Different Responses of Soil Bacterial and Fungal Communities in Three Typical Vegetations following Nitrogen Deposition in an Arid Desert
Abstract
:1. Introduction
2. Materials and Methods
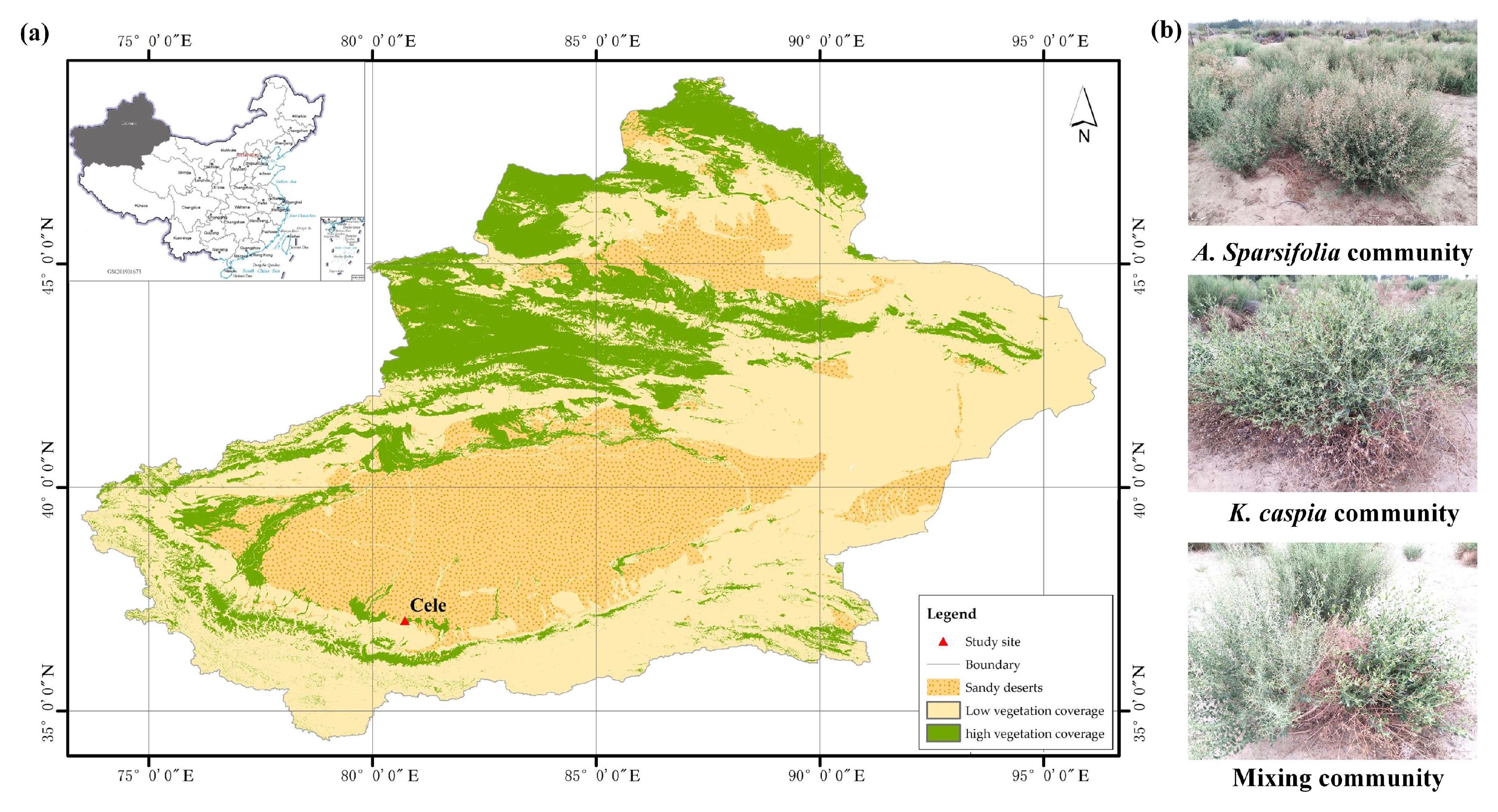
2.1. Site Description
2.2. Experimental Design
2.3. Soil Sampling
2.4. Determination of Soil Properties
2.5. DNA Extraction and Illumina Sequencing
2.6. Statistical Analyses
3. Results
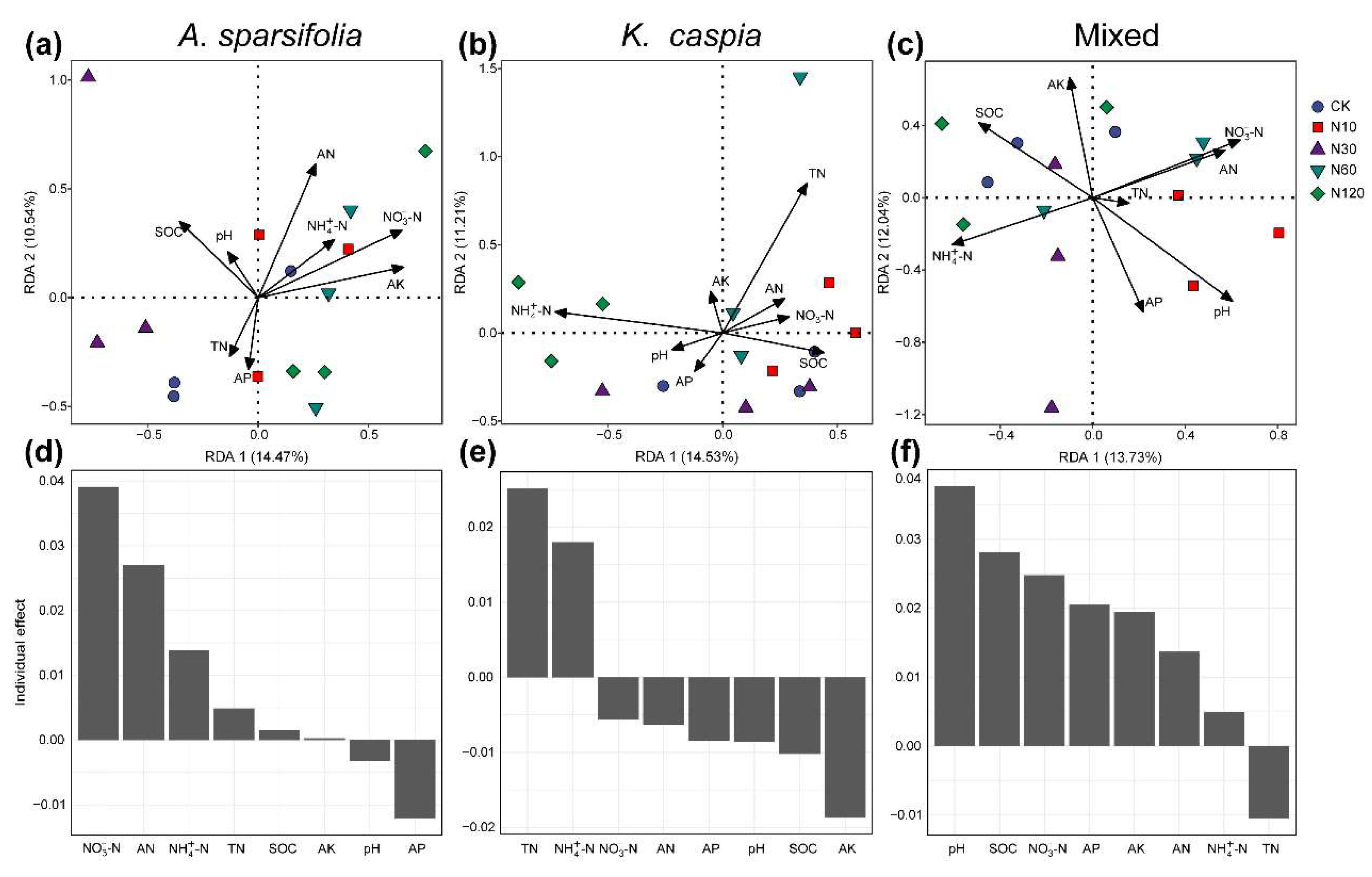
3.1. Soil Properties Affected by N Addition
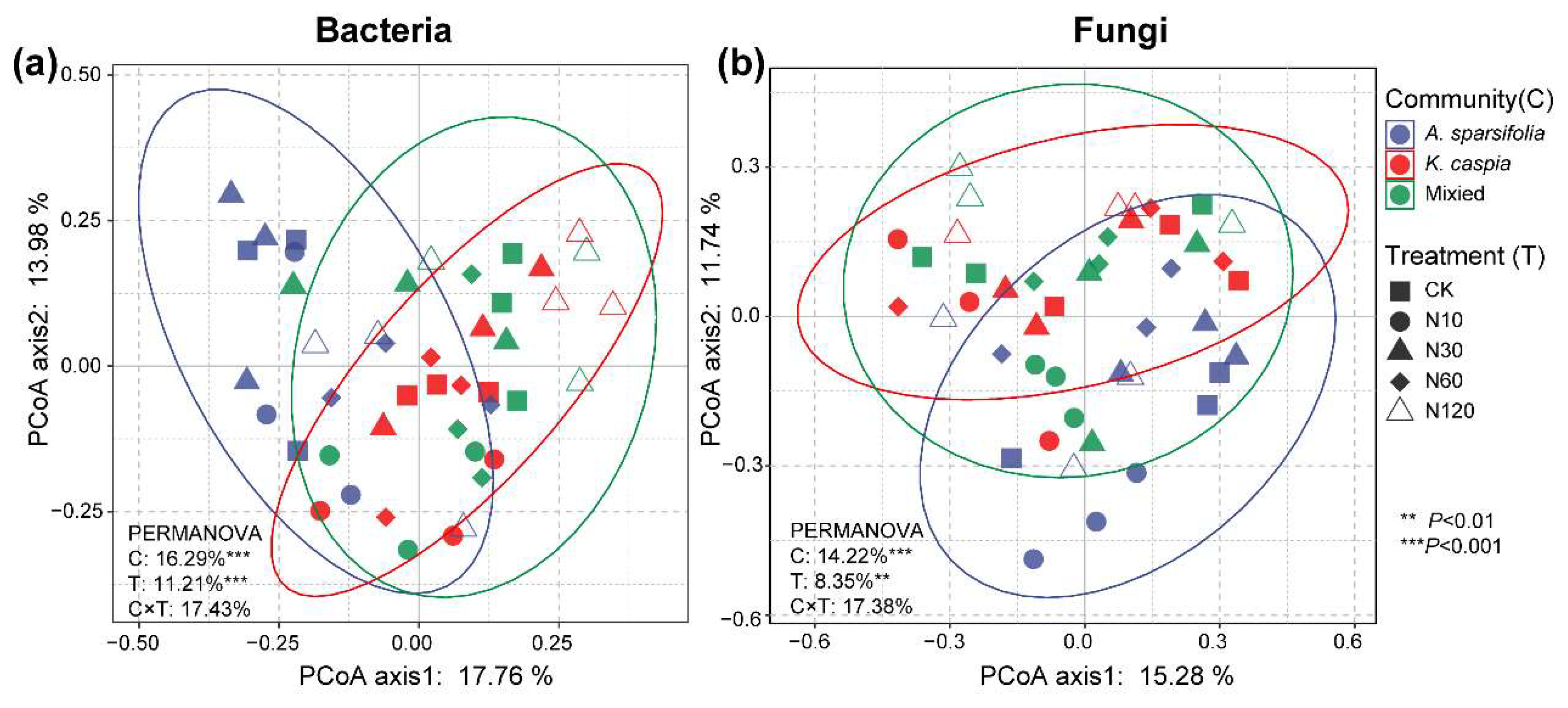
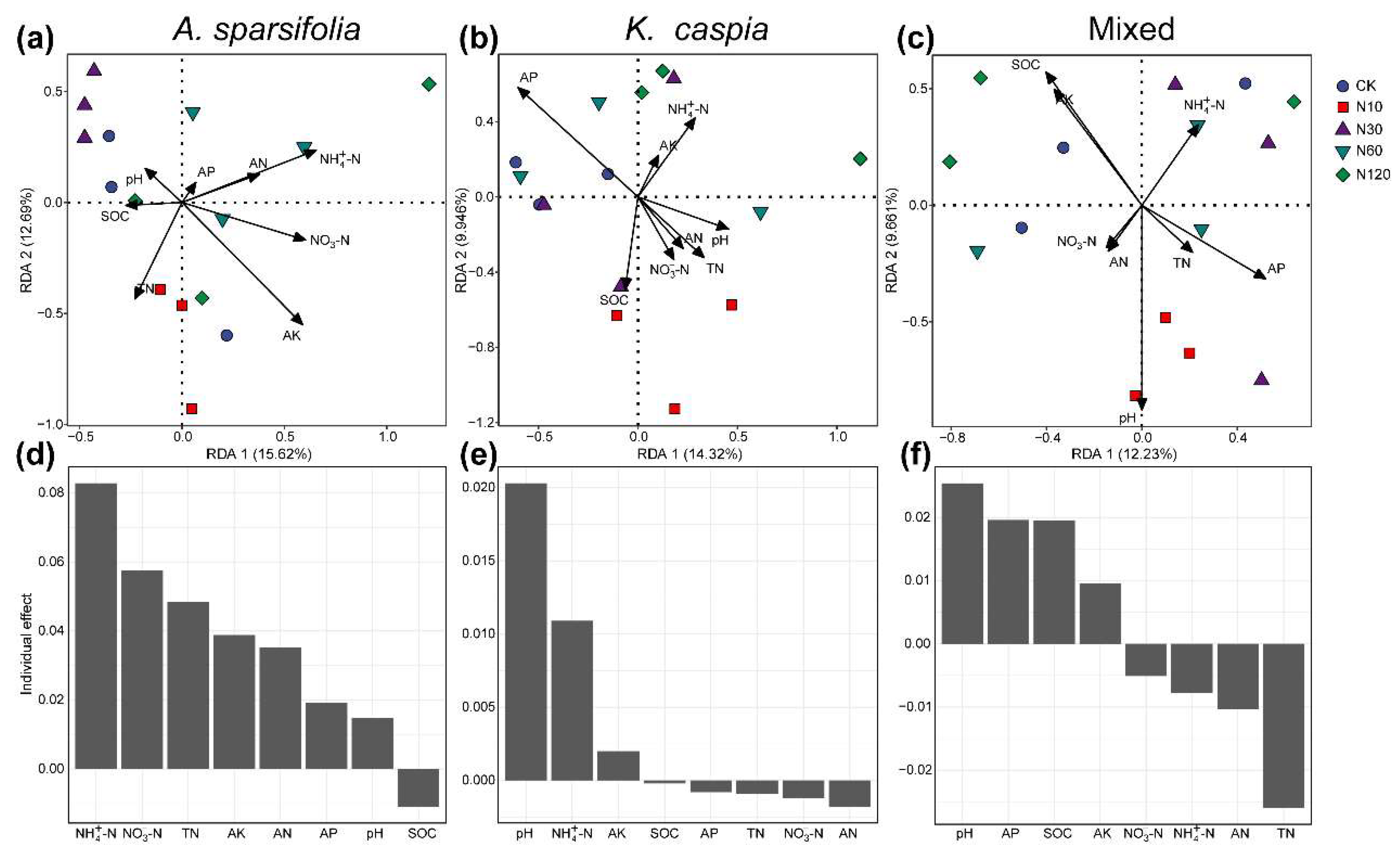
3.2. Soil Bacterial and Fungal Community Composition
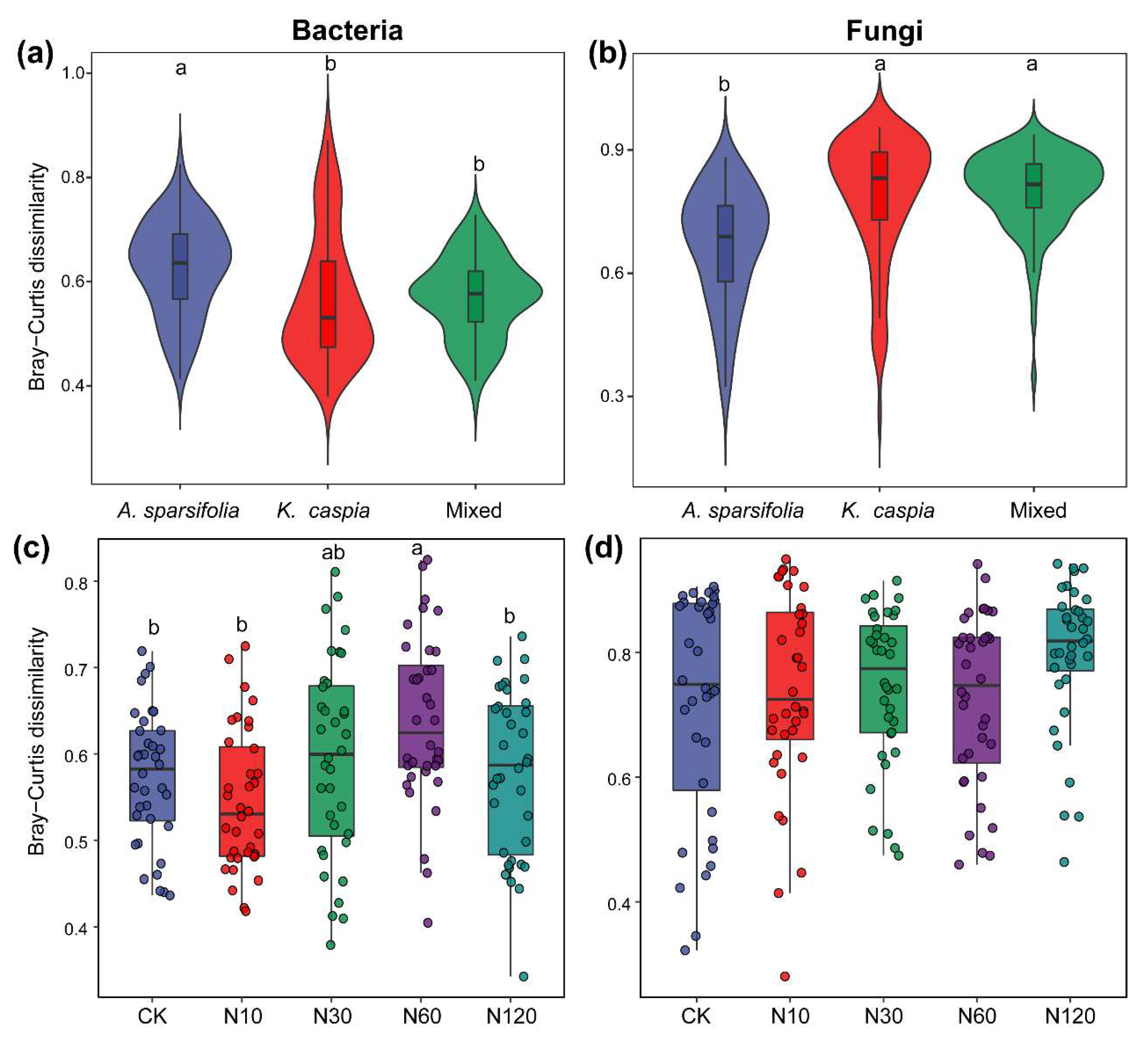
3.3. Diversity of Soil Bacterial and Fungal Communities
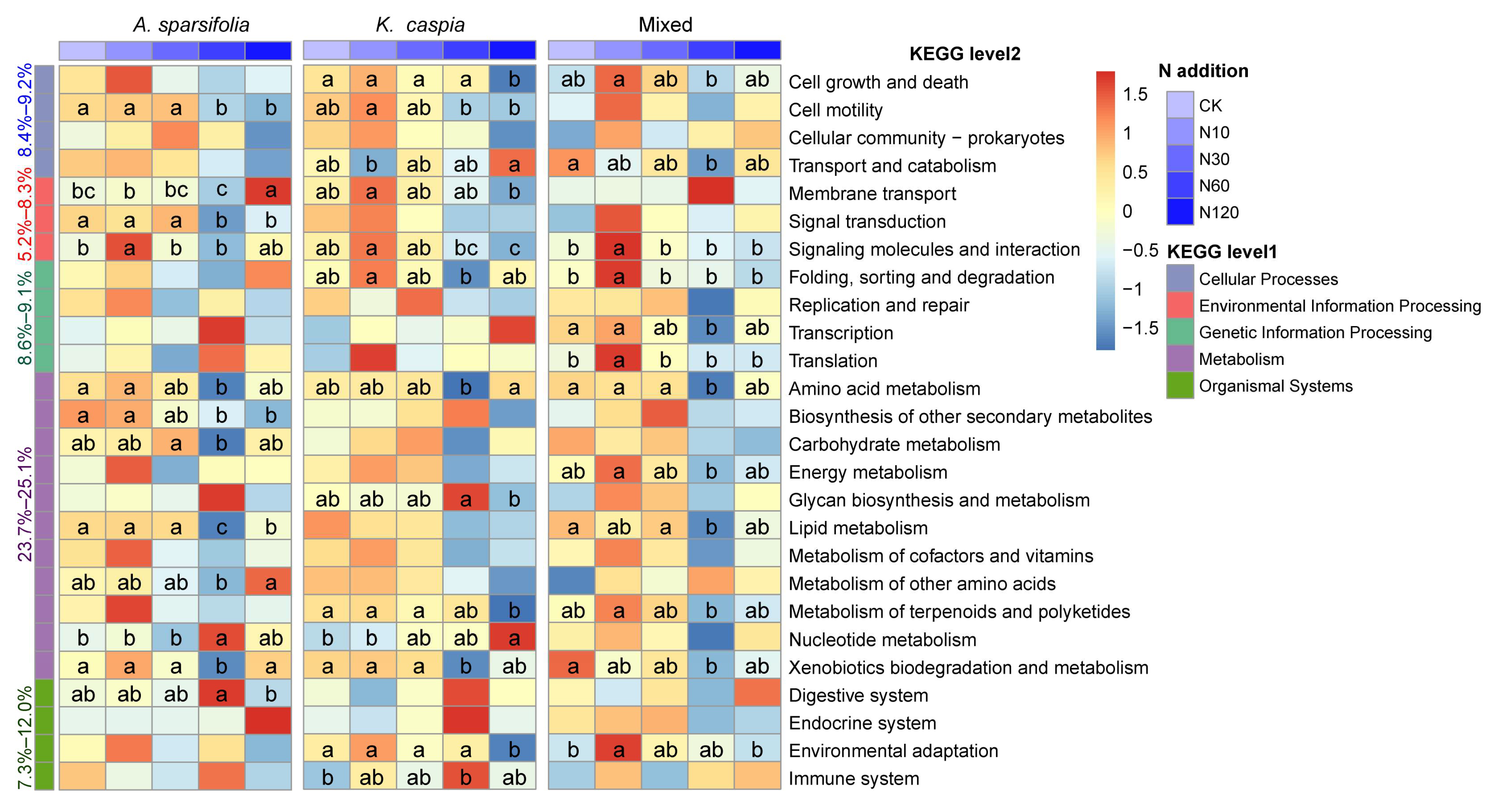
3.4. Function Prediction of Soil Bacterial and Fungal Communities
4. Discussion
4.1. Effects of N Addition on Soil Microbial Community Structure
4.2. Effects of Plant Community Types on Soil Microbial Community Structure
4.3. Effects of N Addition and Plant Community Types on the Function of Soil Microbial Community
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galloway, J.N.; Winiwarter, W.; Leip, A.; Leach, A.M.; Bleeker, A.; Erisman, J.W. Nitrogen Footprints: Past, Present and Future. Environ. Res. Lett. 2014, 9, 115003. [Google Scholar] [CrossRef]
- Galloway, J.N.; Townsend, A.R.; Erisman, J.W.; Bekunda, M.; Cai, Z.; Freney, J.R.; Martinelli, L.A.; Seitzinger, S.P.; Sutton, M.A. Transformation of the Nitrogen Cycle: Recent Trends, Questions, and Potential Solutions. Science 2008, 320, 889–892. [Google Scholar] [CrossRef]
- Grant, T.; Sethuraman, A.; Escobar, M.A.; Vourlitis, G.L. Chronic Dry Nitrogen Inputs Alter Soil Microbial Community Composition in Southern California Semi-Arid Shrublands. Appl. Soil Ecol. 2022, 176, 104496. [Google Scholar] [CrossRef]
- Zhang, Z.; Tariq, A.; Zeng, F.; Graciano, C.; Sun, F.; Chai, X.; Ahmed, Z. Nitrogen and Water Addition Regulate Fungal Community and Microbial Co-Occurrence Network Complexity in the Rhizosphere of Alhagi Sparsifolia Seedlings. Applied Soil Ecology 2021, 164, 103940. [Google Scholar] [CrossRef]
- Li, C.; Liu, L.; Zheng, L.; Yu, Y.; Mushinski, R.M.; Zhou, Y.; Xiao, C. Greater Soil Water and Nitrogen Availability Increase C : N Ratios of Root Exudates in a Temperate Steppe. Soil Biol. Biochem. 2021, 161, 108384. [Google Scholar] [CrossRef]
