Pseudomonas ST1 and Pantoea Paga Strains Cohabit in Olive Knots
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Incidence of Olive Knot Disease in Different Olive Cultivars
2.2. Genetic and Phylogenetic Analysis of Pseudomonas ST1 and Pantoea Paga
2.3. Confirmation of Pathogenicity of Pseudomonas ST1 and Pantoea Paga Isolated from the Trees in a Collection Orchard
The Efficiency of the Primers in Detection of the Bacteria
2.4. Susceptibility of Bacteria to Antibiotic and Chemical Agents Tested by a Standardized Single Disk Method
2.4.1. Bacteria Strains
2.4.2. Growth Media, Antibiotics and Chemical Agents
3. Results
3.1. Susceptibility of Different Olive Cultivars to Olive Knot Disease
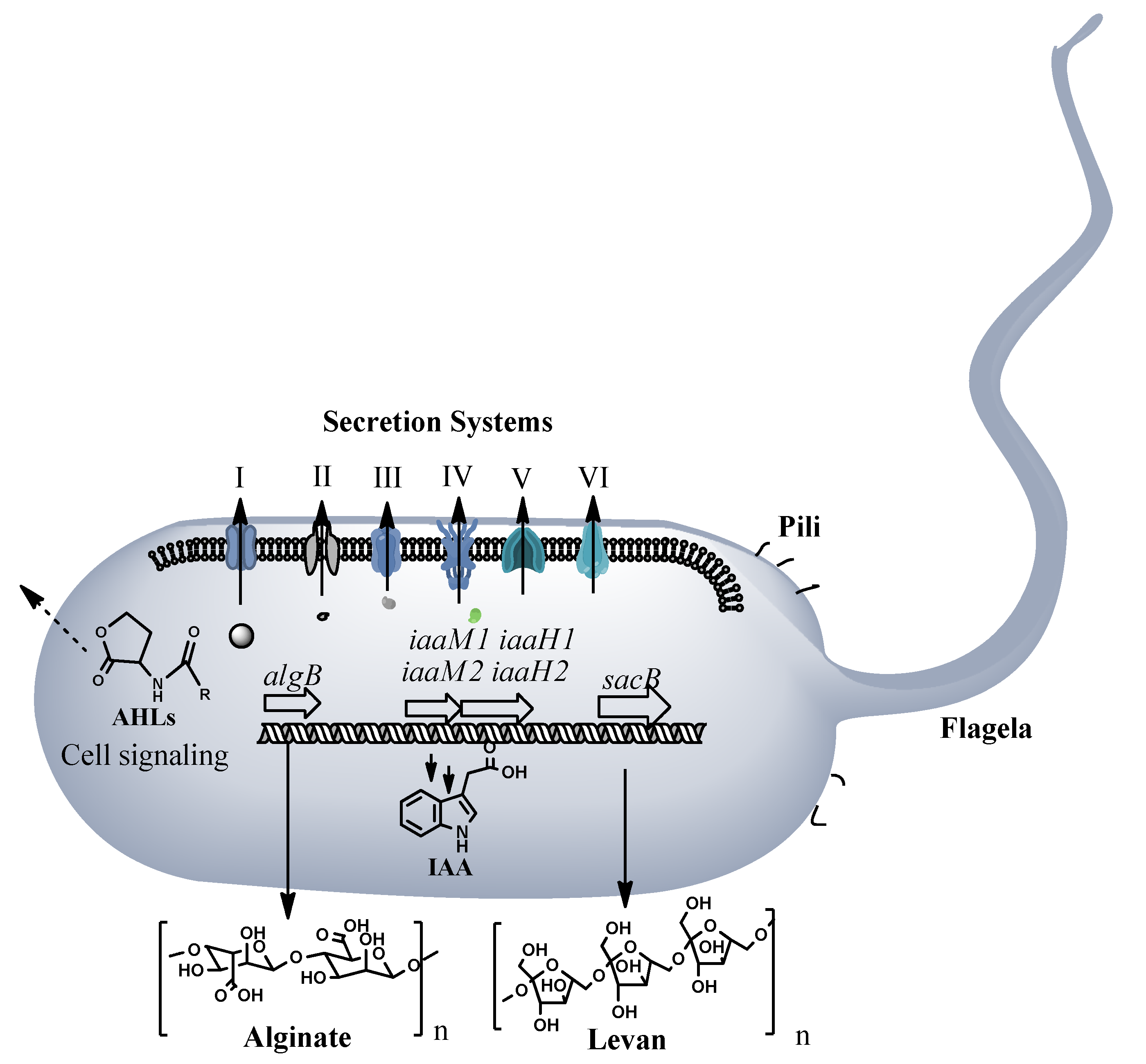
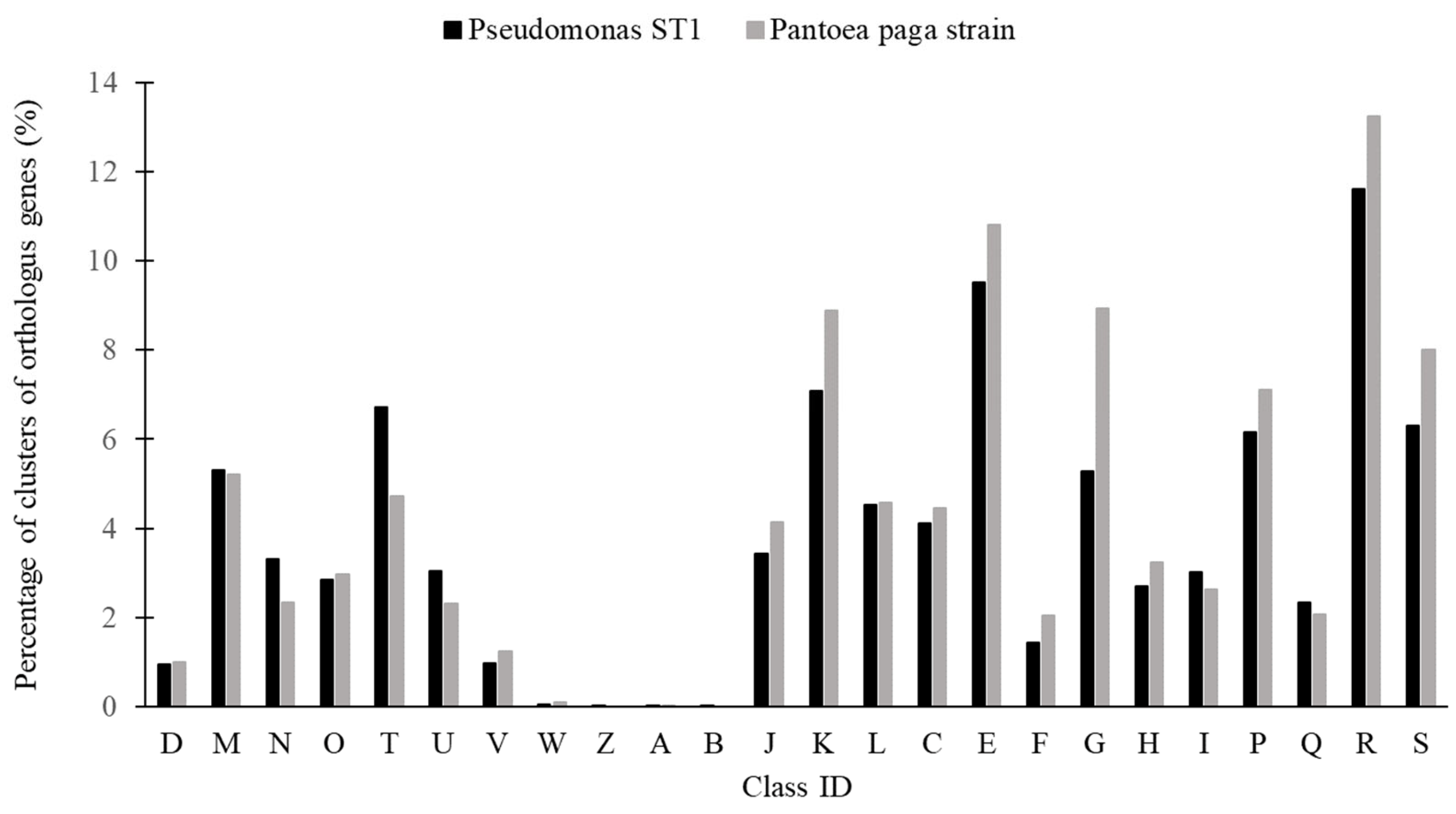
3.2. Genome Analysis of the Pseudomonas ST1 and Pantoea Paga
3.3. Pathogenicity of Pseudomonas ST1 and Pantoea Paga
3.4. Susceptibility of Two Bacteria to Various Chemical Agents and Antibiotics
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Quesada, J.M.; Penyalver, R.; López, M.M. Epidemiology and control of plant diseases caused by phytopathogenic bacteria: The case of olive knot disease caused by Pseudomonas savastanoi pv. savastanoi. In Plant Pathology; Cumagun, C.J., Ed.; Intech: Kayl, Luxembourg, 2012; pp. 299–326. [Google Scholar]
- Vivian, A.; Mansfield, J.A. Proposal for a uniform genetic nomenclature for avirulence genes in phytopathogenic pseudomonads. Mol. Plant-Microbe Interact. 1993, 6, 9–10. [Google Scholar]
- Quesada, J.M.; Penyalver, R.; Pérez-Panades, J.; Salcedo, C.I.; Carbonell, E.A.; López, M.M. Dissemination of Pseudomonas savastanoi pv. savastanoi populations and subsequent appearance of olive knot disease. Plant Pathol. 2010, 59, 262–269. [Google Scholar] [CrossRef]
- Comai, L.; Kosuge, T. Involvement of plasmid deoxyribonucleic acid in indoleacetic acid synthesis in Pseudomonas savastanoi. J. Bacteriol. 1980, 143, 950–957. [Google Scholar]
- Surico, G.; Iacobellis, N.S.; Sisto, A. Studies on the role of indole-3-acetic acid and cytokinins in the formation of knots on olive and oleander plants by Pseudomonas syringae pv. savastanoi. Physiol. Plant Pathol. 1985, 26, 309–320. [Google Scholar] [CrossRef]
- Ramos, C.; Matas, I.M.; Bardaji, L.; Aragon, I.M.; Murillo, J. Pseudomonas savastanoi pv. savastanoi: Some like it knot. Mol. Plant Pathol. 2012, 13, 998–1009. [Google Scholar] [CrossRef]
