Edwardsiella ictaluri T3SS Effector EseN Modulates Expression of Host Genes Involved in the Immune Response
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Infection Procedure
2.3. RNA Extractions and Sequencing
2.4. RT-qPCR
2.5. The mRNAseq Data Analysis
2.6. Prostaglandin Assay
2.7. Statistical Analysis
3. Results
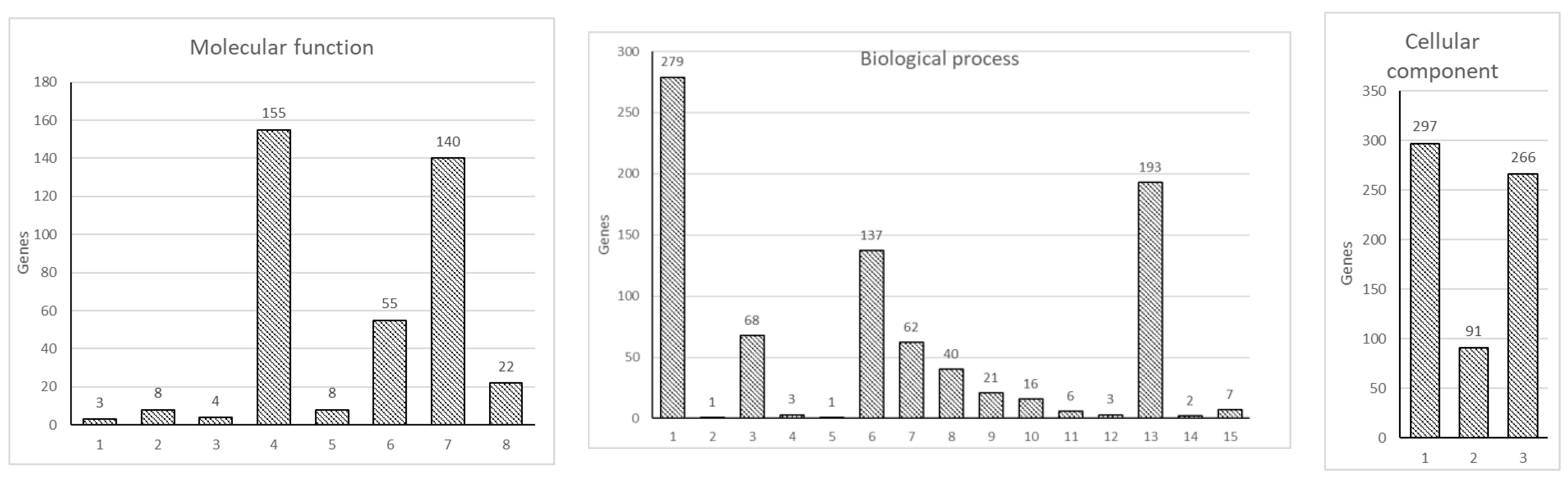
3.1. Edwardsiella ictaluri T3SS Effector EseN Modulates Host Gene Expression
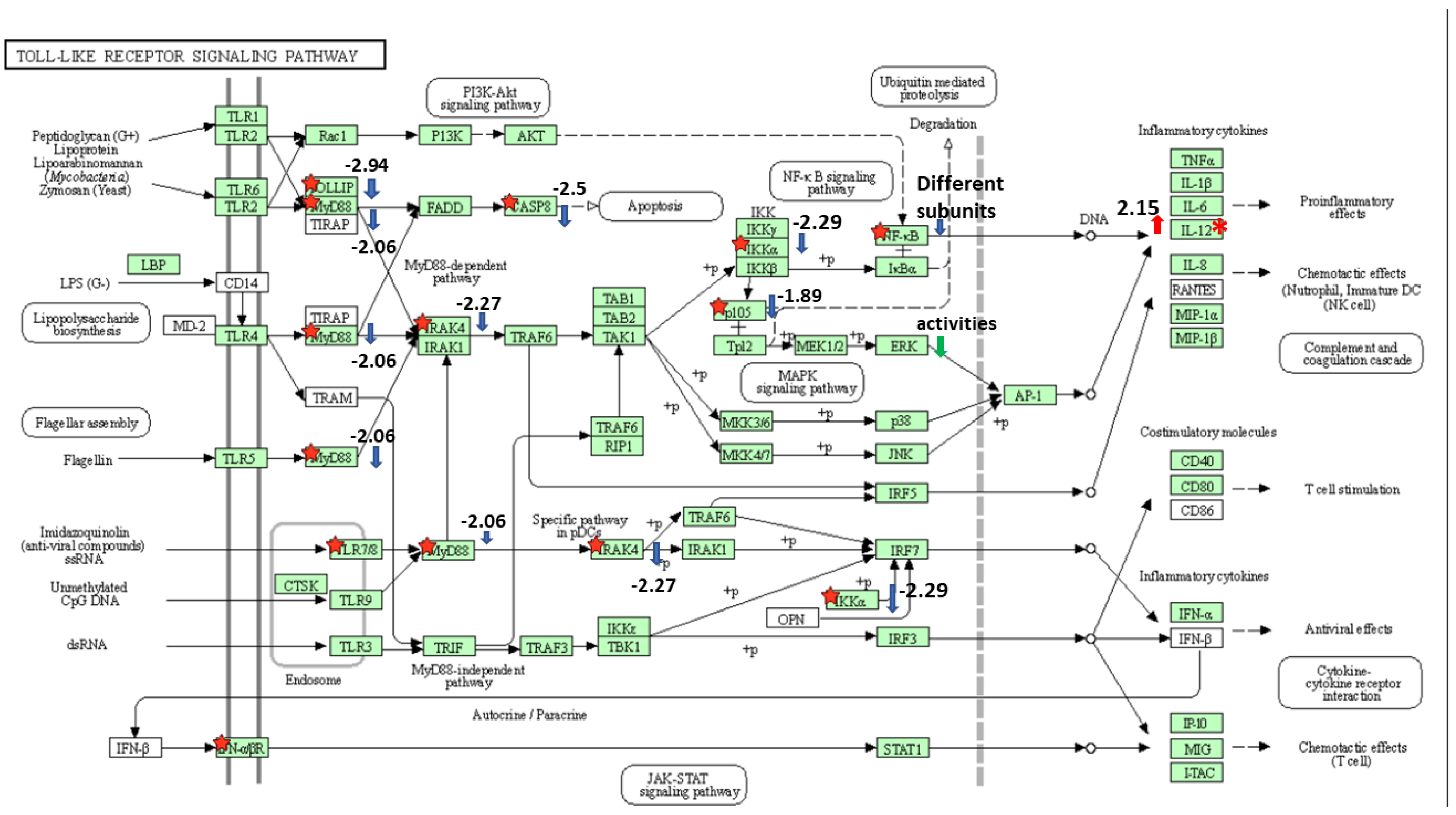
3.2. E. ictaluri Effector EseN Modulates Transcription Factor Expression in HKDM
3.3. Effect of E. ictaluri T3SS Effector EseN on Modulation of Pro and Anti-Inflammatory Cytokine Expression in HKDM
3.4. EseN Modulates COX-2 and Prostaglandin Expression in HKDM
4. Discussions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Thune, R.L.; Fernandez, D.H.; Benoit, J.L.; Kelly-Smith, M.; Rogge, M.L.; Booth, N.J.; Landry, C.A.; Bologna, R.A. Signature-tagged mutagenesis of Edwardsiella ictaluri identifies virulence-related genes, including a salmonella pathogenicity island 2 class of type III secretion systems. Appl. Environ. Microbiol. 2007, 73, 7934–7946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubytska, L.P.; Rogge, M.L.; Thune, R.L. Identification and Characterization of Putative Translocated Effector Proteins of the Edwardsiella ictaluri Type III Secretion System. mSphere 2016, 1, e00039-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornelis, G.R.; Van Gijsegem, F. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 2000, 54, 735–774. [Google Scholar] [CrossRef]
- Galan, J.E.; Wolf-Watz, H. Protein delivery into eukaryotic cells by type III secretion machines. Nature 2006, 444, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Coburn, B.; Sekirov, I.; Finlay, B.B. Type III secretion systems and disease. Clin. Microbiol. Rev. 2007, 20, 535–549. [Google Scholar] [CrossRef] [Green Version]
- Agbor, T.A.; McCormick, B.A. Salmonella effectors: Important players modulating host cell function during infection. Cell. Microbiol. 2011, 13, 1858–1869. [Google Scholar] [CrossRef] [Green Version]
- Dean, P. Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol. Rev. 2011, 35, 1100–1125. [Google Scholar] [CrossRef] [Green Version]
- Figueira, R.; Holden, D.W. Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology 2012, 158, 1147–1161. [Google Scholar] [CrossRef] [Green Version]
- Kaur, J.; Jain, S.K. Role of antigens and virulence factors of Salmonella enterica serovar Typhi in its pathogenesis. Microbiol. Res. 2012, 167, 199–210. [Google Scholar] [CrossRef]
