Current Techniques to Study Beneficial Plant-Microbe Interactions
Abstract
:1. Introduction
2. Techniques to Study Plant-PGPB Interactions
2.1. Omics Techniques
2.1.1. DNA Sequencing
2.1.2. Whole PGPB Genomes
2.1.3. Sequencing of Endophytic Genes
2.1.4. Antibiotic Resistance Genes
2.2. Definitions of Metagenomic, Metaproteomic, Metatranscriptomic, and Metabolomic
2.2.1. Transcriptomics
2.2.2. Proteomics
2.2.3. Metabolomics
2.3. Imaging
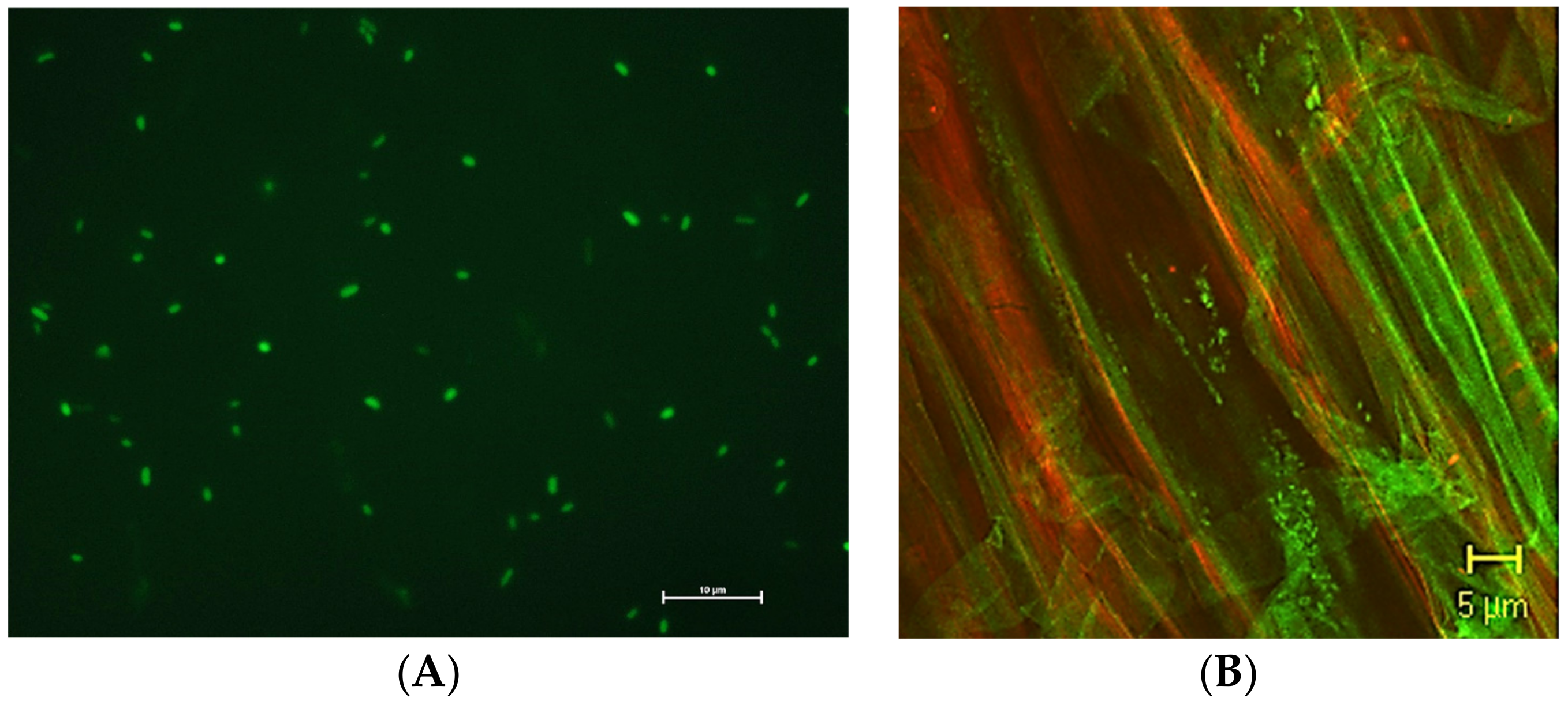
2.3.1. Labeling Bacteria
2.3.2. WinRhizo System
2.3.3. Other Imaging Systems
2.4. Nitrogenase Assays
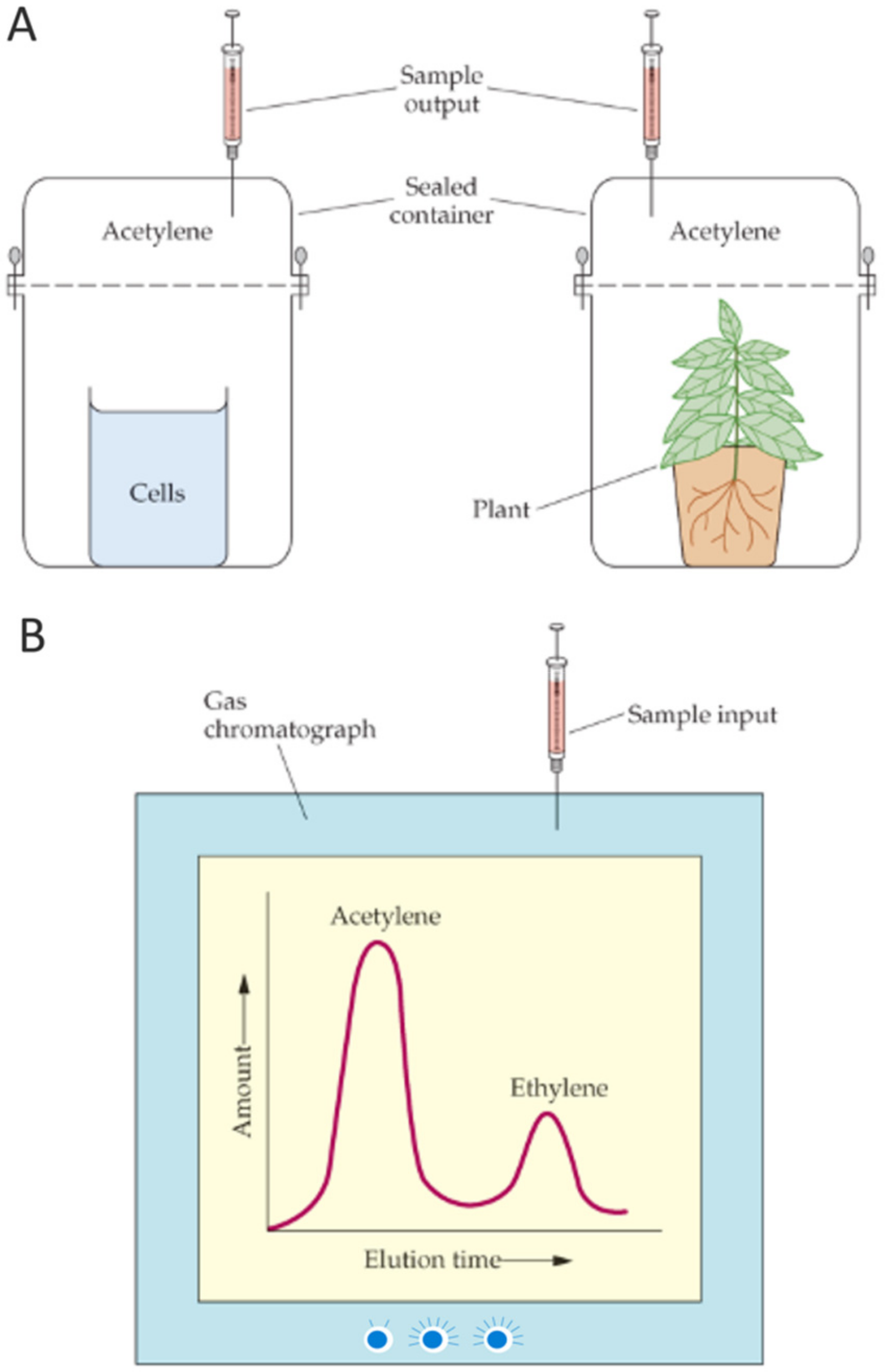
2.4.1. Nitrogen Fixation and Gas Chromatography
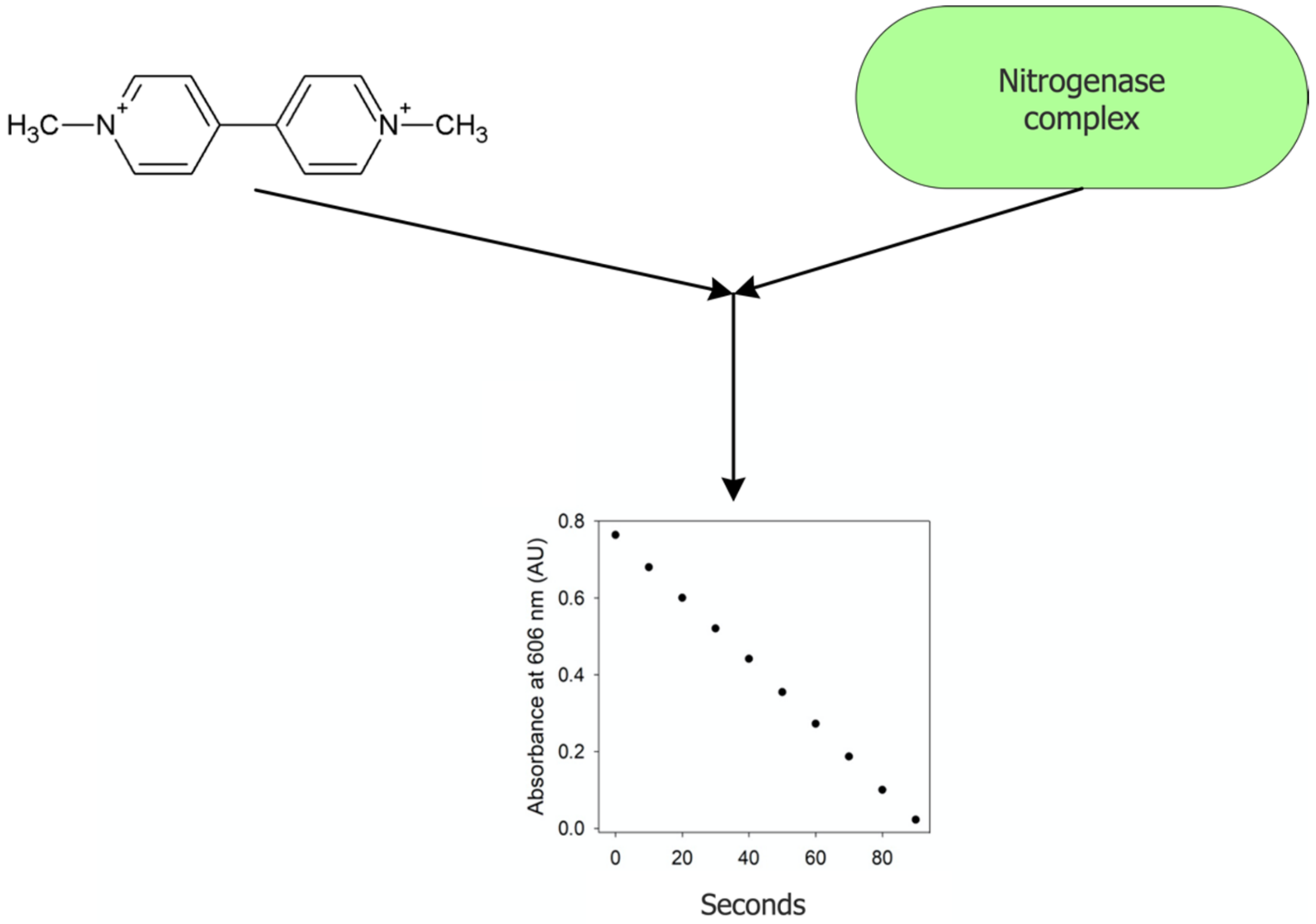
2.4.2. Viologen-Based Assay
2.4.3. 15N Dilution Method
2.4.4. Modified Acetylene Reduction Assay
2.5. Specialized Growth Chambers
2.5.1. Rhizotrons and Mini Rhizotrons
2.5.2. Rhizobox
2.5.3. Split-Root Systems (SRS)
3. Manipulation of the Native Rhizosphere Microbial Community
3.1. Releasing PGPB into the Environment
3.2. CRISPR Fundamentals
3.3. CRISPR Modified PGPB and Phytopathogens
3.4. Inoculant Encapsulation
Brief Overview of Some Encapsulation Techniques
4. Summary and Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Glick, B.R. Beneficial Plant-Bacterial Interactions, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2020; pp. 1–383. [Google Scholar]
- Reed, M.L.E.; Glick, B.R. Applications of plant growth-promoting bacteria for plant and soil systems. In Applications of Microbial Engineering; Gupta, V.K., Schmoll, M., Maki, M., Tuohy, M., Mazutti, M.A., Eds.; Taylor and Francis: Oxford, UK; Enfield: Chennai, India; Hartford, CT, USA, 2013; pp. 181–229. [Google Scholar]
- Levy, A.; Conway, J.M.; Dangl, J.L.; Woyke, T. Elucidating bacterial gene functions in the plant microbiome. Cell Host Microbe 2018, 24, 475–485. [Google Scholar] [CrossRef] [Green Version]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R.; Gamalero, E. Recent developments in the study of plant microbiomes. Microorganisms 2021, 9, 1533. [Google Scholar] [CrossRef] [PubMed]
- Howard, M.M.; Bell, T.H.; Kao-Kniffin, J. Soil microbiome transfer method affects microbiome composition, including dominant microorganisms, in a novel environment. FEMS Microbiol. Lett. 2017, 364, fnx092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, R.; Anand, G.; Gaur, R.; Yadav, D. Plant-microbiome interactions for sustainable agriculture: A review. Physiol. Mol. Biol. Plants 2021, 27, 165–179. [Google Scholar] [CrossRef]
- Korenblum, E.; Massalha, H.; Aharoni, A. Plant–microbe interactions in the rhizosphere via a circular metabolic economy. Plant Cell 2022, koac163. [Google Scholar] [CrossRef]
- Zilber-Rosenberg, I.; Rosenberg, E. Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiol. Rev. 2008, 32, 723–735. [Google Scholar] [CrossRef]
- Cesaro, P.; Gamalero, E.; Zhang, J.; Pivato, B. Editorial: The Plant Holobiont Volume I: Microbiota as Part of the Holobiont; Challenges for Agriculture. Front. Plant Sci. 2021, 12, 799168. [Google Scholar] [CrossRef]
- Bona, E.; Massa, N.; Toumatia, O.; Novello, G.; Cesaro, P.; Todeschini, V.; Boatti, L.; Mignone, F.; Titouah, H.; Zitouni, A.; et al. Climatic zone and soil properties determine the biodiversity of the soil bacterial communities associated to native plants from desert areas of north-central Algeria. Microorganisms 2021, 9, 1359. [Google Scholar] [CrossRef]
- Madsen, J.S.; Sørensen, S.J.; Burmølle, M. Bacterial social interactions and the emergence of community-intrinsic properties. Curr. Opin. Microbiol. 2018, 42, 104–109. [Google Scholar] [CrossRef]
- Pérez-Gutiérrez, R.-A.; López-Ramírez, V.; Islas, Á.; Alcaraz, L.D.; Hernández-González, I.; Olivera, B.C.L.; Santillán, M.; Eguiarte, L.E.; Souza, V.; Travisano, M.; et al. Antagonism influences assembly of a Bacillus guild in a local community and is depicted as a food-chain network. ISME J. 2013, 7, 487–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cairns, J.; Jokela, R.; Hultman, J.; Tamminen, M.; Virta, M.; Hiltunen, T. Construction and characterization of synthetic bacterial community for experimental ecology and evolution. Front. Genet. 2018, 9, 312. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Batista, B.D.; Bazany, K.E.; Singh, B.K. Plant-microbiome interactions under a changing world: Responses, consequences and perspectives. New Phytol. 2022, 234, 1951–1959. [Google Scholar] [CrossRef] [PubMed]
- Fadiji, A.E.; Babalola, O.O. Elucidating mechanisms of endophytes used in plant protection and other bioactivities with multifunctional prospects. Front. Bioengin. Biotechnol. 2020, 8, 467. [Google Scholar] [CrossRef] [PubMed]
- Hugenholtz, P.; Tyson, G.W. Microbiology: Metagenomics. Nature 2008, 455, 481–483. [Google Scholar] [CrossRef]
- Hajibabaei, M.; Singer, G.A.C.; Hebert, P.D.N.; Hickey, D.A. DNA barcoding: How it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet. 2007, 23, 167–172. [Google Scholar] [CrossRef]
- Sogin, M.L.; Morrison, H.G.; Huber, J.A.; Mark Welch, D.; Huse, S.M.; Neal, P.R.; Arrieta, J.M.; Herndl, G.J. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. USA 2006, 103, 12115–12120. [Google Scholar] [CrossRef] [Green Version]
- Andersson, A.F.; Lindberg, M.; Jakobsson, H.; Bäckhed, F.; Nyrén, P.; Engstrand, L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE 2008, 3, e2836. [Google Scholar] [CrossRef]
- Hajibabaei, M.; Shokralla, S.; Zhou, X.; Singer, G.A.; Baird, D.J. Environmental barcoding: A next-generation sequencing approach for biomonitoring applications using river benthos. PLoS ONE 2011, 6, e17497. [Google Scholar] [CrossRef] [Green Version]
- Boessenkool, S.; Epp, L.S.; Haile, J.; Bellemain, E.; Edwards, M.; Coissac, E.; Willerslev, E.; Brochmann, C. Blocking human contaminant DNA during PCR allows amplification of rare mammal species from sedimentary ancient DNA. Mol. Ecol. 2012, 21, 1806–1815. [Google Scholar] [CrossRef]
- Bartram, A.K.; Lynch, M.D.; Stearns, J.C.; Moreno-Hagelsieb, G.; Neufeld, J.D. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl. Environ. Microbiol. 2011, 77, 3846–3852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mason, O.U.; Hazen, T.C.; Borglin, S.; Chain, P.S.; Dubinsky, E.A.; Fortney, J.L.; Han, J.; Holman, H.Y.N.; Hultman, J.; Lamendella, R.; et al. Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J. 2012, 6, 1715–1727. [Google Scholar] [CrossRef] [PubMed]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Li, X.; Yin, L.; Liu, C.; Zou, H.; Wu, Z.; Zhang, Z. Analysis of the complete genome sequence of Brevibacterium frigoritolerans ZB201705 isolated from drought- and salt-stressed rhizosphere soil of maize. Ann. Microbiol. 2019, 69, 1489–1496. [Google Scholar] [CrossRef]
- Berrios, L. Plant-growth-promoting Caulobacter strains isolated from distinct plant hosts share conserved genetic factors involved in beneficial plant-bacteria interactions. Arch. Microbiol. 2021, 204, 43. [Google Scholar] [CrossRef]
- Duan, J.; Jiang, W.; Cheng, Z.; Heikkila, J.J.; Glick, B.R. The complete genome sequence of the Plant Growth-Promoting Bacterium Pseudomonas sp. UW4. PLoS ONE 2013, 8, e58640. [Google Scholar] [CrossRef] [Green Version]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Donati, C.; Medini, D.; Ward, N.L.; Angiuoli, S.V.; Crabtree, J.; Jones, A.L.; Durkin, A.S.; et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef] [Green Version]