- Ma, S.; De Frenne, P.; Boon, N.; Brunet, J.; Cousins, S.A.O.; Decocq, G.; Kolb, A.; Lemke, I.; Liira, J.; Naaf, T.; et al. Plant Species Identity and Soil Characteristics Determine Rhizosphere Soil Bacteria Community Composition in European Temperate Forests. FEMS Microbiol. Ecol. 2019, 95, fiz063. [Google Scholar] [CrossRef] [PubMed]
- Santonja, M.; Rancon, A.; Fromin, N.; Baldy, V.; Hättenschwiler, S.; Fernandez, C.; Montès, N.; Mirleau, P. Plant Litter Diversity Increases Microbial Abundance, Fungal Diversity, and Carbon and Nitrogen Cycling in a Mediterranean Shrubland. Soil Biol. Biochem. 2017, 111, 124–134. [Google Scholar] [CrossRef]
- Zhang, Z.; Chai, X.; Tariq, A.; Zeng, F.; Graciano, C.; Li, X.; Ullah, A. Coordinated Patterns in the Allocation, Composition, and Variability of Multiple Elements Among Organs of Two Desert Shrubs Under Nitrogen Addition and Drought. J. Soil Sci. Plant Nutr. 2022, 22, 47–58. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, C.; Luo, Y. Meta-Analysis of the Impacts of Global Change Factors on Soil Microbial Diversity and Functionality. Nat. Commun. 2020, 11, 3072. [Google Scholar] [CrossRef] [PubMed]
- Brooks, M.L. Effects of Increased Soil Nitrogen on the Dominance of Alien Annual Plants in the Mojave Desert: Nitrogen Effects on Alien Annual Plants. J. Appl. Ecol. 2003, 40, 344–353. [Google Scholar] [CrossRef]
- Fenn, M.E.; Allen, E.B.; Weiss, S.B.; Jovan, S.; Geiser, L.H.; Tonnesen, G.S.; Johnson, R.F.; Rao, L.E.; Gimeno, B.S.; Yuan, F.; et al. Nitrogen Critical Loads and Management Alternatives for N-Impacted Ecosystems in California. J. Environ. Manag. 2010, 91, 2404–2423. [Google Scholar] [CrossRef]
- Laity, J. Deserts and Desert Environments; Wiley: Blackwell, UK, 2009. [Google Scholar]
- Sun, R.; Wang, F.; Hu, C.; Liu, B. Metagenomics Reveals Taxon-Specific Responses of the Nitrogen-Cycling Microbial Community to Long-Term Nitrogen Fertilization. Soil Biol. Biochem. 2021, 156, 108214. [Google Scholar] [CrossRef]
- Wang, Z.; Na, R.; Koziol, L.; Schellenberg, M.P.; Li, X.; Ta, N.; Jin, K.; Wang, H. Response of Bacterial Communities and Plant-Mediated Soil Processes to Nitrogen Deposition and Precipitation in a Desert Steppe. Plant Soil 2020, 448, 277–297. [Google Scholar] [CrossRef]
