Roles of Hcp2, a Hallmark of T6SS2 in Motility, Adhesive Capacity, and Pathogenicity of Vibrio alginolyticus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Media, Plasmids, and Zebrafish Lines and Maintenance
2.2. Construction and Physiology Analysis of the hcp2 Mutation
2.3. Virulence Determination
2.4. Adhesion and Intestinal Colonization Assays
2.5. Construction of hcp2 Complementary Strain and Electron Microscopy Imaging
2.6. Proteomics Analysis of the Proteins Associated with Flagellum Assembly
2.6.1. Protein Extraction, Digestion, TMT/iTRAQ Labeling, and HPLC Fractionation
2.6.2. LC-MS/MS Analysis
2.6.3. Volcano Plot Analysis
2.7. Real-Time Quantitative PCR (RT-qPCR)
2.8. Statistical Analysis
3. Results
3.1. Loss of hcp2 Reduced the Swarming Motility of Vibrio alginolyticus
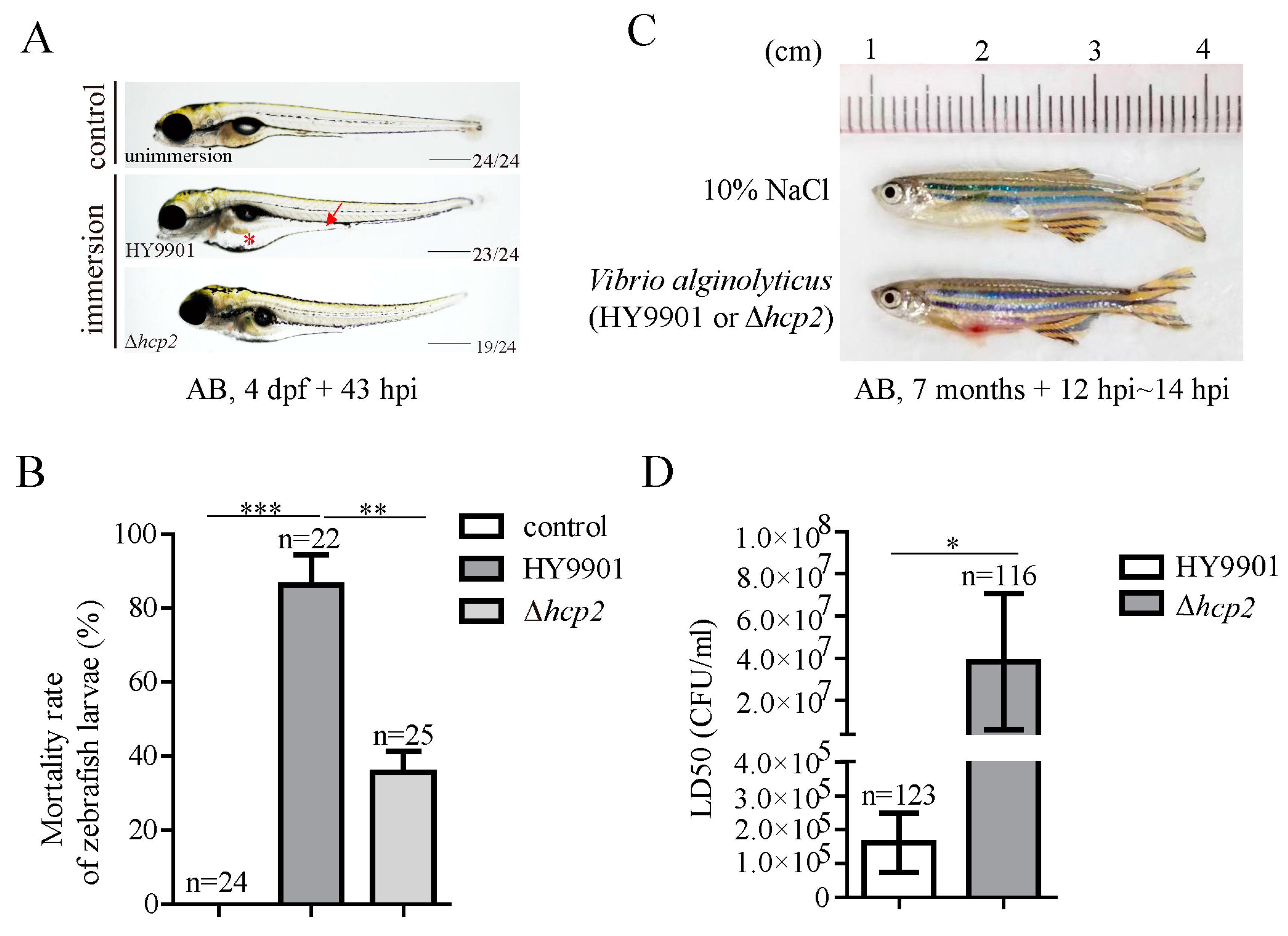
3.2. Attenuated Pathogenicity of Δhcp2 Strain against Zebrafish