- Golicz, A.A.; Bayer, P.E.; Bhalla, P.L.; Batley, J.; Edwards, D. Pangenomics comes of age: From bacteria to plant and animal applications. Trends Genet. 2020, 36, 132–145. [Google Scholar] [CrossRef]
- Costa, S.S.; Guimarães, L.C.; Silva, A.; Soares, S.C.; Baraúna, R.A. First steps in the analysis of Prokaryotic Pan-Genomes. Bioinf. Biol. Insights 2020, 14, 1177932220938064. [Google Scholar] [CrossRef] [PubMed]
- Brockhurst, M.A.; Harrison, E.; Hall, J.P.J.; Richards, T.; McNally, A.; MacLean, C. The ecology and evolution of pangenomes. Curr. Biol. 2019, 21, R1094–R1103. [Google Scholar] [CrossRef]
- Davies, M.R.; McIntyre, L.; Mutreja, A.; Lacey, J.A.; Lees, J.A.; Towers, R.J.; Duchêne, S.; Smeesters, P.R.; Frost, H.R.; Price, D.J.; et al. Atlas of group A streptococcal vaccine candidates compiled using large-scale comparative genomics. Nat. Genet. 2019, 51, 1035–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Land, M.; Hauser, L.; Jun, S.R.; Nookaew, I.; Leuze, M.R.; Ahn, T.H.; Karpinets, T.; Lund, O.; Kora, G.; Wassenaar, T.; et al. Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genom. 2015, 15, 141–161. [Google Scholar] [CrossRef] [Green Version]
- McInerney, J.O.; McNally, A.; O’Connell, M.J. Why prokaryotes have pangenomes. Nat. Microbiol. 2017, 2, 17040. [Google Scholar] [CrossRef]
- Rouli, L.; Merhej, V.; Fournier, P.E.; Raoult, D. The bacterial pangenome as a new tool for analysing pathogenic bacteria. New Microbes New Infect. 2015, 7, 72–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olanrewaju, O.S.; Ayilara, M.S.; Ayangbenro, A.S.; Babalola, O.O. Genome mining of three Plant Growth-Promoting Bacillus species from maize rhizosphere. Appl. Biochem. Biotechnol. 2021, 193, 3949–3969. [Google Scholar] [CrossRef]
- Compant, S.; Cambon, M.C.; Vacher, C.; Mitter, B.; Samad, A.; Sessitsch, A. The plant endosphere world—Bacterial life within plants. Environ. Microbiol. 2021, 23, 1812–1829. [Google Scholar] [CrossRef]
- Adeleke, B.S.; Ayilara, M.S.; Akinola, S.A.; Babalola, O.O. Biocontrol mechanisms of endophytic fungi. Egypt. J. Biol. Pest Control 2022, 32, 46. [Google Scholar] [CrossRef]
- Mahgoub, H.A.; Fouda, A.; Eid, A.M.; Ewais, E.E.-D.; Hassan, S.E.-D. Biotechnological application of plant growth-promoting endophytic bacteria isolated from halophytic plants to ameliorate salinity tolerance of Vicia faba L. Plant Biotechnol. Rep. 2021, 15, 819–843. [Google Scholar] [CrossRef]
- ALKahtani, M.D.F.; Fouda, A.; Attia, K.A.; Al-Otaibi, F.; Eid, A.M.; Ewais, E.E.-D.; Hijri, M.; St-Arnaud, M.; Hassan, S.E.-D.; Khan, N.; et al. Isolation and characterization of Plant Growth Promoting endophytic bacteria from desert plants and their application as bioinoculants for sustainable agriculture. Agronomy 2020, 10, 1325. [Google Scholar] [CrossRef]
- Glick, B.R. Bacterial ACC deaminase and the alleviation of plant stress. Adv. Appl. Microbiol. 2004, 56, 291–312. [Google Scholar] [PubMed]
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial endophyte colonization and distribution within plants. Microorganisms 2017, 25, 77. [Google Scholar] [CrossRef] [Green Version]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar] [CrossRef]
- Sessitsch, A.; Howieson, J.G.; Perret, X.; Antoun, H.; Martinez-Romero, E. Advances in Rhizobium research. Crit. Rev. Plant Sci. 2002, 21, 323–378. [Google Scholar] [CrossRef]
- Weilharter, A.; Mitter, B.; Shin, M.V.; Chain, P.S.; Nowak, J.; Sessitsch, A. Complete genome sequence of the plant growth-promoting endophyte Burkholderia phytofirmans strain PsJN. J. Bacteriol. 2011, 193, 3383–3384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitter, B.; Petric, A.; Shin, M.W.; Chain, P.S.; Hauberg-Lotte, L.; Reinhold-Hurek, B.; Nowak, J.; Sessitsch, A. Comparative genome analysis of Burkholderia phytofirmans PsJN reveals a wide spectrum of endophytic lifestyles based on interaction strategies with host plants. Front. Plant Sci. 2013, 4, 120. [Google Scholar] [CrossRef] [Green Version]
- Zúñiga, A.; Poupin, M.J.; Donoso, R.; Ledger, T.; Guiliani, N.; Gutiérrez, R.A.; González, B. Quorum-sensing and indole-3-acetic acid degradation play a role in colonization and plant growth promotion of Arabidopsis thaliana by Burkholderia phytofirmans PsJN. Mol. Plant Microbe Interact. 2013, 26, 546–553. [Google Scholar] [CrossRef] [Green Version]
- Horswill, A.R.; Stoodley, P.; Stewart, P.S.; Parsek, M.R. The effect of the chemical, biological, and physical environment on quorum sensing in structured microbial communities. Anal. Bioanal. Chem. 2007, 387, 371–380. [Google Scholar] [CrossRef] [Green Version]
- Ali, S.; Duan, J.; Charles, T.C.; Glick, B.R. A bioinformatics approach to the determination of bacterial genes involved in endophytic behavior. J. Theor. Biol 2014, 343, 193–198. [Google Scholar] [CrossRef]
- Orozco-Mosqueda, M.D.; Santoyo, G. Plant-microbial endophytes interactions: Scrutinizing their beneficial mechanisms from genomic explorations. Curr. Plant Biol. 2020, 25, 100189. [Google Scholar] [CrossRef]
- WHO. Antimicrobial Resistance: Global Report on Surveillance; WHO: Geneva, Switzerland, 2014. [Google Scholar]
- Cox, G.; Wright, G.D. Intrinsic antibiotic resistance: Mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 2013, 303, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Demanèche, S.; Sanguin, H.; Poté, J.; Navarro, E.; Bernillon, D.; Mavingui, P.; Wildi, W.; Vogel, T.M.; Simonet, P. Antibiotic-resistant soil bacteria in transgenic plant fields. Proc. Natl. Acad. Sci. USA 2008, 105, 3957–3962. [Google Scholar] [CrossRef] [Green Version]
- Song, J.S.; Jeon, J.H.; Lee, J.H.; Jeong, S.H.; Jeong, B.C.; Kim, S.J. Molecular characterization of TEM-type beta-lactamases identified in cold-seep sediments of Edison Seamount (south of Lihir Island, Papua New Guinea). J. Microbiol. 2005, 43, 172–178. [Google Scholar]
- D’costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R. Antibiotic resistance is ancient. Nature 2011, 477, 457. [Google Scholar] [CrossRef]
- Bhullar, K.; Waglechner, N.; Pawlowski, A.; Koteva, K.; Banks, E.D.; Johnston, M.D.; Barton, H.A.; Wright, G.D. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE 2012, 7, e34953. [Google Scholar] [CrossRef]
- Nesme, J.; Cécillon, S.; Delmont, T.O.; Monier, J.M.; Vogel, T.M.; Simonet, P. Large-scale metagenomic-based study of antibiotic resistance in the environment. Curr. Biol. 2014, 24, 1096–1100. [Google Scholar] [CrossRef] [Green Version]
- Monier, J.M.; Demanèche, S.; Delmont, T.O.; Mathieu, A.; Vogel, T.M.; Simonet, P. Metagenomic exploration of antibiotic resistance in soil. Curr. Opin. Microbiol. 2011, 14, 229–235. [Google Scholar] [CrossRef]
- Torsvik, V.; Øvreås, L. Microbial diversity and function in soil: From genes to ecosystems. Curr. Opin. Microbiol. 2002, 5, 240–245. [Google Scholar] [CrossRef]
- Ranjard, L.; Richaume, A. Quantitative and qualitative microscale distribution of bacteria in soil. Res. Microbiol. 2001, 152, 707–716. [Google Scholar] [CrossRef]
- Nunan, N. Spatial distribution of bacterial communities and their relationships with the micro-architecture of soil. FEMS Microbiol. Ecol. 2003, 44, 203–215. [Google Scholar] [CrossRef] [Green Version]