- Zhang, X.; Song, X.; Wang, T.; Huang, L.; Ma, H.; Wang, M.; Tan, D. The Responses to Long-Term Nitrogen Addition of Soil Bacterial, Fungal, and Archaeal Communities in a Desert Ecosystem. Front. Microbiol. 2022, 13, 1015588. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Li, L.; Su, Y.; Li, Y. Differential Seasonal Effects of Water Addition and Nitrogen Fertilization on Microbial Biomass and Diversity in a Temperate Desert. CATENA 2018, 161, 27–36. [Google Scholar] [CrossRef]
- She, W.; Bai, Y.; Zhang, Y.; Qin, S.; Feng, W.; Sun, Y.; Zheng, J.; Wu, B. Resource Availability Drives Responses of Soil Microbial Communities to Short-Term Precipitation and Nitrogen Addition in a Desert Shrubland. Front. Microbiol. 2018, 9, 186. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Xu, Y.; Yu, H.; Zhu, W.; Wang, P.; Wang, B.; Na, X. Soil Prokaryotic Community Shows No Response to 2 Years of Simulated Nitrogen Deposition in an Arid Ecosystem in Northwestern China. Env. Microbiol. 2021, 23, 1222–1237. [Google Scholar] [CrossRef]
- Dai, Z.; Su, W.; Chen, H.; Barberán, A.; Zhao, H.; Yu, M.; Yu, L.; Brookes, P.C.; Schadt, C.W.; Chang, S.X.; et al. Long-Term Nitrogen Fertilization Decreases Bacterial Diversity and Favors the Growth of Actinobacteria and Proteobacteria in Agro-Ecosystems across the Globe. Glob Change Biol. 2018, 24, 3452–3461. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shi, X.; Zheng, C.; Suter, H.; Huang, Z. Different Responses of Soil Bacterial and Fungal Communities to Nitrogen Deposition in a Subtropical Forest. Sci. Total Environ. 2021, 755, 142449. [Google Scholar] [CrossRef] [PubMed]
- Tariq, A.; Sardans, J.; Peñuelas, J.; Zhang, Z.; Graciano, C.; Zeng, F.; Olatunji, O.A.; Ullah, A.; Pan, K. Intercropping of Leguminous and Non-Leguminous Desert Plant Species Does Not Facilitate Phosphorus Mineralization and Plant Nutrition. Cells 2022, 11, 998. [Google Scholar] [CrossRef]
- Zhang, B.; Zeng, F.; Gao, X.; Shareef, M.; Zhang, Z.; Yu, Q.; Gao, Y.; Li, C.; Yin, H.; Lu, Y.; et al. Groundwater Depth Alters Soil Nutrient Concentrations in Different Environments in an Arid Desert. Front. Environ. Sci. 2022, 10, 939382. [Google Scholar] [CrossRef]
- Wu, Y.; Gao, X.; Liu, X.; Wen, Z.; Liu, J.; Li, X.; Zeng, F. Comparison of Nitrogen Deposition between Two Ecosystems in the Southern Edge of Tarim Basin. Atmos. Environ. 2023, 293, 119452. [Google Scholar] [CrossRef]
- Zhang, Z.; Tariq, A.; Zeng, F.; Graciano, C.; Zhang, B. Nitrogen Application Mitigates Drought-Induced Metabolic Changes in Alhagi Sparsifolia Seedlings by Regulating Nutrient and Biomass Allocation Patterns. Plant Physiol. Biochem. 2020, 155, 828–841. [Google Scholar] [CrossRef]