- Schroth, M.N.; Osgood, J.W.; Miller, T.D. Quantitative assessment of the effect of the olive knot disease on olive yield and quality. Phytopathology 1973, 63, 1064–1065. [Google Scholar]
- Wilson, E.E. The olive knot disease: Its inception, development and control. Hilgardia 1935, 9, 231–264. [Google Scholar]
- Passos da Silva, D.; Castaneda-Ojeda, P.M.; Moretti, C.; Buonaurio, R.; Ramos, C.; Venturi, V. Bacterial multispecies studies and microbiome analysis of a plant disease. Microbiology 2014, 160, 556–566. [Google Scholar] [CrossRef]
- Marchi, G.; Sisto, A.; Cimmino, A.; Andolfi, A.; Cipriani, M.G.; Evidente, A.; Surico, G. Interaction between Pseudomonas savastanoi pv. savastanoi and Pantoea agglomerans in olive knots. Plant Pathol. 2006, 55, 614–624. [Google Scholar] [CrossRef]
- Penyalver, R.; García, A.; Ferrer, A.; Bertolini, E.; Quesada, J.M.; Salcedo, C.I.; Piquer, J.; Pérez-Panadés, J.; Carbonell, E.A.; del Río, C.; et al. Factors affecting Pseudomonas savastanoi pv. savastanoi plant inoculations and their use for evaluation of olive cultivar susceptibility. Phytopathology 2006, 96, 313–319. [Google Scholar] [CrossRef]
- Hosni, T.; Moretti, C.; Devescovi, G.; Suarez-Moreno, Z.R.; Fatmi, M.B.; Guarnaccia, C.; Pongor, S.; Onofri, A.; Buonaurio, R.; Venturi, V.; et al. Sharing of quorum-sensing signals and role of interspecies communities in a bacterial plant disease. ISME J. 2011, 5, 1857–1870. [Google Scholar] [CrossRef]
- Buonaurio, R.; Moretti, C.; Cortese, C.; da Silva, D.P.; Ramos, C.; Venturi, V. The olive knot disease as a model to study the role of interspecies bacterial communities in plant disease. Front. Plant Sci. 2015, 6, 434. [Google Scholar] [CrossRef] [Green Version]
- Bodenhausen, N.; Bortfeld-Miller, M.; Ackermann, M.; Vorholt, J.A. A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLOS Genet. 2014, 10, e1004283. [Google Scholar] [CrossRef] [Green Version]
- Gomes, T.; Pereira, J.A.; Lino-Neto, T.; Bennett, A.E.; Baptista, P. Bacterial disease induced changes in fungal communities of olive tree twigs depend on host genotype. Sci. Rep. 2019, 9, 5882. [Google Scholar] [CrossRef] [Green Version]