- Kline, T.; Felise, H.B.; Sanowar, S.; Miller, S.I. The type III secretion system as a source of novel antibacterial drug targets. Curr. Drug Targets 2012, 13, 338–351. [Google Scholar] [CrossRef]
- Marteyn, B.; Gazi, A.; Sansonetti, P. Shigella: A model of virulence regulation in vivo. Gut Microbes 2012, 3, 104–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubytska, L.P.; Thune, R.L. Edwardsiella ictaluri type III secretion system (T3SS) effector EseN is a phosphothreonine lyase that inactivates ERK1/2. Dis. Aquat. Organ. 2018, 130, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Mazurkiewicz, P.; Thomas, J.; Thompson, J.A.; Liu, M.; Arbibe, L.; Sansonetti, P.; Holden, D.W. SpvC is a Salmonella effector with phosphothreonine lyase activity on host mitogen-activated protein kinases. Mol. Microbiol. 2008, 67, 1371–1383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zurawski, D.V.; Mitsuhata, C.; Mumy, K.L.; McCormick, B.A.; Maurelli, A.T. OspF and OspC1 are Shigella flexneri type III secretion system effectors that are required for postinvasion aspects of virulence. Infect. Immun. 2006, 74, 5964–5976. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.L.; Dong, C. MAP kinases in immune responses. Cell. Mol. Immunol. 2005, 2, 20–27. [Google Scholar]
- Lawrence, T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009, 1, a001651. [Google Scholar] [CrossRef] [Green Version]
- Skaug, B.; Jiang, X.; Chen, Z.J. The role of ubiquitin in NF-kappaB regulatory pathways. Annu. Rev. Biochem. 2009, 78, 769–796. [Google Scholar] [CrossRef]
- Zhu, J.; Mohan, C. Toll-like receptor signaling pathways-therapeutic opportunities. Mediat. Inflamm. 2010, 2010, 781235. [Google Scholar] [CrossRef] [Green Version]
- Bhavsar, A.P.; Guttman, J.A.; Finlay, B.B. Manipulation of host-cell pathways by bacterial pathogens. Nature 2007, 449, 827–834. [Google Scholar] [CrossRef]
- Arthur, J.S.; Ley, S.C. Mitogen-activated protein kinases in innate immunity. Nat. Rev. Immunol. 2013, 13, 679–692. [Google Scholar] [CrossRef]
- Arbibe, L.; Kim, D.W.; Batsche, E.; Pedron, T.; Mateescu, B.; Muchardt, C.; Parsot, C.; Sansonetti, P.J. An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat. Immunol. 2007, 8, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Kramer, R.W.; Slagowski, N.L.; Eze, N.A.; Giddings, K.S.; Morrison, M.F.; Siggers, K.A.; Starnbach, M.N.; Lesser, C.F. Yeast functional genomic screens lead to identification of a role for a bacterial effector in innate immunity regulation. PLoS Pathog. 