- Segata, N.; Börnigen, D.; Morgan, X.; Huttenhower, C. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat. Commun. 2013, 4, 2304. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, P.; Bond, P.L. Metaproteomics: Studying functional gene expression in microbial ecosystems. Trends Microbiol. 2006, 14, 92–97. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Hughes, M. Gene expression profiling: Metatranscriptomics. Meth. Mol. Biol. 2011, 733, 195–205. [Google Scholar]
- Dollive, S.; Peterfreund, G.L.; Sherrill-Mix, S.; Bittinger, K.; Sinha, R.; Hoffmann, C.; Nabel, C.; Hill, D.A.; Artis, D.; Bachman, M.A. A tool kit for quantifying eukaryotic rRNA gene sequences from human microbiome samples. Genome Biol. 2012, 13, R60. [Google Scholar] [CrossRef] [Green Version]
- Nicora, G.; Zucca, S.; Limongelli, I.; Bellazzi, R.; Magni, P. A machine learning approach based on ACMG/AMP guidelines for genomic variant classification and prioritization. Sci. Rep. 2022, 12, 2517. [Google Scholar] [CrossRef]
- Coutinho, B.G.; Licastro, D.; Mendonca-Previato, L.; Camara, M.; Venturi, V. Plant-influenced gene expression in the rice endophyte Burkholderia kururiensis M130. Mol. Plant Microbe Interact. 2015, 28, 10–21. [Google Scholar] [CrossRef] [Green Version]
- Levy, A.; Salas Gonzalez, I.; Mittelviefhaus, M.; Clingenpeel, S.; Herrera Paredes, S.; Miao, J.; Wang, K.; Devescovi, G.; Stillman, K.; Monteiro, F.; et al. Genomic features of bacterial adaptation to plants. Nat. Genet. 2018, 50, 138–150. [Google Scholar] [CrossRef] [Green Version]
- Stearns, J.C.; Woody, O.Z.; McConkey, B.J.; Glick, B.R. Effects of bacterial ACC deaminase on Brassica napus gene expression. Mol. Plant-Microbe Interact. 2012, 25, 668–676. [Google Scholar] [CrossRef] [Green Version]
- Suchan, D.M.; Bergsveinson, J.; Manzon, L.; Pierce, A.; Kryachko, Y.; Korber, D.; Tan, Y.; Tambalo, D.D.; Khan, N.H.; Whiting, M.; et al. Transcriptomics reveal core activities of the plant growth-promoting bacterium Delftia acidovorans RAY209 during interaction with canola and soybean roots. Microb. Genom. 2020, 6, mgen000462. [Google Scholar] [CrossRef] [PubMed]
- Samaras, A.; Kamou, N.; Tzelepis, G.; Karamanoli, K.; Menkissoglu-Spiroudi, U.; Karaoglanidis, G.S. Root transcriptional and metabolic dynamics induced by the Plant Growth Promoting Rhizobacterium (PGPR) Bacillus subtilis Mbi600 on cucumber plants. Plants 2022, 11, 1218. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014, 8, 790–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapelle, E.; Mendes, R.; Bakker, P.; Raaijmakers, J.M. Fungal invasion of the rhizosphere microbiome. ISME J 2016, 10, 265–268. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Z.; Duan, J.; Hao, Y.; McConkey, B.J.; Glick, B.R. Identification of bacterial proteins mediating the interaction between the plant growth-promoting bacterium Pseudomonas putida UW4 and Brassica napus (canola). Mol. Plant-Microbe Interact. 2009, 22, 686–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambais, M.R.; Barrera, S.E.; Santos, E.C.; Crowley, D.E.; Jumpponen, A. Phyllosphere metaproteomes of trees from the brazilian atlantic forest show high levels of functional redundancy. Microb. Ecol. 2017, 73, 123–134. [Google Scholar] [CrossRef]
- Kierul, K.; Voigt, B.; Albrecht, D.; Chen, X.H.; Carvalhais, L.C.; Borriss, R. Influence of root exudates on the extracellular proteome of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Microbiology 2015, 161, 131–147. [Google Scholar] [CrossRef] [Green Version]
- Bao, Y.Y.; Qin, X.; Yu, B.; Chen, L.B.; Wang, Z.C.; Zhang, C.X. Genomic insights into the serine protease gene family and expression profile analysis in the planthopper, Nilaparvata lugens. BMC Genom. 2014, 15, 507. [Google Scholar] [CrossRef] [Green Version]
- Bona, E.; Massa, N.; Novello, G.; Boatti, L.; Cesaro, P.; Todeschini, V.; Magnelli, V.; Manfredi, M.; Marengo, E.; Mignone, F.; et al. Metaproteomic characterization of the Vitis vinifera rhizosphere. FEMS Microbiol. Ecol. 2019, 95, fiy204. [Google Scholar] [CrossRef]
- Novello, G.; Gamalero, E.; Bona, E.; Boatti, L.; Mignone, F.; Massa, N.; Cesaro, P.; Lingua, G.; Berta, G. The rhizosphere bacterial microbiota of Vitis vinifera cv. Pinot Noir in an integrated pest management vineyard. Front. Microbiol. 2017, 8, 1528. [Google Scholar] [CrossRef]
- Domon, B.; Aebersold, R. Options and considerations when selecting a quantitative proteomics strategy. Nat. Biotechnol. 2010, 28, 710–721. [Google Scholar] [CrossRef] [PubMed]
- Nakayasu, E.S.; Nicora, C.D.; Sims, A.C.; Burnum-Johnson, K.E.; Kim, Y.M.; Kyle, J.E.; Matzke, M.M.; Shukla, A.K.; Chu, R.K.; Schepmoes, A.A.; et al. MPLEx: A robust and universal protocol for single-sample integrative proteomic, metabolomic, and lipidomic analyses. mSystems 2016, 1, e00043-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rochfort, S. Metabolomics reviewed: A new “omics” platform technology for systems biology and implications for natural products research. J. Nat. Prod. 2005, 68, 1813–1820. [Google Scholar] [CrossRef]
- Rochfort, S.; Ezernieks, V.; Mele, P.; Kitching, M. NMR metabolomics for soil analysis provide complementary, orthogonal data to MIR and traditional soil chemistry approaches—A land use study. Magn. Reson. Chem. 2015, 53, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Schillaci, M.; Roessner, U. Metabolomics as an emerging tool to study plant–microbe interactions. Emerg. Top. Life Sci. 2022, 6, 175–183. [Google Scholar] [CrossRef]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loqué, D.; Bowen, B.P.; et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018, 3, 470–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valette, M.; Rey, M.; Doré, J.; Gerin, F.; Wisniewski-Dyé, F. Identification of a small set of genes commonly regulated in rice roots in response to beneficial rhizobacteria. Physiol. Mol. Biol. Plants 2020, 26, 2537–2551. [Google Scholar] [CrossRef] [PubMed]
- Gamalero, E.; Lingua, G.; Berta, G.; Lemanceau, P. Methods for studying root colonization by introduced beneficial bacteria. Agronomie 2003, 23, 407–418. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, F.; Husain, F.M.; Ahmad, I. Rhizosphere and Root Colonization by bacterial inoculants and their monitoring methods: A critical area in PGPR research. In Microbes and Microbial Technology; Ahmad, I., Ahmad, F., Pichtel, J., Eds.; Springer: New York, NY, USA, 2011. [Google Scholar]
- Kang, Y.; Shen, M.; Xia, D.; Ye, K.; Zhao, Q.; Hu, J. Caution of intensified spread of antibiotic resistance genes by inadvertent introduction of beneficial bacteria into soil. Acta Agric. Scand. Sect. B Soil Plant Sci. 2017, 67, 576–582. [Google Scholar] [CrossRef]
- Miller, J.; Ferreira, P.; LeJeune, J.T. Antimicrobial Use and Resistance in Plant Agriculture: A One Health Perspective. Agriculture 2022, 12, 289. [Google Scholar] [CrossRef]
- Nescerecka, A.; Hammes, F.; Juhna, T. A pipeline for developing and testing staining protocols for flow cytometry, demonstrated with SYBR Green I and propidium iodide viability staining. J. Microbiol. Meth. 2016, 131, 172–180. [Google Scholar] [CrossRef]
- Kragelund, L.; Nybroe, O. Competition between Pseudomonas fluorescens Ag1 and Alcaligenes eutrophus JMP134 (pJP4) during colonization of barley roots. FEMS Microbiol. Ecol. 1996, 20, 41–51. [Google Scholar] [CrossRef]
- Sørensen, J.; Jensen, L.E.; Nybroe, O. Soil and rhizosphere as habitats for Pseudomonas inoculants: New knowledge on distribution, activity and physiological state derived from micro-scale and single-cell studies. Plant Soil 2001, 232, 97–108. [Google Scholar] [CrossRef]