- Mina, D.; Pereira, J.A.; Lino-Neto, T.; Baptista, P. Impact of plant genotype and plant habitat in shaping bacterial pathobiome: A comparative study in olive tree. Sci. Rep. 2020, 10, 3475. [Google Scholar] [CrossRef] [Green Version]
- Sisto, A.; Cipriani, M.G.; Morea, M. Knot formation caused by Pseudomonas syringae subsp. savastanoi on olive plants is hrp-dependent. Phytopathology 2004, 94, 484–489. [Google Scholar] [CrossRef] [Green Version]
- Cunnac, S.; Lindeberg, M.; Collmer, A. Pseudomonas syringae type III secretion system effectors: Repertoires in search of functions. Curr. Opin. Microbiol. 2009, 12, 53–60. [Google Scholar] [CrossRef]
- Mansfield, J.W. From bacterial avirulence genes to effector functions via the hrp delivery system: An overview of 25 years of progress in our understanding of plant innate immunity. Mol. Plant Pathol. 2009, 10, 721–734. [Google Scholar] [CrossRef]
- Pérez-Martínez, I.; Rodríguez-Moreno, L.; Lambertsen, L.; Matas, I.M.; Murillo, J.; Tegli, S.; Jiménez, A.J.; Ramos, C. Fate of a Pseudomonas savastanoi pv. savastanoi type III secretion system mutant in olive plants (Olea europaea L.). Appl. Environ. Microbiol. 2010, 76, 3611–3619. [Google Scholar] [CrossRef] [Green Version]
- Grant, S.R.; Fisher, E.J.; Chang, J.H.; Mole, B.M.; Dangl, J.L. Subterfuge and manipulation: Type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 2006, 60, 425–449. [Google Scholar] [CrossRef] [Green Version]
- Heath, M.C. Hypersensitive response-related death. Plant Mol. Biol. 2000, 44, 312–334. [Google Scholar] [CrossRef]
- Glickmann, E.; Gardan, L.; Jacquet, S.; Hussain, S.; Elasri, M.; Petit, A.; Dessaux, Y. Auxin production is a common feature of most pathovars of Pseudomonas syringae. Mol. Plant Microbe Interact. 1998, 11, 156–162. [Google Scholar]
- Bender, C.L.; Alarcón-Chaidez, F.; Gross, D.C. Pseudomonas syringae phytotoxins: Mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 1999, 63, 266–292. [Google Scholar] [CrossRef] [Green Version]
- Caballo-Ponce, E.; Murillo, J.; Martínez-Gil, M.; Moreno-Pérez, A.; Pintado, A.; Ramos, C. Knots untie: Molecular determinants involved in knot formation induced by Pseudomonas savastanoi in woody hosts. Front. Plant Sci. 2017, 8, 1089. [Google Scholar] [CrossRef] [Green Version]
- Dillon, M.M.; Almeida, R.N.D.; Laflamme, B.; Martel, A.; Weir, B.S.; Desveaux, D.; Guttman, D.S. Molecular evolution of Pseudomonas syringae type III secreted effector proteins. Front. Plant Sci. 2019, 10, 418. [Google Scholar] [CrossRef] [Green Version]