- Claesson, M.J.; O’Sullivan, O.; Wang, Q.; Nikkilä, J.; Marchesi, J.R.; Smidt, H.; de Vos, W.M.; Ross, R.P.; O’Toole, P.W. Comparative Analysis of Pyrosequencing and a Phylogenetic Microarray for Exploring Microbial Community Structures in the Human Distal Intestine. PLoS ONE 2009, 4, e6669. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal Rna Genes for Phylogenetics. In PCR Protocols; Elsevier: Amsterdam, The Netherlands, 1990; pp. 315–322. ISBN 978-0-12-372180-8. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact Sequence Variants Should Replace Operational Taxonomic Units in Marker-Gene Data Analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a Unified Paradigm for Sequence-Based Identification of Fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Lai, J.; Zou, Y.; Zhang, J.; Peres-Neto, P.R. Generalizing Hierarchical and Variation Partitioning in Multiple Regression and Canonical Analyses Using the Rdacca.Hp R Package. Methods Ecol. Evol. 2022, 13, 782–788. [Google Scholar] [CrossRef]
- Sha, M.; Xu, J.; Zheng, Z.; Fa, K. Enhanced Atmospheric Nitrogen Deposition Triggered Little Change in Soil Microbial Diversity and Structure in a Desert Ecosystem. Glob. Ecol. Conserv. 2021, 31, e01879. [Google Scholar] [CrossRef]
- Li, J.; Xie, J.; Zhang, Y.; Dong, L.; Shangguan, Z.; Deng, L. Interactive Effects of Nitrogen and Water Addition on Soil Microbial Resource Limitation in a Temperate Desert Shrubland. Plant Soil 2022, 475, 361–378. [Google Scholar] [CrossRef]
- Liu, W.; Jiang, L.; Yang, S.; Wang, Z.; Tian, R.; Peng, Z.; Chen, Y.; Zhang, X.; Kuang, J.; Ling, N.; et al. Critical Transition of Soil Bacterial Diversity and Composition Triggered by Nitrogen Enrichment. Ecology 2020, 101, e03053. [Google Scholar] [CrossRef]
- Nie, Y.; Wang, M.; Zhang, W.; Ni, Z.; Hashidoko, Y.; Shen, W. Ammonium Nitrogen Content Is a Dominant Predictor of Bacterial Community Composition in an Acidic Forest Soil with Exogenous Nitrogen Enrichment. Sci. Total Environ. 2018, 624, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Manzoni, S.; Schimel, J.P.; Porporato, A. Responses of Soil Microbial Communities to Water Stress: Results from a Meta-analysis. Ecology 2012, 93, 930–938. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Ródenas, A.; Lozano-Carrillo, M.C.; Pérez-Gilabert, M.; Morte, A. Effect of Water Stress on in Vitro Mycelium Cultures of Two Mycorrhizal Desert Truffles. Mycorrhiza 2011, 21, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yu, K.; Wu, Q.; Cheng, X.; Li, Z.; Wang, Z.; Zhao, M.; Wilkes, A.; Bisselling, T.; Han, G.; et al. Seasonal Precipitation and Soil Microbial Community Influence Plant Growth Response to Warming and N Addition in a Desert Steppe. Plant Soil 2022, 482, 245–259. [Google Scholar] [CrossRef]
