, A.C.M.; Wong, J.T.Y. Knockdown of dinoflagellate cellulose synthase CesA1 resulted in malformed in-tracellular cellulosic thecal plates and severely impeded cyst-to-swarmer transition. Front. Microbiol. 2019, 10, 546. [Google Scholar] [CrossRef] [PubMed]
- Akbar, M.A.; Ahmad, A.; Usup, G.; Bunawan, H. Current knowledge and recent advances in marine dinoflagellate tran-scriptomic research. J. Mar. Sci. Eng. 2018, 6, 13. [Google Scholar] [CrossRef] [Green Version]
- Baumgarten, S.; Bayer, T.; Aranda, M.; Liew, Y.J.; Carr, A.; Micklem, G.; Voolstra, C.R. Integrating microRNA and mRNA expression profiling in Symbiodinium microadriaticum, a dinoflagellate symbiont of reef-building corals. BMC Genom. 2013, 14, 704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Summerton, J.E. Endo-Porter: A Novel Reagent for Safe, Effective Delivery of Substances into Cells. Ann. N. Y. Acad. Sci. 2005, 1058, 62–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.; Jagus, R.; Morse, D. Translation and translational control in dinoflagellates. Microorganisms 2018, 6, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berges, J.A.; Franklin, D.J.; Harrison, P.J. Evolution of an artificial seawater medium: Improvements in enriched seawater, artificial water over the last two decades. J. Phycol. 2001, 37, 1138–1145. [Google Scholar] [CrossRef]
- Liu, C.L.; Place, A.R.; Jagus, R. Use of antibiotics for maintenance of axenic cultures of Amphidinium carterae for the analy-sis of translation. Mar. Drugs 2017, 15, 242. [Google Scholar] [CrossRef] [Green Version]
- Todaro, A.M.; Hackeng, T.M.; Castoldi, E. Antisense-mediated down-regulation of factor v-short splicing in a liver cell line model. Appl. Sci. 2021, 11, 9621. [Google Scholar] [CrossRef]
- The R Development Core Team. R: A Language and Environment for Statistical Computing. 2013. Available online: http://www.R-Project.org/ (accessed on 24 May 2022).
- Mellert, K.; Lamla, M.; Scheffzek, K.; Wittig, R.; Kaufmann, D. Enhancing Endosomal Escape of Transduced Proteins by Photochemical Internalisation. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [Green Version]
- Stephens, T.G.; Ragan, M.A.; Bhattacharya, D.; Chan, C.X. Core genes in diverse dinoflagellate lineages include a wealth of conserved dark genes with unknown functions. Sci. Rep. 2018, 8, 17175. [Google Scholar] [CrossRef]
- Sarai, C.; Tanifuji, G.; Nakayama, T.; Kamikawa, R.; Takahashi, K.; Yazaki, E.; Matsuo, E.; Miyashita, H.; Ishida, K.-I.; Iwataki, M.; et al. Dinoflagellates with relic endosymbiont nuclei as models for elucidating organellogenesis. Proc. Natl. Acad. Sci. USA 2020, 117, 5364–5375. [Google Scholar] [CrossRef] [PubMed]
- Morden, C.W.; Sherwood, A.R. Continued evolutionary surprises among dinoflagellates. Proc. Natl. Acad. Sci. USA 2002, 99, 11558–11560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mackiewicz, P.; Bodył, A.; Moszczyński, K. The case of horizontal gene transfer from bacteria to the peculiar dinoflagellate plastid genome. Mob. Genet. Elem. 2013, 3, e25845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wisecaver, J.H.; Brosnahan, M.L.; Hackett, J.D. Horizontal gene transfer is a significant driver of gene innovation in dino-flagellates. Genome Biol. Evol. 2013, 5, 2368–2381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gavelis, G.S.; Herranz, M.; Wakeman, K.C.; Ripken, C.; Mitarai, S.; Gile, G.H.; Keeling, P.J.; Leander, B.S. Dinoflagellate nu-cleus contains an extensive endomembrane network, the nuclear net. Sci. Rep. 2019, 9, 839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gornik, S.G.; Hu, I.; Lassadi, I.; Waller, R.F. The biochemistry and evolution of the dinoflagellate nucleus. Microorganisms 2019, 7, 245. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.Y.S.; Putnam, A.; Lu, T.; He, S.; Ouyang, J.P.T.; Seydoux, G. Recruitment of mRNAs to P granules by condensation with intrinsically-disordered proteins. eLife 2020, 9, e52896. [Google Scholar] [CrossRef]
- Soyer-Gobillard, M.O.; Geraud, M.L. Nucleolus behaviour during the cell cycle of a primitive dinoflagellate eukaryote, Prorocentrum micans Ehr., seen by light microscopy and electron microscopy. J. Cell Sci. 1992, 102, 475–485. [Google Scholar] [CrossRef]
- Sheng, J.; Malkiel, E.; Katz, J.; Adolf, J.; Belas, R.; Place, A.R. Digital holographic microscopy reveals prey-induced changes in swimming behavior of predatory dinoflagellates. Proc. Natl. Acad. Sci. USA 2007, 104, 17512–17517. [Google Scholar] [CrossRef] [Green Version]



| Morpholino Concentration | 1 μM | 10 μM | |
|---|---|---|---|
| Relative eIF4E-1a Production | Percent Change | ||
| Antisense Morpholino Only | 78.8% * | 78.3% | 0.5% |
| Antisense Morpholino + Endo Porter | 70.1% * | 58.3% * | 11.8% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Judd, M.; Place, A.R. A Strategy for Gene Knockdown in Dinoflagellates. Microorganisms 2022, 10, 1131. https://doi.org/10.3390/microorganisms10061131
Judd M, Place AR. A Strategy for Gene Knockdown in Dinoflagellates. Microorganisms. 2022; 10(6):1131. https://doi.org/10.3390/microorganisms10061131
Chicago/Turabian StyleJudd, Miranda, and Allen R. Place. 2022. "A Strategy for Gene Knockdown in Dinoflagellates" Microorganisms 10, no. 6: 1131. https://doi.org/10.3390/microorganisms10061131