- Glass, N.L.; Kosuge, T. Role of indoleacetic acid-lysine synthetase in regulation of indoleacetic acid pool size and virulence of Pseudomonas syringae subsp. savastanoi. J. Bacteriol. 1988, 170, 2367–2373. [Google Scholar] [CrossRef] [Green Version]
- Quesada, J.M.; Penyalver, R.; Pérez-Panadés, J.; Salcedo, C.I.; Carbonell, E.A.; López, M.M. Comparison of chemical treatments for reducing epiphytic Pseudomonas savastanoi pv. savastanoi populations and for improving subsequent control of olive knot disease. Crop Prot. 2010, 29, 1413–1420. [Google Scholar] [CrossRef]
- Marcelo, A.; Fernandes, M.; Fatima Potes, M.; Serrano, J.F. Reactions of some cultivars of Olea europaea L. to experimental inoculation with Pseudomonas syringae pv. savastanoi. Acta Hortic. 1999, 474, 581–584. [Google Scholar]
- Hassani, D.; Buonaurio, R.; Tombesi, A. Response of some olive cultivars, hybrid and open pollinated seedling to Pseudomonas savastanoi pv. Savastanoi. In Pseudomonas syringae and Related Pathogens; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2003; pp. 489–494. [Google Scholar]
- Valverde, P.; Zucchini, M.; Polverigiani, S.; Lodolini, E.M.; López-Escudero, F.J.; Neri, D. Olive knot damages in ten olive cultivars after late-winter frost in central Italy. Sci. Hortic. 2020, 266, 109274. [Google Scholar] [CrossRef]
- EPPO. Pathogen-tested olive trees and rootstocks. EPPO Bull. 2006, 36, 77–83. [Google Scholar]
- Gavini, F.; Mergaert, J.; Beji, A.; Mielcarek, C.; Izard, D.; Kersters, K.; De Ley, J. Transfer of Enterobacter agglomerans (Beijerinck 1888) Ewing and Fife 1972 to Pantoea gen. nov. as Pantoea agglomerans comb. nov. and description of Pantoea dispersa sp. nov. Int. J. Syst. Bacteriol. 1989, 39, 337–345. [Google Scholar]
- Manulis, S.; Gafni, Y.; Clark, E.; Zutra, D.; Ophir, Y.; Barash, I. Identification of a plasmid DNA probe for detection of Erwinia herbicola pathogenic on Gypsophila paniculata. Phytopathology 1991, 81, 54–57. [Google Scholar]
- Manulis, M.; Barash, I. The molecular basis for transformation of an epiphyte into a gall-forming pathogen as exemplified by Erwinia herbicola pv. gypsophilae. In Plant-Microbe Interactions; Stacey, G., Keen, N., Eds.; American Phytopathological Society: St. Paul, MN, USA, 2003; pp. 9–52. [Google Scholar]