- García-Palacios, P.; Vandegehuchte, M.L.; Shaw, E.A.; Dam, M.; Post, K.H.; Ramirez, K.S.; Sylvain, Z.A.; de Tomasel, C.M.; Wall, D.H. Are There Links between Responses of Soil Microbes and Ecosystem Functioning to Elevated CO2, N Deposition and Warming? A Global Perspective. Glob Change Biol. 2015, 21, 1590–1600. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Gan, P.; Chen, A. Environmental Controls on Soil pH in Planted Forest and Its Response to Nitrogen Deposition. Environ. Res. 2019, 172, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.; Yu, L.; Xu, J.; Brookes, P.C. pH, Nitrogen Mineralization, and KCl-Extractable Aluminum as Affected by Initial Soil pH and Rate of Vetch Residue Application: Results from a Laboratory Study. J. Soils Sediments 2014, 14, 1513–1525. [Google Scholar] [CrossRef]
- Yang, Y.; Ji, C.; Ma, W.; Wang, S.; Wang, S.; Han, W.; Mohammat, A.; Robinson, D.; Smith, P. Significant Soil Acidification across Northern China’s Grasslands during 1980s–2000s. Glob. Change Biol. 2012, 18, 2292–2300. [Google Scholar] [CrossRef]
- Hartikainen, H.; Yli-Halla, M. Solubility of Soil Phosphorus as Influenced by Urea. Z. Für Pflanzenernährung Und Bodenkd. 1996, 159, 327–332. [Google Scholar] [CrossRef]
- Rochette, P.; Angers, D.A.; Chantigny, M.H.; Gasser, M.-O.; MacDonald, J.D.; Pelster, D.E.; Bertrand, N. NH3 Volatilization, Soil Concentration and Soil pH Following Subsurface Banding of Urea at Increasing Rates. Can. J. Soil. Sci. 2013, 93, 261–268. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an Ecological Classification of Soil Bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Ho, A.; Lonardo, D.P.D.; Bodelier, P.L.E. Revisiting Life Strategy Concepts in Environmental Microbial Ecology. Microbiol. Ecol. 2017, 93, fix006. [Google Scholar] [CrossRef]
- Beskrovnaya, P.; Fakih, D.; Morneau, I.; Hashimi, A.; Bello, D.G.; Xing, S.; Nanci, A.; Huan, T.; Tocheva, E.I. No Endospore Formation Confirmed in Members of the Phylum. Appl. Environ. Microbiol. 2021, 87, 11. [Google Scholar] [CrossRef] [PubMed]