- Troxler, J.; Zala, M.; Natsch, A.; Moënne-Loccoz, Y.; Défago, G. Autecology of the biocontrol strain Pseudomonas fluorescens CHA0 in the rhizosphere and inside roots at later stages of plant development. FEMS Microbiol. Ecol. 1997, 23, 119–130. [Google Scholar] [CrossRef]
- Gamalero, E.; Lingua, G.; Giusy Caprì, F.; Fusconi, A.; Berta, G.; Lemanceau, P. Colonization pattern of primary tomato roots by Pseudomonas fluorescens A6RI characterized by dilution plating, flow cytometry, fluorescence, confocal and scanning electron microscopy. FEMS Microbiol. Ecol. 2004, 48, 79–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalfie, M.; Tu, Y.; Euskirchen, G.; Ward, W.W.; Prasher, D.C. Green fluorescent protein as a marker for gene expression. Science 1994, 263, 802–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bloemberg, G.V.; Wijfjes, A.H.M.; Lamers, G.E.M.; Stuurman, N.; Lugtenberg, B.J.J. Simultaneous imaging of Pseudomonas fluorescens WCS365 populations expressing three different autofluorescent proteins in the rhizosphere: New perspectives for studying microbial communities. Mol. Plant Microbe Interact. 2000, 13, 1170–1176. [Google Scholar] [CrossRef] [Green Version]
- Matz, M.V.; Fradkov, A.F.; Labas, Y.A.; Savitsky, A.P.; Zaraisky, A.G.; Markelov, M.L.; Lukyanov, S.A. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 1999, 17, 969–973. [Google Scholar] [CrossRef]
- Daims, H.; Wagner, M. Quantification of uncultured microorganisms by fluorescence microscopy and digital image analysis. Appl. Microbiol. Biotechnol. 2007, 75, 237–248. [Google Scholar] [CrossRef]
- Tombolini, R.; Van Der Gaag, D.J.; Gerhardson, B.; Jansson, J.K. Colonization pattern of the biocontrol strain Pseudomonas chlororaphis MA 342 on barley seeds vizualized by using green fluorescent protein. Appl. Environ. Microbiol. 1999, 65, 3674–3680. [Google Scholar] [CrossRef] [Green Version]
- Tombolini, R.; Unge, A.; Davey, M.E.; de Bruijn, F.J.; Jansson, J.K. Flow cytometric and microscopic analysis of GFP-tagged Pseudomonas fluorescens bacteria. FEMS Microbiol. Ecol. 1997, 22, 17–28. [Google Scholar] [CrossRef]
- Bloemberg, G.V. Microscopic analysis of plant–bacterium interactions using auto fluorescent proteins. Eur. J. Plant Pathol. 2007, 119, 301–309. [Google Scholar] [CrossRef] [Green Version]
- Gamalero, E.; Lingua, G.; Tombolini, R.; Avidano, L.; Pivato, B.; Berta, G. Colonization of tomato root seedling by Pseudomonas fluorescens 92 rkG5: Spatio-temporal dynamics, localization, organization, viability, and culturability. Microb. Ecol. 2005, 50, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Monmeyran, A.; Thomen, P.; Jonquière, H.; Sureau, F.; Li, C.; Plamont, M.A.; Douarche, C.; Casella, J.F.; Gautier, A.; Henry, N. The inducible chemical-genetic fluorescent marker FAST outperforms classical fluorescent proteins in the quantitative reporting of bacterial biofilm dynamics. Sci. Rep. 2018, 8, 10336. [Google Scholar] [CrossRef] [Green Version]
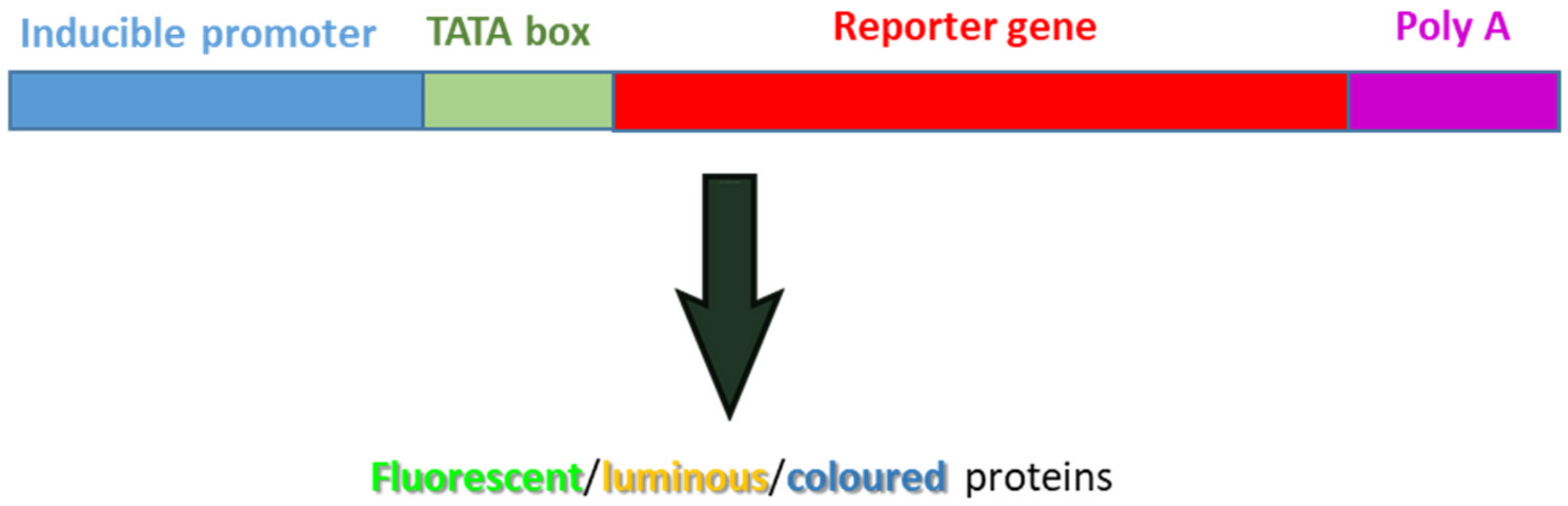
- Lindow, S.E. The use of reporter genes in the study of microbial ecology. Mol. Ecol. 1995, 4, 555–566. [Google Scholar] [CrossRef]
- Sørensen, J.; Nybroe, O. Reporter Genes in Bacterial Inoculants Can Monitor Life Conditions and Functions in Soil. In Nucleic Acids and Proteins; Nannipieri, P., Smalla, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 8. [Google Scholar]
- De Wet, J.R.; Wood, K.V.; DeLuca, M.; Helinski, D.R.; Subramani, S. Firefly luciferase gene:structure and expression in mammalian cells. Mol. Cell. Biol. 1987, 7, 725. [Google Scholar]
- Wood, K.V.; DeLuca, M. Photographic detection of luminescence in Escherichia coli containing the gene for firefly luciferase. Anal. Biochem. 1987, 161, 501–507. [Google Scholar] [CrossRef]
- Björklöf, K.; Jørgensen, K.S. Applicability of non-antibiotic resistance marker genes in ecological studies of introduced bacteria in forest soil. FEMS Microbiol. Ecol. 2001, 38, 179–188. [Google Scholar] [CrossRef]
- Kumar, S.S.; Ghosh, A.R. Assessment of bacterial viability: A comprehensive review on recent advances and challenges. Microbiology 2019, 165, 593–610. [Google Scholar] [CrossRef]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef]
- Harris, R.L.; Vetter, M.C.Y.L.; van Heerden, E.; Cason, E.; Vermeulen, J.G.; Taneja, A.; Kieft, T.L.; DeCoste, C.J.; Laevsky, G.S.; Onstott, T.C. FISH-TAMB, a fixation-free mRNA fluorescent labeling technique to target transcriptionally active members in microbial communities. Microb. Ecol. 2022, 84, 182–197. [Google Scholar] [CrossRef] [PubMed]
- Lukumbuzya, M.; Schmid, M.; Pjevac, P.; Daims, H. A Multicolor fluorescence in situ hybridization approach using an extended set of fluorophores to visualize microorganisms. Front. Microbiol. 2019, 10, 1383. [Google Scholar] [CrossRef]
- Musat, N.; Foster, R.; Vagner, T.; Adam, B.; Kuypers, M.M. Detecting metabolic activities in single cells, with emphasis on nanoSIMS. FEMS Microbiol. Rev. 2012, 36, 486–511. [Google Scholar] [CrossRef] [Green Version]
- McBratney, A.; Field, D.J.; Koch, A. The dimensions of soil security. Geoderma 2014, 213, 203–213. [Google Scholar] [CrossRef] [Green Version]
- Lynch, J. Root architecture and plant productivity. Plant Physiol. 1995, 109, 7–13. [Google Scholar] [CrossRef]
- Arsenault, J.-L.; Pouleur, S.; Messier, C.; Guay, R. WinRHIZO™, a root-measuring system with a unique overlap correction method. Hort Sci. 1995, 30, 906. [Google Scholar]
- Gamalero, E.; Martinotti, M.G.; Trotta, A.; Lemanceau, P.; Berta, G. Morphogenetic modifications induced by Pseudomonas fluorescens A6RI and Glomus mosseae BEG12 in the root system of tomato differ according to plant growth conditions. New Phytol. 2002, 155, 293–300. [Google Scholar] [CrossRef]
- Gamalero, E.; Trotta, A.; Massa, N.; Copetta, A.; Martinotti, M.G.; Berta, G. Impact of two fluorescent pseudomonads and an arbuscular mycorrhizal fungus on tomato plant growth, root architecture and P acquisition. Mycorrhiza 2004, 14, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Gamalero, E.; Berta, G.; Massa, N.; Glick, B.R.; Lingua, G. Synergistic interactions between the ACC deaminase-producing bacterium Pseudomonas putida UW4 and the AM fungus Gigaspora rosea positively affect cucumber plant growth. FEMS Microbiol. Ecol. 2008, 64, 459–467. [Google Scholar] [CrossRef]
- Gamalero, E.; Berta, G.; Massa, N.; Glick, B.R.; Lingua, G. Interactions between Pseudomonas putida UW4 and Gigaspora rosea BEG9 and their consequences on the growth of cucumber under salt stress conditions. J. Appl. Microbiol. 2010, 108, 236–245. [Google Scholar] [CrossRef]
- Mougel, C.; Offre, P.; Ranjard, L.; Corberand, T.; Gamalero, E.; Robin, C.; Lemanceau, P. Dynamic of the genetic structure of bacterial and fungal communities at different developmental stages of Medicago truncatula Gaertn. cv. Jemalong line J5. New Phytol. 2006, 170, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Cesaro, P.; Massa, N.; Cantamessa, S.; Todeschini, V.; Bona, E.; Berta, G.; Barbato, R.; Lingua, G. Tomato responses to Funneliformis mosseae during the early stages of arbuscular mycorrhizal symbiosis. Mycorrhiza 2020, 30, 601–610. [Google Scholar] [CrossRef]
- Berta, G.; Fusconi, A.; Trotta, A.; Scannerini, S. Morphogenetic modifications induced by the mycorrhizal fungus Glomus strain E3 in the root system of Allium porrum L. New Phytol. 1990, 114, 207–215. [Google Scholar] [CrossRef]