- Volcani, Z. Bacterial Diseases of Plants in Israel; Agricultural Research Organization, the Volcani Center: Bet Dagan, Israel, 1985; p. 388. [Google Scholar]
- Vuletin Selak, G.; Raboteg, M.; Fournier, P.; Dubost, A.; Abrouk, D.; Žanić, K.; Perica, S.; Normand, P.; Pujić, P. Genome sequence of Pseudomonas sp. Strain ST1, Isolated from Olive (Olea europaea L.) Knot Galls in Croatia. Microbiol. Resour. Announc. 2019, 8, e00986-19. [Google Scholar] [CrossRef] [Green Version]
- Vuletin Selak, G.; Raboteg, M.; Dubost, A.; Abrouk, D.; Žanić, K.; Normand, P.; Pujić, P. Whole-genome sequence of a Pantoea sp. strain isolated from an olive (Olea europaea L.) Knot. Microbiol. Resour. Announc. 2019, 8, e00978-19. [Google Scholar] [CrossRef] [Green Version]
- Vallenet, D.; Calteau, A.; Cruveiller, S.; Gachet, M.; Lajus, A.; Josso, A.; Mercier, J.; Renaux, A.; Rolli, J.; Rouy, Z.; et al. MicroScope in 2017: An expanding and evolving integrated resource for community expertise of microbial genomes. Nucleic Acids Res. 2017, 45, D517–D528. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Natale, D.A.; Garkavtsev, I.V.; Tatusova, T.A.; Shankavaram, U.T.; Rao, B.S.; Kiryutin, B.; Galperin, M.Y.; Fedorova, N.D.; Koonin, E.V. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001, 29, 22–28. [Google Scholar] [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. AntiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar]
- Liu, B.; Zheng, D.D.; Zhou, S.Y.; Chen, L.H.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: Hoboken, NJ, USA, 1991; pp. 115–175. [Google Scholar]
- Olofsson, T.C.; Ahrné, S.; Molin, G. Composition of the bacterial population of refrigerated beef, identified with direct 16S rRNA gene analysis and pure culture technique. Int. J. Food Microbiol. 2007, 18, 233–240. [Google Scholar]
- Edwards, U.; Rogall, T.; Böcker, H.; Emde, M.; Böttger, E. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal DNA. Nucleic Acids Res. 1989, 17, 7843–7853. [Google Scholar]
- Lane, D.J.; Pace, B.; Olson, G.J.; Stahl, D.A.; Sagin, M.L.; Pace, N.R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analysis. Proc. Natl. Acad. Sci. USA 1985, 82, 6955–6959. [Google Scholar]