- Zak, D.R.; Holmes, W.E.; White, D.C.; Peacock, A.D.; Tilman, D. Plant Diversity, Soil Microbial Communities, and Ecosystem Function: Are There Any Links? Ecology 2003, 84, 2042–2050. [Google Scholar] [CrossRef]
- Bakker, M.G.; Schlatter, D.C.; Otto-Hanson, L.; Kinkel, L.L. Diffuse Symbioses: Roles of Plant–Plant, Plant–Microbe and Microbe–Microbe Interactions in Structuring the Soil Microbiome. Mol. Ecol. 2014, 23, 1571–1583. [Google Scholar] [CrossRef]
- Xie, H.; Tang, Y.; Yu, M.; Geoff Wang, G. The Effects of Afforestation Tree Species Mixing on Soil Organic Carbon Stock, Nutrients Accumulation, and Understory Vegetation Diversity on Reclaimed Coastal Lands in Eastern China. Glob. Ecol. Conserv. 2021, 26, e01478. [Google Scholar] [CrossRef]
- Fujii, K.; Funakawa, S.; Hayakawa, C.; Kosaki, T. Contribution of Different Proton Sources to Pedogenetic Soil Acidification in Forested Ecosystems in Japan. Geoderma 2008, 144, 478–490. [Google Scholar] [CrossRef]
- Mueller, K.E.; Eissenstat, D.M.; Hobbie, S.E.; Oleksyn, J.; Jagodzinski, A.M.; Reich, P.B.; Chadwick, O.A.; Chorover, J. Tree Species Effects on Coupled Cycles of Carbon, Nitrogen, and Acidity in Mineral Soils at a Common Garden Experiment. Biogeochemistry 2012, 111, 601–614. [Google Scholar] [CrossRef]
- Guckland, A.; Jacob, M.; Flessa, H.; Thomas, F.M.; Leuschner, C. Acidity, Nutrient Stocks, and Organic-matter Content in Soils of a Temperate Deciduous Forest with Different Abundance of European Beech (Fagus Sylvatica L.). Z. Pflanzenernähr. Bodenk. 2009, 172, 500–511. [Google Scholar] [CrossRef]
- Mastný, J.; Kaštovská, E.; Bárta, J.; Chroňáková, A.; Borovec, J.; Šantrůčková, H.; Urbanová, Z.; Edwards, K.R.; Picek, T. Quality of DOC Produced during Litter Decomposition of Peatland Plant Dominants. Soil Biol. Biochem. 2018, 121, 221–230. [Google Scholar] [CrossRef]
- Thoms, C.; Gattinger, A.; Jacob, M.; Thomas, F.M.; Gleixner, G. Direct and Indirect Effects of Tree Diversity Drive Soil Microbial Diversity in Temperate Deciduous Forest. Soil Biol. Biochem. 2010, 42, 1558–1565. [Google Scholar] [CrossRef]
- Deng, Q.; Hui, D.; Dennis, S.; Reddy, K.C.; Xu, X. Responses of Terrestrial Ecosystem Phosphorus Cycling to Nitrogen Addition: A Meta-analysis. Glob. Ecol. Biogeogr. 2017, 26, 713–728. [Google Scholar] [CrossRef]