2007, 3, e21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Xu, H.; Zhou, Y.; Zhang, J.; Long, C.; Li, S.; Chen, S.; Zhou, J.M.; Shao, F. The phosphothreonine lyase activity of a bacterial type III effector family. Science 2007, 315, 1000–1003. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Deng, Q.; Porath, J.A.; Williams, C.L.; Pederson-Gulrud, K.J.; Barbieri, J.T. Plasma membrane localization affects the RhoGAP specificity of Pseudomonas ExoS. Cell. Microbiol. 2007, 9, 2192–2201. [Google Scholar] [CrossRef]
- He, Q.Z.; Zeng, H.C.; Huang, Y.; Hu, Y.Q.; Wu, Y.M. The type III secretion system (T3SS) of Chlamydophila psittaci is involved in the host inflammatory response by activating the JNK/ERK signaling pathway. Biomed. Res. Int. 2015, 2015, 652416. [Google Scholar] [CrossRef] [Green Version]
- Bogdan, C.; Nathan, C. Modulation of macrophage function by transforming growth factor beta, interleukin-4, and interleukin-10. Ann. N. Y. Acad. Sci. 1993, 685, 713–739. [Google Scholar] [CrossRef]
- Smith, W.L.; Marnett, L.J. Prostaglandin endoperoxide synthase: Structure and catalysis. Biochim. Biophys. Acta (BBA)-Lipids Lipid Metab. 1991, 1083, 1–17. [Google Scholar] [CrossRef]
- Lee, S.H.; Soyoola, E.; Chanmugam, P.; Hart, S.; Sun, W.; Zhong, H.; Liou, S.; Simmons, D.; Hwang, D. Selective expression of mitogen-inducible cyclooxygenase in macrophages stimulated with lipopolysaccharide. J. Biol. Chem. 1992, 267, 25934–25938. [Google Scholar] [CrossRef]
- Booth, N.J.; Elkamel, A.; Thune, R.L. Thune. Intracellular Replication of Edwardsiella ictaluri in Channel Catfish Macrophages. J. Aquat. Anim. Health 2006, 18, 101–108. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, S.; Yao, J.; Bao, L.; Zhang, J.; Li, Y.; Jiang, C.; Sun, L.; Wang, R.; Zhang, Y.; et al. The channel catfish genome sequence provides insights into the evolution of scale formation in teleosts. Nat. Commun. 2016, 7, 11757. [Google Scholar] [CrossRef] [Green Version]
- Leng, N.; Dawson, J.A.; Thomson, J.A.; Ruotti, V.; Rissman, A.I.; Smits, B.M.; Haag, J.D.; Gould, M.N.; Stewart, R.M.; Kendziorski, C. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 2013, 29, 1035–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Booth, N.J.; Beekman, J.B.; Thune, R.L. Edwardsiella ictaluri encodes an acid-activated urease that is required for intracellular replication in channel catfish (Ictalurus punctatus) macrophages. Appl. Environ. Microbiol. 2009, 75, 6712–6720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cirillo, D.M.; Valdivia, R.H.; Monack, D.M.; Falkow, S. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 1998, 30, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Hensel, M. Whole genome scan for habitat-specific genes by signature-tagged mutagenesis. Electrophoresis 1998, 19, 608–612. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, W.A.; Dubytska, L.; Rogge, M.L.; Mottram, P.J.; Thune, R.L. Modulation of vacuolar pH is required for replication of Edwardsiella ictaluri in channel catfish macrophages. Infect. Immun. 2014, 82, 2329–2336. [Google Scholar] [CrossRef] [Green Version]