- Rodríguez-Palenzuela, P.; Matas, I.M.; Murillo, J.; López-Solanilla, E.; Bardaji, L.; Pérez-Martínez, I.; Rodríguez-Moskera, M.E.; Penyalver, R.; López, M.M.; Quesada, J.M.; et al. Annotation and overview of the Pseudomonas savastanoi pv. savastanoi NCPPB 3335 draft genome reveals the virulence gene complement of atumour-inducing pathogen of woody hosts. Environ. Microbiol. 2010, 12, 1604–1620. [Google Scholar]
- Bartoli, C.; Carrere, S.; Lamichhane, J.R.; Varvaro, L.; Morris, C.E. Whole-genome sequencing of 10 Pseudomonas syringae strains representing different host range spectra. Genome Announc. 2015, 3, e00379-15. [Google Scholar]
- Turco, S.; Drais, M.I.; Rossini, L.; Chaboteaux, E.; Rahi, Y.J.; Balestra, G.M.; Iacobellis, N.S.; Mazzaglia, A. Complete genome assembly of the levan-positive strain PVFi1 of Pseudomonas savastanoi pv. savastanoi isolated from olive knots in Central Italy. Environ. Microbiol. Rep. 2022, 14, 274–285. [Google Scholar] [CrossRef]
- Barash, I.; Manulis-Sasson, S. Virulence mechanisms and host specificity of gall-forming Pantoea agglomerans. Trends Microbiol. 2007, 15, 538–545. [Google Scholar] [CrossRef]
- Matas, I.M.; Lambertsen, L.; Rodríguez-Moreno, L.; Ramos, C. Identification of novel virulence genes and metabolic pathways required for full fitness of Pseudomonas savastanoi pv. savastanoi in olive (Olea europaea) knots. New Phytol. 2012, 196, 1182–1196. [Google Scholar] [CrossRef] [Green Version]
- Matas, I.M.; Castañeda-Ojeda, M.P.; Aragón, I.M.; Antúnez-Lamas, M.; Murillo, J.; Rodríguez-Palenzuela, P.; López-Solanilla, E.; Ramos, C. Translocation and functional analysis of Pseudomonas savastanoi pv. savastanoi NCPPB 3335 type III secretion system effectors reveals two novel effector families of the Pseudomonas syringae complex. Mol. Plant-Microbe Interact. 2014, 27, 424–436. [Google Scholar] [CrossRef] [Green Version]
- Sinha Roy, R.; Gehring, A.M.; Milne, J.C.; Belshaw, P.J.; Walsh, C.T. Thiazole and oxazole peptides: Biosynthesis and molecular machinery. Nat. Prod. Rep. 1999, 16, 249–263. [Google Scholar] [CrossRef]







| Category | |||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 |
| Cultivar | |||||
| Coratina | Fasolina | Canino | Drobnica | Frantoio | Chemlali |
| Favarol | Grignan | Koroneiki | Pendolino | Lastovka | |
| Leccino | Moraiolo | Levantinka | P. marocaine | S. Catarina | |
| Oblica | Maurino | Taggiasca | |||
| Sigoise | Rosciola | ||||