- Wei, Y.; Zhao, Y.; Shi, M.; Cao, Z.; Lu, Q.; Yang, T.; Fan, Y.; Wei, Z. Effect of Organic Acids Production and Bacterial Community on the Possible Mechanism of Phosphorus Solubilization during Composting with Enriched Phosphate-Solubilizing Bacteria Inoculation. Bioresour. Technol. 2018, 247, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Bell, C.W.; Tissue, D.T.; Loik, M.E.; Wallenstein, M.D.; Acosta-Martinez, V.; Erickson, R.A.; Zak, J.C. Soil Microbial and Nutrient Responses to 7 Years of Seasonally Altered Precipitation in a Chihuahuan Desert Grassland. Glob. Change Biol. 2014, 20, 1657–1673. [Google Scholar] [CrossRef] [PubMed]
- Yang, L. Bacterial Communities Drive the Resistance of Soil Multifunctionality to Land-Use Change in Karst Soils. Eur. J. Soil Biol. 2021, 9, 103313. [Google Scholar] [CrossRef]
- Zhu, J.-T.; Li, X.-Y.; Zhang, X.-M.; Yu, Q.; Lin, L.-S. Leaf Nitrogen Allocation and Partitioning in Three Groundwater-Dependent Herbaceous Species in a Hyper-Arid Desert Region of North-Western China. Aust. J. Bot. 2012, 60, 61–67. [Google Scholar] [CrossRef]
- Li, M.; Petrie, M.D.; Tariq, A.; Zeng, F. Response of Nodulation, Nitrogen Fixation to Salt Stress in a Desert Legume Alhagi Sparsifolia. Environ. Exp. Bot. 2021, 183, 104348. [Google Scholar] [CrossRef]
- Florencio-Ortiz, V.; Sellés-Marchart, S.; Zubcoff-Vallejo, J.; Jander, G.; Casas, J.L. Changes in the Free Amino Acid Composition of Capsicum Annuum (Pepper) Leaves in Response to Myzus Persicae (Green Peach Aphid) Infestation. A Comparison with Water Stress. PLoS ONE 2018, 13, e0198093. [Google Scholar] [CrossRef]
- Corrigan, R.M.; Bowman, L.; Willis, A.R.; Kaever, V.; Gründling, A. Cross-Talk between Two Nucleotide-Signaling Pathways in Staphylococcus Aureus. J. Biol. Chem. 2015, 290, 5826–5839. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Sanders, N.J.; Shi, Y.; Chu, H.; Classen, A.T.; Zhao, K.; Chen, L.; Shi, Y.; Jiang, Y.; He, J.-S. The Links between Ecosystem Multifunctionality and Above- and Belowground Biodiversity Are Mediated by Climate. Nat. Commun. 2015, 6, 8159. [Google Scholar] [CrossRef] [PubMed]


| N-Addition Treatment × Plant Community Type | |||||
|---|---|---|---|---|---|
| TN | SOC | AP | AK | ||
| A. sparsifolia | CK | 0.290 ± 0.040 a | 1.71 ± 0.10 b | 6.11 ± 0.71 a | 201.13 ± 8.37 a |
| N10 | 0.186 ± 0.041 def | 1.65 ± 0.35 b | 3.81 ± 0.39 ab | 244.15 ± 16.95 a | |
| N30 | 0.160 ± 0.033 ef | 1.88 ± 0.35 ab | 4.38 ± 1.35 ab | 178.83 ± 21.65 a | |
| N60 | 0.150 ± 0.046 f | 1.38 ± 0.24 bc | 4.45 ± 0.82 ab | 228.80 ± 33.56 a | |