- Sturite, I.; Herkensen, T.M.; Breland, T.A. Distinguishing between metabolically active and inactive roots by combined staining with 2,3,5 triphenultetrazolium chloride and image colour analysis. Plant Soil 2005, 271, 75–82. [Google Scholar] [CrossRef]
- Seitaro Deguchi, S.; Matsuda, Y.; Takenaka, C.; Hajime, Y.S.; Ogata, Y. Proposal of a new estimation method of colonization rate of arbuscular mycorrhizal fungi in the roots of Chengiopanax sciadophylloides. Mycobiology 2017, 45, 15–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, K.; Kikuchi, S.; Yamasaki, S.I. Accurate root length measurement by image analysis. Plant Soil 1999, 216, 117–127. [Google Scholar] [CrossRef]
- Tajima, R.; Kato, Y. Comparison of threshold algorithms for automatic image processing of rice roots using freeware ImageJ. Field Crops Res. 2011, 121, 460–463. [Google Scholar] [CrossRef]
- Pierret, A.; Gonkhamdee, S.; Jourdan, C.; Maeght, J.L. IJ_Rhizo: An open-source software to measure scanned images of root samples. Plant Soil 2013, 373, 531–539. [Google Scholar] [CrossRef]
- Delory, B.M.; Weidlich, E.W.A.; Meder, L.; Lutje, A.; Van Duijnen, R.; Weidlich, R.; Temperton, V.M. Accuracy and bias of methods used for root length measurements in functional root research. Meth. Ecol. Evol. 2017, 8, 1594–1606. [Google Scholar] [CrossRef] [Green Version]
- Tennant, D. A test of modified line intersect method of estimating root length. J. Ecol. 1975, 63, 995–1001. [Google Scholar] [CrossRef]
- Seethepalli, A.; Dhakal, K.; Griffiths, M.; Guo, H.; Freschet, G.T.; York, L.M. RhizoVision Explorer: Open-source software for root image analysis and measurement standardization. AoB Plants 2021, 13, plab056. [Google Scholar] [CrossRef] [PubMed]
- Daglio, G.; Cesaro, P.; Todeschini, V.; Lingua, G.; Lazzari, M.; Berta, G.; Massa, N. Potential field detection of Flavescence dorée and Esca diseases using a ground sensing optical system. Biosyst. Eng. 2022, 215, 203–214. [Google Scholar] [CrossRef]
- Gresshoff, P.M.; Roth, L.E.; Stacey, G.; Newton, W.E. (Eds.) Nitrogen Fixation: Achievements and Objectives; Chapman & Hall: New York, NY, USA, 1990; pp. 1–869. [Google Scholar]
- Thamdrop, B. Pathways and processes in the global nitrogen cycle. Annu. Rev. Ecol. Evol. Syst. 2012, 43, 407–428. [Google Scholar] [CrossRef]
- Mylona, P.; Pawlowski, K.; Bisseling, T. Symbiotic nitrogen fixation. Plant Cell 1995, 7, 869–885. [Google Scholar] [CrossRef]
- Burris, R.; Roberts, G. Biological nitrogen fixation. Annu. Rev. Nutr. 1993, 13, 317–335. [Google Scholar] [CrossRef] [PubMed]
- Badalyan, A.; Yang, Z.-Y.; Hu, B.; Luo, J.; Hu, M.; Liu, T.L.; Seefeldt, L.C. An efficient violgen-based electron donor to nitrogenase. Biochemistry 2019, 58, 4590–4595. [Google Scholar] [CrossRef]
- Glick, B.R.; Martin, W.G.; Giroux, J.J.; Williams, R.E. The interaction of polymeric viologens with hydrogenases from Desulfovibrio desulfuricans and Clostridium pasteurianum. Can. J. Biochem. 1979, 57, 1093–1098. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.M.; Glick, B.R.; Martin, W.G. Factors affecting the production of hydrogenase by Desulfovibrio desulfuricans. Can. J. Microbiol. 1980, 26, 1209–1213. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.L.; Hume, D.J. Comparison of assay methods for nitrogen fixation utilizing white bean and soybean. Can. J. Plant Sci. 1987, 67, 11–19. [Google Scholar] [CrossRef]
- Haskett, T.L.; Knights, H.E.; Jorrin, B.; Mendes, M.D.; Poole, P.S. A simple in situ assay to assess plant-associative bacterial nitrogenase activity. Front. Microbiol. 2021, 12, 690439. [Google Scholar] [CrossRef]
- Kloepper, B.; Kaspar, T.C. Rhizotrons: Their development and use in agricultural research. Agron. J. 1994, 86, 745–753. [Google Scholar] [CrossRef]
- Vamerali, T.; Ganis, A.; Bona, S.; Mosca, G. An approach to minirhizotron root image analysis. Plant Soil 1999, 217, 183–193. [Google Scholar] [CrossRef]
- Rogers, W.S. The East Mailing root-observation laboratories. In Root Growth; Whittington, J.W., Ed.; Butterworth: London, UK, 1969; pp. 361–376. [Google Scholar]
- Mohamed, A.; Monnier, Y.; Mao, Z.; Lobet, G.; Maeght, J.L.; Ramel, M.; Stokes, A. An evaluation of inexpensive methods for root image acquisition when using rhizotrons. Plant Meth. 2017, 13, 11. [Google Scholar] [CrossRef] [PubMed]
- Van As, H.; van Duynhoven, J. MRI of plants and foods. J. Magn. Reson. 2013, 229, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Metzner, R.; Eggert, A.; van Dusschoten, D.; Pflugfelder, D.; Gerth, S.; Schurr, U.; Uhlmann, N.; Jahnke, S. Direct comparison of MRI and X-ray CT technologies for 3D imaging of root systems in soil: Potential and challenges for root trait quantification. Plant Meth. 2015, 11, 17. [Google Scholar] [CrossRef]
- Moradi, A.B.; Carminati, A.; Vetterlein, D.; Vontobel, P.; Lehmann, E.; Weller, U.; Hopmans, J.W.; Vogel, H.J.; Oswald, S.E. Three-dimensional visualization and quantification of water content in the rhizosphere. New Phytol. 2011, 192, 653–663. [Google Scholar] [CrossRef] [Green Version]
- Liao, R.W.; Liu, J.M.; An, S.Q.; Niu, J.; Liang, H.; Ren, S.; Le, Z.; Cao, Y.; Li, W. Monitor of corn root growth in soil based on minirhizotron technique. Trans. Chin. Soc. Agric. Eng. 2010, 26, 156–161. [Google Scholar]
- Abramoff, M.D.; Magalhaes, P.J.; Ram, S.J. Image processing with Image. J. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Le Bot, J.; Serra, V.; Fabre, J.; Draye, X.; Adamowicz, S.; Pages, L. DART: A software to analyse root system architecture and development from captured images. Plant Soil 2010, 326, 261–273. [Google Scholar] [CrossRef]
- Krzyzaniak, Y.; Cointault, F.; Loupiac, C.; Bernaud, E.; Ott, F.; Salon, C.; Laybros, A.; Han, S.; Héloir, M.-C.; Adrian, M.; et al. In situ phenotyping of grapevine root system architecture by 2D or 3D imaging: Advantages and limits of three cultivation methods. Front. Plant Sci. 2021, 12, 638688. [Google Scholar] [CrossRef]
- Maskova, T.; Klimes, A. The effects of rhizoboxes on plant growth and root:shoot biomass partitioning. Front. Plant Sci. 2020, 10, 1693. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, S.T.; Burr, A.A.; Reeb, R.A.; Melero Pardo, A.L.; Woods, K.D.; Wood, C.W. Using clear plastic CD cases as low-cost mini-rhizotrons to phenotype root traits. Appl. Plant Sci. 2020, 8, e11340. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.G.; Tingey, D.T.; Phillips, D.L.; Storm, M.J. Advancing fine root research with minirhizotrons. Environ. Exp. Bot. 2001, 45, 263–289. [Google Scholar] [CrossRef]
- Xu, S.; Gong, P.; Ding, W.; Wu, S.; Yu, X.; Liang, P. Mercury uptake by Paspalum distichum L. in relation to the mercury distribution pattern in rhizosphere soil. Environ. Sci. Pollut. Res. Int. 2021, 28, 66990–66997. [Google Scholar] [CrossRef]
- Li, N.; Możdżeń, K.; Zhang, Z.; Liu, C.; Zandi, P.; Sardar, M.F.; Zhu, C. Rhizosphere effect on removal and bioavailability of PAHs in contaminated agricultural soil. Biologia 2021, 76, 841–851. [Google Scholar] [CrossRef]
- Vahedi, R.; Rasouli-Sadaghiani, M.; Barin, M.; Vetukuri, R.R. Interactions between biochar and compost treatment and mycorrhizal fungi to improve the qualitative properties of a calcareous soil under rhizobox conditions. Agriculture 2021, 11, 993. [Google Scholar] [CrossRef]