- Campoy, E.; Colombo, M.I. Autophagy in intracellular bacterial infection. Biochim. Biophys. Acta 2009, 1793, 1465–1477. [Google Scholar] [CrossRef] [Green Version]
- DeLeo, F.R. Modulation of phagocyte apoptosis by bacterial pathogens. Apoptosis 2004, 9, 399–413. [Google Scholar] [CrossRef]
- Dabbs, E.R. Mapping of the genes for Bacillus subtilis ribosomal proteins S6 and S16: Comparison of the chromosomal distribution of ribosomal protein genes in this bacterium with the distribution in Escherichia coli. Mol. Gen. Genet. 1983, 192, 386–390. [Google Scholar] [CrossRef]
- Diacovich, L.; Gorvel, J.P. Bacterial manipulation of innate immunity to promote infection. Nat. Rev. Microbiol. 2010, 8, 117–128. [Google Scholar] [CrossRef]
- Zurawski, D.V.; Mumy, K.L.; Faherty, C.S.; McCormick, B.A.; Maurelli, A.T. Shigella flexneri type III secretion system effectors OspB and OspF target the nucleus to downregulate the host inflammatory response via interactions with retinoblastoma protein. Mol. Microbiol. 2009, 71, 350–368. [Google Scholar] [CrossRef] [Green Version]
- Rogge, M.L.; Thune, R.L. Regulation of the Edwardsiella ictaluri type III secretion system by pH and phosphate concentration through EsrA, EsrB, and EsrC. Appl. Environ. Microbiol. 2011, 77, 4293–4302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Liang, H.; Zen, K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front. Immunol. 2014, 5, 614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vijayan, V.; Srinu, T.; Karnati, S.; Garikapati, V.; Linke, M.; Kamalyan, L.; Mali, S.R.; Sudan, K.; Kollas, A.; Schmid, T.; et al. A New Immunomodulatory Role for Peroxisomes in Macrophages Activated by the TLR4 Ligand Lipopolysaccharide. J. Immunol. 2017, 198, 2414–2425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benoit, M.; Desnues, B.; Mege, J.L. Macrophage polarization in bacterial infections. J. Immunol. 2008, 181, 3733–3739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [Green Version]
- Bianco, C.; Mohr, I. Ribosome biogenesis restricts innate immune responses to virus infection and DNA. Elife 2019, 8, e49551. [Google Scholar] [CrossRef]