| Antibiotic | Pseudomonas ST1 Sensitivity | Pantoea PAGA Sensitivity |
|---|---|---|
| Ampicillin | 2 | 2 |
| Apramycin | 1 | 1 |
| Carbenicillin | 2 | 2 |
| Chloramphenicol | 0.4 | 0.4 |
| Ciprofloxacin | 0.05 | 0.05 |
| G418 | 2 | 2 |
| Geneticin | 0.8 | 0.8 |
| Hygromycin | 2 | 10 |
| Kanamycin | 0.8 | 0.8 |
| Neomycin | 2 | 2 |
| Novobiocin | 0.748 | 0.748 |
| Rifampicin | 1.8 | 1.8 |
| Spectinomycin | 6 | 6 |
| Streptomycin | 0.6 | 0.6 |
| Tetracycline | 0.5 | 0.5 |
| Ticarcillin | 250 | 250 |
| Trimethoprim | 0.5 | 0.5 |
| Vancomycin | 0.58 | 28.86 |
| Chemical Agent | ||
| Benzoic acid (C7H6O2) | 50 | 10 |
| Copper (II) acetate | 5 | 5 |
| Copper (II) sulfate pentahydrate (CuSO4*5H2O) | 5 | 5 |
| Ethylenediaminetetraacetic acid (EDTA; C10H16N2O8) | 25 | 10 |
| Hydrogen peroxide (H2O2) | 0.6% | 0.6% |
| Iron (II) citrate (C12H30Fe3O24) | Resistant | Resistant |
| Iron (III) chloride (FeCl3) | 1 | 1 |
| Magnesium carbonate (MgCO3) | Resistant | Resistant |
| Manganese sulphate monohydrate (MnSO4*H2O) | 10 | 50 |
| Mineral oil | Resistant | Resistant |
| p-hydroxybenzoic acid (C7H6O3) | Resistant | Resistant |
| Polyoxyethylenesorbitan monolaurate (tween; C58H114O26) | Resistant | Resistant |
| Potassium iodide (KI) | Resistant | Resistant |
| Potassium sulfate (K2SO4) | Resistant | Resistant |
| Sodium dodecyl sulfate(SDS; C12H25NaO4S) | 0.004% | 20% |
| Sodium molybdate (Na2MoO4) | 50 | Resistant |
| Zinc sulfate heptahydrate(ZnSO4*7 H2O) | 1 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vuletin Selak, G.; Raboteg Božiković, M.; Abrouk, D.; Bolčić, M.; Žanić, K.; Perica, S.; Normand, P.; Pujic, P. Pseudomonas ST1 and Pantoea Paga Strains Cohabit in Olive Knots. Microorganisms 2022, 10, 1529. https://doi.org/10.3390/microorganisms10081529
Vuletin Selak G, Raboteg Božiković M, Abrouk D, Bolčić M, Žanić K, Perica S, Normand P, Pujic P. Pseudomonas ST1 and Pantoea Paga Strains Cohabit in Olive Knots. Microorganisms. 2022; 10(8):1529. https://doi.org/10.3390/microorganisms10081529
Chicago/Turabian StyleVuletin Selak, Gabriela, Marina Raboteg Božiković, Danis Abrouk, Marija Bolčić, Katja Žanić, Slavko Perica, Philippe Normand, and Petar Pujic. 2022. "Pseudomonas ST1 and Pantoea Paga Strains Cohabit in Olive Knots" Microorganisms 10, no. 8: 1529. https://doi.org/10.3390/microorganisms10081529