- Zhou, J.; Chai, X.; Zhang, L.; George, T.S.; Wang, F.; Feng, G. Different arbuscular mycorrhizal fungi co-colonizing on a single plant root system recruit distinct microbiomes. Systems 2020, 5, e00929-20. [Google Scholar]
- Moro, H.; Park, H.-D.; Kunito, T. Organic phosphorus substantially contributes to crop plant nutrition in soils with low phosphorus availability. Agronomy 2021, 11, 903. [Google Scholar] [CrossRef]
- Romain, M.; Gloaguen, Z.; Brym, T.; Peeples, J.; Xu, W.; Chun, H.C.; Rowland, D.L. The plasticity of early root development in Sesamum indicum L. as influenced by genotype, water, and nutrient availability. Rhizosphere 2022, 21, 100457. [Google Scholar]
- Bontpart, T.; Concha, C.; Giuffrida, M.V.; Robertson, I.; Admkie, K.; Degefu, T.; Girma, N.; Tesfaye, K.; Haileselassie, T.; Fikre, A.; et al. Affordable and robust phenotyping framework to analyse root system architecture of soil-grown plants. Plant J. 2020, 103, 2330–2343. [Google Scholar] [CrossRef]
- Wu, J.; Wu, Q.; Pagès, L.; Yuan, Y.; Zhang, X.; Du, M.; Tian, X.; Li, Z. RhizoChamber-Monitor: A robotic platform and software enabling characterization of root growth. Plant Meth. 2018, 14, 44. [Google Scholar] [CrossRef] [PubMed]
- Alsalem, M.; Salehi, A.; Zhao, J.; Rewald, B.; Bodner, G. Combining image analyses tools for comprehensive characterization of root systems from soil-filled rhizobox phenotyping platforms. Int. Agrophys. 2021, 35, 257–268. [Google Scholar] [CrossRef]
- Kassaw, T.K.; Frugoli, J.A. Simple and efficient methods to generate split roots and grafted plants useful for long-distance signaling studies in Medicago truncatula and other small plants. Plant Meth. 2012, 8, 38. [Google Scholar] [CrossRef] [Green Version]
- Marino, D.; Damiani, I.; Gucciardo, S.; Mijangos, I.; Pauly, N.; Puppo, A. Inhibition of nitrogen fixation in symbiotic Medicago truncatula upon Cd exposure is a local process involving leghemoglobin. J. Exp. Bot. 2013, 64, 5651–5660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larrainzar, E.; Gil-Quintana, E.; Arrese-Igor, C.; González, E.M.; Marino, D. Split-root systems applied to the study of the legume-rhizobial symbiosis: What have we learned? J. Integr. Plant Biol. 2014, 56, 1118–1124. [Google Scholar] [CrossRef] [PubMed]
- Thilakarathna, M.S.; Cope, K.R. Split-root assays for studying legume–rhizobia symbioses, rhizodeposition, and belowground nitrogen transfer in legumes. J. Exp. Bot. 2021, 72, 5285–5299. [Google Scholar] [CrossRef] [PubMed]
- Long, E.M. The effect of salt additions to the substrate on intake of water and nutrients by roots of approach-grafted tomato plants. Am. J. Bot. 1943, 30, 594–601. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, L.; Wu, R. Plant grafting: How genetic exchange promotes vascular reconnection. New Phytol. 2017, 214, 56–65. [Google Scholar] [CrossRef] [Green Version]
- Alexander, M. Ecological consequences: Reducing the uncertainties. Issues Sci. Technol. 1985, 1, 57–68. [Google Scholar]
- Glick, B.R.; Skof, Y.C. Environmental implications of recombinant DNA technology. Biotechnol. Adv. 1986, 4, 261–277. [Google Scholar] [CrossRef]
- De Leij, F.A.A.M.; Sutton, E.J.; Whipps, J.M.; Fenlon, J.S.; Lynch, J.M. Field release of a genetically modified Pseudomonas fluorescens on wheat: Establishment, survival and dissemination. Bio/Technology 1985, 13, 1488–1492. [Google Scholar] [CrossRef]
- Tiedje, J.M.; Colwell, R.K.; Grossman, Y.L.; Hodson, R.E.; Lenski, R.E.; Mack, R.N.; Regal, P.J. The planned introduction of genetically engineered organisms: Ecological considerations and recommendations. Ecology 1989, 70, 298–315. [Google Scholar] [CrossRef]
- Prosser, J.I. Molecular marker systems for detection of genetically engineered micro-organisms in the environment. Microbiology 1994, 140, 5–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glick, B.R. Metabolic load and heterologous gene expression. Biotechnol. Adv. 1995, 13, 247–261. [Google Scholar] [CrossRef]
- Lynch, J.M. The potential for gene exchange between rhizosphere bacteria. In Bacterial Genetics in Natural Environments; Fry, J.C., Day, M.J., Eds.; Chapman and Hall: London, UK, 1990; pp. 172–181. [Google Scholar]
- Natsch, A.; Troxler, J.; Défago, G. Assessment of risks associated with the release of wild-type and genetically modified plant growth promoting rhizobacteria. In Plant Growth-Promoting Rhizobacteria: Present Status and Future Prospects; Ogoshi, A., Kobayashi, K., Homma, Y., Kodama, F., Kondo, N., Akino, S., Eds.; OECD: Paris, France, 1997; pp. 87–92. [Google Scholar]
- Wilson, M.; Lindow, S.E. Release of recombinant organisms. Annu. Rev. Microbiol. 1993, 47, 913–944. [Google Scholar] [CrossRef]
- Eriksson, D. Recovering the original intentions of risk assessment and management of genetically modified organisms in the European Union. Front. Bioeng. Biotechnol. 2018, 6, 52. [Google Scholar] [CrossRef]
- Dolgin, E. GM microbes created that can’t escape the lab. Nature 2015, 517, 423. [Google Scholar] [CrossRef] [Green Version]
- Hirsch, P.R. Release of transgenic bacterial inoculants—Rhizobia as a case study. Plant Soil 2004, 266, 1–10. [Google Scholar] [CrossRef]
- Ronson, C.W.; Bosworth, A.H.; Genova, M.; Gudbrandsen, S.; Hankinson, T.; Kwiatowski, R.; Ratcliffe, H.; Robie, C.; Sweeney, P.; Szeto, W.; et al. Field release of genetically engineered Rhizobium meliloti and Bradyrhizobium japonicum strains. In Nitrogen Fixation: Achievements and Objectives; Gresshoff, L.E., Stacey, G., Newton, W.E., Eds.; Chapman and Hall: New York, NY, USA, 1990; pp. 397–403. [Google Scholar]
- O’Flaherty, S.; Moënne-Loccoz, Y.; Boesten, B.; Higgins, P.; Dowling, D.N.; Condon, S.; O’Gara, F. Greenhouse and field evaluations of an autoselective system based on an essential thymidylate synthase gene for improved maintenance of plasmid vectors in modified Rhizobium meliloti. Appl. Environ. Microbiol. 1995, 61, 4051–4056. [Google Scholar] [CrossRef] [Green Version]
- Van Dillewijn, P.; Soto, M.J.; Villadas, P.J.; Toro, N. Construction and environmental release of a Sinorhizobium meliloti strain genetically modified to be more competitive for alfalfa nodulation. Appl. Environ. Microbiol. 2001, 67, 3860–3865. [Google Scholar] [CrossRef] [Green Version]
- Robleto, E.A.; Kmiecik, K.; Oplinger, E.S.; Nienhuis, J.; Triplett, E.W. Trifolitoxin production increases nodulation competitiveness of Rhizobium etli CE3 under agricultural conditions. Appl. Environ. Microbiol. 1998, 64, 2630–2633. [Google Scholar] [CrossRef] [Green Version]
- Hirsch, P.R.; Spokes, J.D. Survival and dispersion of genetically-modified rhizobia in the field and genetic interactions with native strains. FEMS Microbiol. Ecol. 1994, 15, 147–159. [Google Scholar] [CrossRef]
- Pitkäjärvi, J.; Räsänen, L.; Langenskiöld, J.; Wallenius, K.; Niem, M.; Lindström, K. Persistence, population dynamics and competitiveness for nodulation of marker gene tagged Rhizobium galegae strains in field lysimeters in the boreal climatic zone. FEMS Microbiol. Ecol. 2003, 46, 91–104. [Google Scholar] [CrossRef] [Green Version]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A. A programmable dual-RNA-guided endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.L.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.B.; Jiang, W.Y.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 5, 1173–1183. [Google Scholar] [CrossRef] [Green Version]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [Green Version]