- Asrat, S.; Dugan, A.S.; Isberg, R.R. The frustrated host response to Legionella pneumophila is bypassed by MyD88-dependent translation of pro-inflammatory cytokines. PLoS Pathog. 2014, 10, e1004229. [Google Scholar] [CrossRef]
- Rolhion, N.; Furniss, R.C.; Grabe, G.; Ryan, A.; Liu, M.; Matthews, S.A.; Holden, D.W. Inhibition of Nuclear Transport of NF-kB p65 by the Salmonella Type III Secretion System Effector SpvD. PLoS Pathog. 2016, 12, e1005653. [Google Scholar] [CrossRef]
- Evans, S.M.; Rodino, K.G.; Adcox, H.E.; Carlyon, J.A. Orientia tsutsugamushi uses two Ank effectors to modulate NF-kappaB p65 nuclear transport and inhibit NF-kappaB transcriptional activation. PLoS Pathog. 2018, 14, e1007023. [Google Scholar] [CrossRef] [Green Version]
- Burette, M.; Allombert, J.; Lambou, K.; Maarifi, G.; Nisole, S.; Di Russo Case, E.; Blanchet, F.P.; Hassen-Khodja, C.; Cabantous, S.; Samuel, J.; et al. Modulation of innate immune signaling by a Coxiella burnetii eukaryotic-like effector protein. Proc. Natl. Acad. Sci. USA 2020, 117, 13708–13718. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Ghosh, S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef] [PubMed]
- Baeuerle, P.A.; Henkel, T. Function and activation of NF-kappa B in the immune system. Annu. Rev. Immunol. 1994, 12, 141–179. [Google Scholar] [CrossRef] [PubMed]
- Jordan, K.A.; Dupont, C.D.; Tait, E.D.; Liou, H.C.; Hunter, C.A. Role of the NF-κB transcription factor c-Rel in the generation of CD8+ T-cell responses to Toxoplasma gondii. Int. Immunol. 2010, 22, 851–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murli, S.; Watson, R.O.; Galán, J.E. Role of tyrosine kinases and the tyrosine phosphatase SptP in the interaction of Salmonella with host cells. Cell. Microbiol. 2001, 3, 795–810. [Google Scholar] [CrossRef] [Green Version]
- Miao, E.A.; Brittnacher, M.; Haraga, A.; Jeng, R.L.; Welch, M.D.; Miller, S.I. Salmonella effectors translocated across the vacuolar membrane interact with the actin cytoskeleton. Mol. Microbiol. 2003, 48, 401–415. [Google Scholar] [CrossRef]
- Wall, E.C.; Everett, D.B.; Mukaka, M.; Bar-Zeev, N.; Feasey, N.; Jahn, A.; Moore, M.; van Oosterhout, J.J.; Pensalo, P.; Baguimira, K.; et al. Bacterial meningitis in Malawian adults, adolescents, and children during the era of antiretroviral scale-up and Haemophilus influenzae type b vaccination, 2000–2012. Clin. Infect. Dis. 2014, 58, e137–e145. [Google Scholar] [CrossRef] [Green Version]
- Uchiya, K.I.; Isono, S.; Yoshimura, M.; Wajima, T.; Nikai, T. Salmonella fimbrial protein StcD induces cyclooxygenase-2 expression via Toll-like receptor 4. J. Microbiol. Immunol. Infect. 2021, in press. [Google Scholar] [CrossRef]