| N120 | 0.185 ± 0.033 def | 1.40 ± 0.27 bc | 4.24 ± 0.90 ab | 205.40 ± 17.45 a | |
| K. caspia | CK | 0.273 ± 0.048 ab | 1.34 ± 0.02 bc | 5.41 ± 1.59 ab | 226.03 ± 18.94 a |
| N10 | 0.264 ± 0.017 abc | 1.81 ± 0.12 ab | 2.99 ± 0.41 b | 210.06 ± 18.60 a | |
| N30 | 0.197 ± 0.036 cdef | 1.77 ± 0.22 ab | 4.06 ± 0.49 ab | 221.66 ± 24.23 a | |
| N60 | 0.238 ± 0.014 abcd | 1.48 ± 0.14 bc | 5.01 ± 1.39 ab | 246.43 ± 34.71 a | |
| N120 | 0.229 ± 0.035 abcde | 1.46 ± 0.38 bc | 3.77 ± 1.03 ab | 234.03 ± 41.39 a | |
| Mixing | CK | 0.185 ± 0.029 def | 1.44 ± 0.23 bc | 3.69 ± 0.60 ab | 243.66 ± 35.22 a |
| N10 | 0.238 ± 0.030 abcd | 1.00 ± 0.16 c | 4.77 ± 0.19 ab | 216.40 ± 40.17 a | |
| N30 | 0.182 ± 0.022 def | 1.34 ± 0.20 bc | 5.34 ± 1.56 ab | 206.76 ± 27.73 a | |
| N60 | 0.232 ± 0.044 abcde | 1.61 ± 0.33 b | 5.28 ± 0.75 ab | 231.56 ± 11.71 a | |
| N120 | 0.215 ± 0.024 bcdef | 2.29 ± 0.31 a | 4.67 ± 0.63 ab | 240.13 ± 7.82 a | |
| N-addition treatment | |||||
| AN | NH4+−N | NO3−−N | pH | ||
| CK | 28.9 ± 8.5 b | 3.8 ± 0.8 a | 19.7 ± 7.9 b | 7.11 ± 0.06 c | |
| N10 | 37.6 ± 13.9 ab | 3.1 ± 0.2 b | 30.1 ± 11.8 ab | 7.51 ± 0.13 a | |
| N30 | 31.0 ± 9.70 b | 3.6 ± 0.8 ab | 18.6 ± 9.9 b | 7.49 ± 0.20 a | |
| N60 | 46.9 ± 16.8 a | 3.5 ± 0.4 ab | 34.0 ± 16.5 a | 7.28 ± 0.09 b | |
| N120 | 25.4 ± 15.0 b | 4.2 ± 1.0 a | 18.8 ± 13.9 b | 7.38 ± 0.21 ab | |
| Plant community type | |||||
| pH | |||||
| A. sparsifolia | 7.37 ± 0.19 b | ||||
| K. caspia | 7.41 ± 0.20 a | ||||
| Mixing | 7.29 ± 0.21 c | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Tang, G.; Chai, X.; Liu, B.; Gao, X.; Zeng, F.; Wang, Y.; Zhang, B. Different Responses of Soil Bacterial and Fungal Communities in Three Typical Vegetations following Nitrogen Deposition in an Arid Desert. Microorganisms 2023, 11, 2471. https://doi.org/10.3390/microorganisms11102471
Zhang Z, Tang G, Chai X, Liu B, Gao X, Zeng F, Wang Y, Zhang B. Different Responses of Soil Bacterial and Fungal Communities in Three Typical Vegetations following Nitrogen Deposition in an Arid Desert. Microorganisms. 2023; 11(10):2471. https://doi.org/10.3390/microorganisms11102471
Chicago/Turabian StyleZhang, Zhihao, Gangliang Tang, Xutian Chai, Bo Liu, Xiaopeng Gao, Fanjiang Zeng, Yun Wang, and Bo Zhang. 2023. "Different Responses of Soil Bacterial and Fungal Communities in Three Typical Vegetations following Nitrogen Deposition in an Arid Desert" Microorganisms 11, no. 10: 2471. https://doi.org/10.3390/microorganisms11102471