- Yi, Y.; Li, Z.; Song, C.; Kuipers, O.P. Exploring plant-microbe interactions of the rhizobacteria Bacillus subtilis and Bacillus mycoides by use of the CRISPR-Cas9 system. Environ. Microbiol. 2018, 20, 4245–4260. [Google Scholar] [CrossRef] [Green Version]
- Wenderoth, M.; Pinecker, C.; Voß, B.; Fischer, R. Establishment of CRISPR/Cas9 in Alternaria alternata. Fungal Genet. Biol. 2017, 101, 55–60. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Zhang, Y.; Yu, P.L.; Pan, H.; Rollins, J.A. Introduction of large sequence inserts by CRISPR/Cas9 to create pathogenicity mutants in the multinucleate filamentous pathogen Sclerotinia sclerotiorum. MBio 2018, 9, e00567-18. [Google Scholar] [CrossRef] [Green Version]
- Miao, J.; Chi, Y.; Lin, D.; Tyler, B.M.; Liu, X. Mutations in ORP1 conferring oxathiapiprolin resistance by genome editing using CRISPR/Cas9 in Phytophthora capsica and P. sojae. Phytopathology 2018, 108, 412–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, M.; Okamura, Y.; Iwai, H. Plasmid-based and -free methods using CRISPR/Cas9 system for replacement of targeted genes in Colletotrichum samsevieriae. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Cobine, P.A.; Coleman, J.J. Efficient genome editing in Fusarium oxysporum based on CRISPR/Cas9 ribonucleoprotein complexes. Fungal Genet. Biol. 2018, 117, 21–29. [Google Scholar] [CrossRef]
- Fang, C.; Chen, X. Potential biocontrol efficacy of Trichoderma atroviride with cellulase expression regulator ace1 gene knock-out. Biotech 2018, 8, 302. [Google Scholar] [CrossRef]
- Kong, G.; Wan, L.; Deng, Y.Z.; Yang, W.; Li, W.; Jiang, L.; Situ, J.; Xi, P.; Li, M.; Jiang, Z. Pectin acetylesterase PAE5 is associated with the virulence of plant pathogenic oomycete Peronophythora litchi. Physiol. Mol. Plant Pathol. 2019, 106, 16–22. [Google Scholar] [CrossRef]
- Rilling, J.I.; Maruyama, F.; Sadowsky, M.J.; Acuna, J.J.; Jorquera, M.A. CRISPR loci-PCR as tool for tracking Azospirillum sp. strain B510. Microorganisms 2021, 9, 1351. [Google Scholar] [CrossRef]
- Hwang, I.S.; Oh, E.J.; Lee, H.B.; Oh, C.S. Functional characterization of two cellulase genes in the Gram-positive pathogenic bacterium Clavibacter michiganensis for wilting in tomato. Mol. Plant-Microbe Interact. 2019, 32, 491–501. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Tan, Q.; Lyu, Q.; Yu, C.; Jiang, N.; Li, J.; Luo, L. Unmarked gene editing in Clavibacter michiganensis using CRISPR/Cas9and 5-fluorocytosine counterselection. Mol. Plant Microbe Interact. 2022, 35, 4–14. [Google Scholar] [CrossRef]
- Glick, B.R. Phytoremediation: Synergistic use of plants and bacteria to clean up the environment. Biotechnol. Adv. 2003, 21, 383–393. [Google Scholar] [CrossRef]
- Glick, B.R. Using soil bacteria to facilitate phytoremediation. Biotechnol. Adv. 2010, 28, 367–374. [Google Scholar] [CrossRef]
- Basharat, Z.; Novo, L.; Yasmin, A. Genome editing weds CRISPR: What is in it for phytoremediation? Plants 2018, 7, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, V.C.; Gajic, G.; Sharma, P.; Roy, M. Designer plant for climate-resilient phytoremediation. In Adaptive Phytoremediation Practices: Resilience to Climate Change; Elsevier: Amsterdam, The Netherlands, 2022; pp. 227–274. [Google Scholar]
- Bashan, Y.; de-Bashan, L.E.; Prabhu, S.R.; Hernandez, J.-P. Advances in plant growth-promoting bacterial inoculant technology: Formulations and practical perspectives (1998–2013). Plant Soil 2014, 378, 1–33. [Google Scholar] [CrossRef] [Green Version]
- Vassilev, N.; Vassileva, M.; Lopez, D.; Martos, V.; Reyes, A.; Maksimivich, I.; Eichler-Löbermann, B.; Malusa, E. Unexploited potential of some biotechnological techniques for biofertilizer production and formulation. Appl. Microbiol. Biotechnol. 2015, 99, 4983–4996. [Google Scholar] [CrossRef] [PubMed]
- Arora, N.K.; Mishra, J. Prospecting the roles of metabolites and additives in future bioformulations for sustainable agriculture. Appl. Soil Ecol. 2016, 107, 405–407. [Google Scholar] [CrossRef]
- Malusá, E.; Pinzari, F.; Canfora, L. Efficacy of biofertilizers: Challenges to improve crop production. In Microbial Inoculants in Sustainable Agricultural Productivity; Singh, D., Singh, H., Prabha, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 17–40. [Google Scholar]
- Martinez-Viveros, O.A.; Jorquera, M.A.; Crowley, D.E.; Gajardo, G.; Mora, M.D. Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. J. Soil Sci. Plant Nutr. 2010, 10, 293–319. [Google Scholar] [CrossRef] [Green Version]
- Bashan, Y.; de-Bashan, L.E.; Prabhu, S.R. Superior polymeric formulations and emerging innovative products of bacterial inoculants for sustainable agricultura and the environment. In Agriculturally Important Microorganisms—Commercialization and Regulatory Requirements in Asia; Singh, H.B., Sarma, B.K., Keswani, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 15–46. [Google Scholar]
- Baez-Rogelio, A.; Morales-Garcia, Y.E.; Quintero-Hernandez, V.; Muños-Rojas, J. Next generation of microbial inoculants for agriculture and bioremediation. Microb. Biotechnol. 2017, 10, 19–21. [Google Scholar] [CrossRef]
- Stamenkovic, S.; Beskoski, V.; Karabegovic, I.; Lazic, M.; Nikolic, N. Microbial fertilizers: A comprehensive review on current findings and future perspectives. Span. J. Agric. Res. 2018, 16, e09R01. [Google Scholar] [CrossRef] [Green Version]
- John, R.P.; Tyagi, R.D.; Brar, S.K.; Surmpalli, R.Y.; Prevost, D. Bio-encapsulation of microbial cells for target agricultural delivery. Crit. Rev. Biotechnol. 2011, 31, 211–226. [Google Scholar] [CrossRef]
- Schoebitz, M.; López, M.; Roldán, A. Bioencapsulation of microbial inoculants for better soil–plant fertilization. A review. Agron. Sustain. Dev. 2013, 33, 751–765. [Google Scholar] [CrossRef]
- Mattos, B.D.; Tardy, B.L.; Magalhaes, W.L.E.; Rojas, O.J. Controlled release for crop and wood protection: Recent progress toward sustainable and safe nanostructured biocidal systems. J. Control. Release 2017, 262, 139–150. [Google Scholar] [CrossRef]
- Mejri, D.; Gamalero, E.; Souissi, T. Formulation development of the deleterious rhizobacterium Pseudomonas trivialis X33d for biocontrol of brome (Bromus diandrus) in durum wheat. J. Appl. Microbiol. 2013, 114, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Vejan, P.; Khadiran, T.; Abdullah, R.; Ismail, S.; Dadrasnia, A. Encapsulation of plant growth promoting Rhizobacteria—prospects and potential in agricultural sector: A review. J. Plant Nutr. 2019, 42, 2600–2623. [Google Scholar] [CrossRef]
- Rekha, P.D.; Lai, W.A.; Arun, A.B.; Young, C.C. Effect of free and encapsulated Pseudomonas putida CC-FR2-4 and Bacillus subtilis CC-pg104 on plant growth under gnotobiotic conditions. Bioresour. Technol. 2007, 98, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Varankovich, N.V.; Khan, N.H.; Nickerson, M.T.; Kalmokoff, M.; Korber, D.R. Evaluation of pea protein–polysaccharide matrices for encapsulation of acid-sensitive bacteria. Food Res. Int. 2015, 70, 118–124. [Google Scholar] [CrossRef]
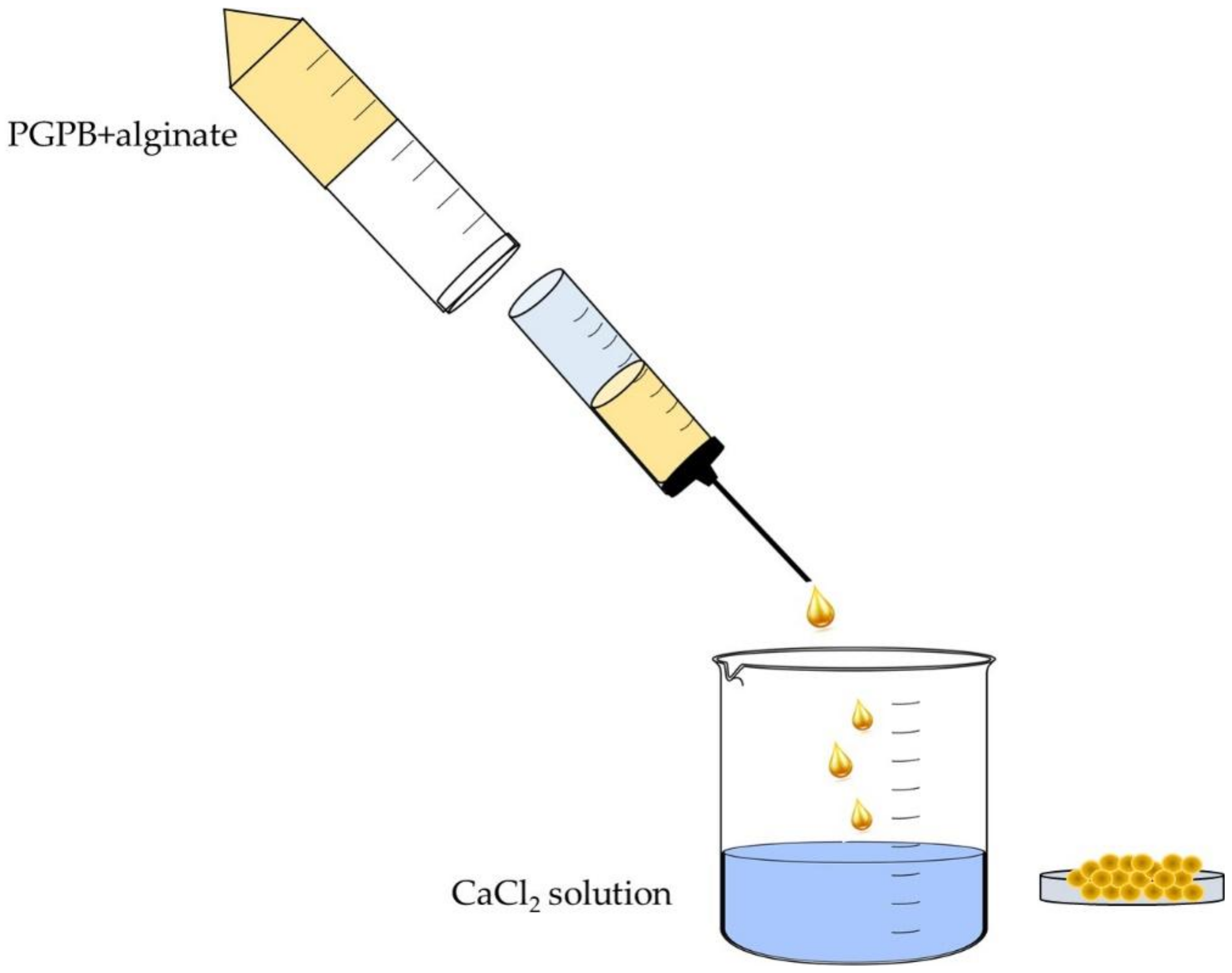
- Saberi Riseh, R.; Skorik, Y.A.; Thakur, V.K.; Moradi Pour, M.; Tamanadar, E.; Noghabi, S.S. Encapsulation of Plant Biocontrol Bacteria with Alginate as a Main Polymer Material. Int. J. Mol. Sci. 2021, 22, 11165. [Google Scholar] [CrossRef]