- Kunkel, S.L.; Spengler, M.; May, M.A.; Spengler, R.; Larrick, J.; Remick, D. Prostaglandin E2 regulates macrophage-derived tumor necrosis factor gene expression. J. Biol. Chem. 1988, 263, 5380–5384. [Google Scholar] [CrossRef]
- Strassmann, G.; Patil-Koota, V.; Finkelman, F.; Fong, M.; Kambayashi, T. Evidence for the involvement of interleukin 10 in the differential deactivation of murine peritoneal macrophages by prostaglandin E2. J. Exp. Med. 1994, 180, 2365–2370. [Google Scholar] [CrossRef]
- Williams, J.A.; Shacter, E. Regulation of macrophage cytokine production by prostaglandin E2. Distinct roles of cyclooxygenase-1 and -2. J. Biol. Chem. 1997, 272, 25693–25699. [Google Scholar] [CrossRef] [Green Version]
- Agard, M.; Asakrah, S.; Morici, L. PGE2 suppression of innate immunity during mucosal bacterial infection. Front. Cell. Infect. Microbiol. 2013, 3, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.F.; Pandey, S.; Day, C.H.; Chen, Y.F.; Jiang, A.Z.; Ho, T.J.; Chen, R.J.; Padma, V.V.; Kuo, W.W.; Huang, C.Y. Synergistic effect of HIF-1α and FoxO3a trigger cardiomyocyte apoptosis under hyperglycemic ischemia condition. J. Cell. Physiol. 2018, 233, 3660–3671. [Google Scholar] [CrossRef] [PubMed]
- McClelland Descalzo, D.L.; Satoorian, T.S.; Walker, L.M.; Sparks, N.R.; Pulyanina, P.Y.; Zur Nieden, N.I. Glucose-Induced Oxidative Stress Reduces Proliferation in Embryonic Stem Cells via FOXO3A/β-Catenin-Dependent Transcription of p21(cip1). Stem Cell Rep. 2016, 7, 55–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGowan, S.E.; McCoy, D.M. Platelet-derived growth factor-A regulates lung fibroblast S-phase entry through p27(kip1) and FoxO3a. Respir. Res. 2013, 14, 68. [Google Scholar] [CrossRef] [Green Version]
- Joseph, J.; Ametepe, E.S.; Haribabu, N.; Agbayani, G.; Krishnan, L.; Blais, A.; Sad, S. Inhibition of ROS and upregulation of inflammatory cytokines by FoxO3a promotes survival against Salmonella typhimurium. Nat. Commun. 2016, 7, 12748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fluteau, A.; Ince, P.G.; Minett, T.; Matthews, F.E.; Brayne, C.; Garwood, C.J.; Ratcliffe, L.E.; Morgan, S.; Heath, P.R.; Shaw, P.J.; et al. The nuclear retention of transcription factor FOXO3a correlates with a DNA damage response and increased glutamine synthetase expression by astrocytes suggesting a neuroprotective role in the ageing brain. Neurosci. Lett. 2015, 609, 11–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Primers | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| CanX | GCTGTTAAACCGGAGGACTG | GCAGGTCCTCGAAGTAGTCAG |
| SDHA | CTCCAGGGATGTGGTTTCAC | GCATACAACCCTGGCACTAC |