- Khan, N.H.; Korber, D.R.; Low, N.H.; Nickerson, M.T. Development of extrusion-based legume protein isolate–alginate capsules for the protection and delivery of the acid sensitive probiotic, Bifidobacterium adolescentis. Food Res. Int. 2013, 54, 730–737. [Google Scholar] [CrossRef]
- Nesterenko, A.; Alric, I.; Silvestre, F.; Durrieu, V. Vegetable proteins in microencapsulation: A review of recent interventions and their effectiveness. Ind. Crops Prod. 2013, 42, 469–479. [Google Scholar] [CrossRef] [Green Version]
- Kosaraju, S.L. Colon targeted delivery systems: Review of polysaccharides for encapsulation and delivery. Crit. Rev. Food Sci. Nutr. 2005, 45, 251–258. [Google Scholar] [CrossRef]
- Yeung, T.W.; Üçok, E.F.; Tiani, K.A.; McClements, D.J.; Sela, D.A. Microencapsulation in alginate and chitosan microgels to enhance viability of Bifidobacterium longum for oral delivery. Front. Microbiol. 2016, 7, 494. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Ma, X.; Lang, D.; Zhang, X.; Zhou, L.; Wang, L.; Zhang, X. Encapsulation of Bacillus pumilus G5 from polyvinyl alcohol-sodium alginate (PVA-SA) and its implications in improving plant growth and soil fertility under drought and salt soil conditions. Int. J. Biol. Macromol. 2022, 209 Pt A, 231–243. [Google Scholar] [CrossRef]
- Zhi, J.; Zhang, B.; Wu, Y.; Feng, Z. Study on a series of main-chain liquid–crystalline ionomers containing sulfonate groups. J. Appl. Polym. Sci. 2001, 81, 2210–2218. [Google Scholar] [CrossRef]
- Korus, J. Microencapsulation of flavours in starch matrix by coacervation method. Pol. J. Food Nutr. Sci. 2001, 51, 17–23. [Google Scholar]
- Immordino, M.L.; Dosio, F.; Cattel, L. Stealth liposomes: Review of the basic science, rationale, and clinical applications, existing and potential. Int. J. Nanomed. 2006, 1, 297–315. [Google Scholar]
- Lim, F.; Sun, A.M. Microencapsulated islets as bioartificial endocrine pancreas. Science 1980, 210, 908–910. [Google Scholar] [CrossRef]
- Rosas-Flores, W.; Ramos-Ramırez, E.G.; Salazar-Montoya, J.A. Microencapsulation of Lactobacillus helveticus and Lactobacillus delbrueckii using alginate and gellan gum. Carbohydr. Polym. 2013, 98, 1011–1017. [Google Scholar] [CrossRef]
- Anandhakumar, S.; Krishnamoorthy, G.; Ramkumar, K.M.; Raichur, A.M. Preparation of collagen peptide functionalized chitosan nanoparticles by ionic gelation method: An effective carrier system for encapsulation and release of doxorubicin for cancer drug delivery. Mat. Sci. Eng. C 2017, 70, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Koyama, K.; Seki, M. Evaluation of mass-transfer characteristics in alginate-membrane liquid-core capsules prepared using polyethylene glycol. J. Biosci. Bioeng. 2004, 98, 114–121. [Google Scholar] [CrossRef]
- Sasaki, E.; Kuruyama, F.; Ida, J.; Matsuyama, T.; Yamamoto, H. Preparation of microcapsules by electrostatic atomization. J. Electrostat. 2008, 66, 312–318. [Google Scholar] [CrossRef]
- Gagne-Bourque, F.; Xu, M.; Dumont, M.J.; Jabaji, S. Pea protein alginate encapsulated Bacillus subtilis B26, a plant biostimulant, provides controlled release and increased storage survival. J. Biofertil. Biopest. 2015, 6. [Google Scholar] [CrossRef]
- Pacheco-Aguirre, J.; Ruiz-Sanchez, E.; Reyes-Ramırez, A.; Cristobal-Alejo, J.; Tun-Suarez, J.; Borges-Gomez, L. Polymer-based encapsulation of Bacillus subtilis and its effect on Meloidogyne incognita in tomato. Phyton Int. J. Exp. Bot. 2015, 85, 1–6. [Google Scholar]
- He, Y.; Wu, Z.; Tu, L.; Shan, C. Effect of encapsulated Pseudomonas putida Rs-198 strain on alleviating salt stress of cotton. J. Plant Nutr. 2017, 40, 1180–1189. [Google Scholar] [CrossRef]
- Vassilev, N.; Vassileva, M.; Martos, V.; Garcia Del Moral, L.F.; Kowalska, J.; Tylkowski, B.; Malusá, E. Formulation of microbial inoculants by encapsulation in natural polysaccharides: Focus on beneficial properties of carrier additives and derivatives. Front. Plant Sci. 2020, 11, 270. [Google Scholar] [CrossRef] [PubMed]
- Dixshit, A.; Shukla, S.K.; Mishra, R.K. Exploring Nanomaterials with PGPR in Current Agriculture Scenario PGPR with Special Reference to Nanomaterials; Lab Lambert Acedamic Publication: Saarbrücken, Germany, 2013; p. 51. [Google Scholar]
- Panichikkal, J.; Edayileveetil Krishnankutty, R. Rhizobacterial biofilm and plant growth promoting trait enhancement by organic acids and sugars. Biofouling 2020, 36, 990–999. [Google Scholar] [CrossRef] [PubMed]
- Pour, M.M.; Saberi-Riseh, R.; Mohammadinejad, R.; Hosseini, A. Nano-encapsulation of Plant Growth-Promoting Rhizobacteria and their metabolites using alginate-silica nanoparticles and carbon nanotube improves UCB1 pistachio micropropagation. J. Microbiol. Biotechnol. 2019, 28, 1096–1103. [Google Scholar]





| Advantages | Limitations | |
|---|---|---|
| Next-generation sequencing (NGS) | Possibility to identify in the same analysis all the species present in the sample and so also the unculturable strains. | No information regarding the activity of the identified bacteria and also the role of each species/interaction with other species only predictive data |
| Whole metagenome shotgun sequencing | Information regarding all the genomes present in the microbial communities | No information regarding the expression of the different genes |
| Metatranscriptomic | Detailed information regarding the transcripted genes (RNAs). | Low content of RNA in soil. RNA is a molecule with low stability. No information regarding the enzyme translation. |
| Metaproteomic | Detailed information regarding proteins and so the effective microbial work in the soil. Information regarding the species that produce the protein and so the role of each species. | Low amount of proteins in the soil sample/difficulties in the protein extraction and purification due to contaminant molecules present in soil. Problems with species attribution/necessity of an in-house database produced by NGS analysis. |
| Metabolomics | Targeted or untargeted metabolomics can be used to measure changes in specific metabolite levels in response to a given treatment. Detailed information regarding the produced metabolite. | Difficulties to attribute the species that produce the identified metabolite. Difficulties in the purification of the metabolites present in soil and the quantification. |
| Bacterium | Method of Application | Test Country | References |
|---|---|---|---|
| Ensifer meliloti | Alfalfa spray inoculation | USA | [188] |
| E. meliloti | Alfalfa seed coating | Ireland, Spain | [189,190] |
| Bradyrhizobium japonicum | Soybean seed coating | USA | [188] |
| Rhizobium etli | Bean seed coating | USA | [191] |
| Rhizobium leguninosarum | Liquid seed coating | France | [192] |
| Rhizobium galegae | Liquid seed coating | Finland | [193] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gamalero, E.; Bona, E.; Glick, B.R. Current Techniques to Study Beneficial Plant-Microbe Interactions. Microorganisms 2022, 10, 1380. https://doi.org/10.3390/microorganisms10071380
Gamalero E, Bona E, Glick BR. Current Techniques to Study Beneficial Plant-Microbe Interactions. Microorganisms. 2022; 10(7):1380. https://doi.org/10.3390/microorganisms10071380
Chicago/Turabian StyleGamalero, Elisa, Elisa Bona, and Bernard R. Glick. 2022. "Current Techniques to Study Beneficial Plant-Microbe Interactions" Microorganisms 10, no. 7: 1380. https://doi.org/10.3390/microorganisms10071380