| COX2 | CAGGTCGAGATGCACTACC | GTAGTAGCCGCTCAGGTG |
| IL8 | GATTGCTCAACCTTTTCGCATTAC | CATGGCCTGTGATTTAGCTGTG |
| IL6 | ACAGCGGAGACACGGA | GGTAGATCCGCTGCAGAC |
| IL15 | CGGCGATTTGTTCGATGCAG | CTCCTGGTTCAAGGGTCAC |
| IL12a | GAAGACCGTAAGGACCTGTG | TTGCAAAGTAGTAGTGCGGAG |
| IL16 | CATCAGTCGACACCCTGAC | CTGTGGCTTCGCTCCATAC |
| SOCS6 | CGTTGACCTCATCGAGCACT | TCAATCGAACCGGGTACGTG |
| CREB3L4 | GTGGAGTTCTCCGATGCTCA | TCAAGATCAGAGTGGGCGTG |
| FOXO3a set 1 | TGGAGGGACAAGTTGTGTCG | TTGATTAGTGCAGCCAACGGA |
| FOXO3a set 2 | GGAGGGACAAGTTGTGTCGT | AGCCAACGGAAGGCTATTCA |
| NFKβ1 | CGACCCCACTTAACATGGCT | TTGTGGTGGGATGATGACCG |
| Rel | ACCCTGGGGCTAGTAATGGA | GGGAGGCTGTTTCCACTCTC |
| RelB | AGACGAGGTCATGAGCACAC | GAGCCACTGTCCTCGTAGTG |
| TRIM24 | GTGCATGGAAGTCGAGGTCT | TTCTCTACGGTGCTGCTTGG |
| Category | Term | Gene Count | % | p Value |
|---|---|---|---|---|
| KEGG_PATHWAY | Ribosome biogenesis in eukaryotes | 11 | 1.644245 | 8.21 × 10−5 |
| KEGG_PATHWAY | Nucleocytoplasmic transport | 13 | 1.943199 | 1.05 × 10−4 |
| KEGG_PATHWAY | Herpes simplex virus 1 infection | 19 | 2.84006 | 0.001088 |
| KEGG_PATHWAY | Lysine degradation | 10 | 1.494768 | 0.003896 |
| KEGG_PATHWAY | Cell cycle | 13 | 1.943199 | 0.00414 |
| KEGG_PATHWAY | Measles | 12 | 1.793722 | 0.009788 |
| KEGG_PATHWAY | Epstein-Barr virus infection | 15 | 2.242152 | 0.01266 |
| KEGG_PATHWAY | One carbon pool by folate | 4 | 0.597907 | 0.020781 |
| KEGG_PATHWAY | Toll and Imd signaling pathway | 6 | 0.896861 | 0.02225 |
| KEGG_PATHWAY | Toll-like receptor signaling pathway | 9 | 1.345291 | 0.022977 |
| KEGG_PATHWAY | Peroxisome | 8 | 1.195815 | 0.026114 |
| KEGG_PATHWAY | NOD-like receptor signaling pathway | 11 | 1.644245 | 0.029469 |
| KEGG_PATHWAY | Alcoholic liver disease | 10 | 1.494768 | 0.043176 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dubytska, L.P.; Koirala, R.; Sanchez, A.; Thune, R. Edwardsiella ictaluri T3SS Effector EseN Modulates Expression of Host Genes Involved in the Immune Response. Microorganisms 2022, 10, 1334. https://doi.org/10.3390/microorganisms10071334
Dubytska LP, Koirala R, Sanchez A, Thune R. Edwardsiella ictaluri T3SS Effector EseN Modulates Expression of Host Genes Involved in the Immune Response. Microorganisms. 2022; 10(7):1334. https://doi.org/10.3390/microorganisms10071334
Chicago/Turabian StyleDubytska, Lidiya P., Ranjan Koirala, Azhia Sanchez, and Ronald Thune. 2022. "Edwardsiella ictaluri T3SS Effector EseN Modulates Expression of Host Genes Involved in the Immune Response" Microorganisms 10, no. 7: 1334. https://doi.org/10.3390/microorganisms10071334