3.3. Reduced Adhesive Ability of Δhcp2 Strain on the Zebrafish larval Surface and Intestinal Tract
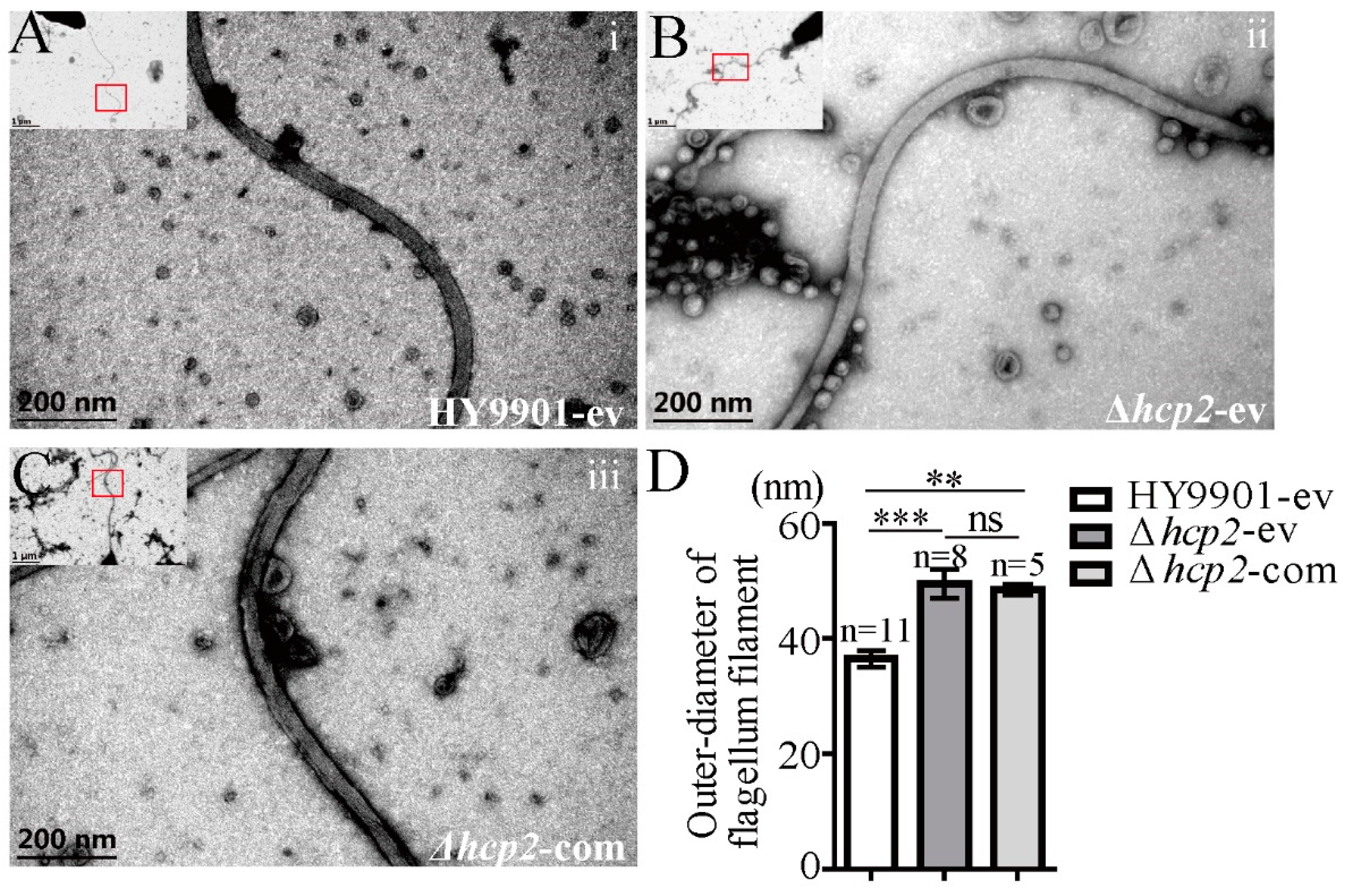
3.4. A Malformed Morphology of Flagellar Filaments in hcp2 Mutation of Vibrio alginolyticus
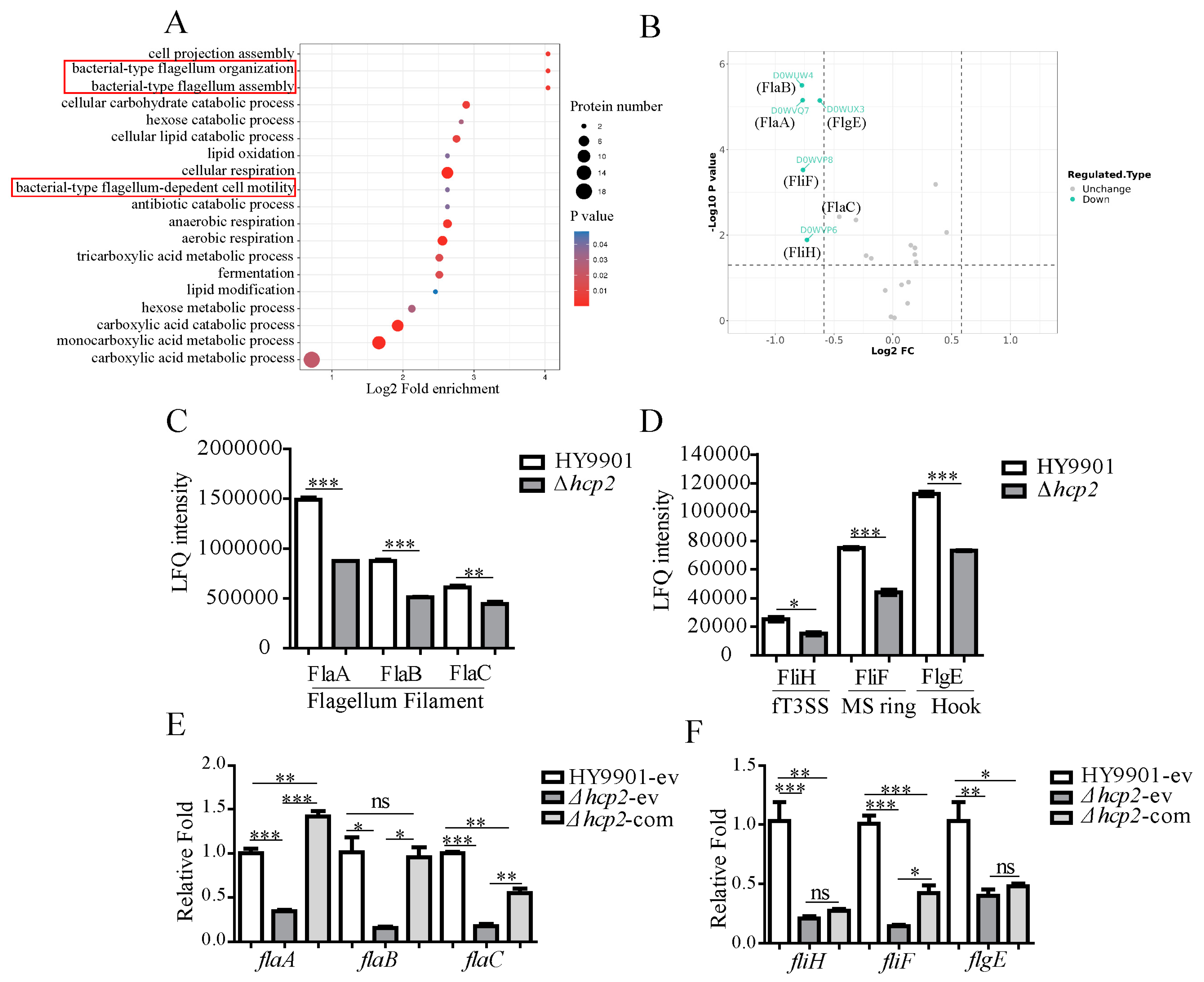
3.5. Expression and Transcription Levels Analysis of Flagellin and Flagellum Export Apparatus by Quantitative Proteomics and RT-qPCR, Respectively
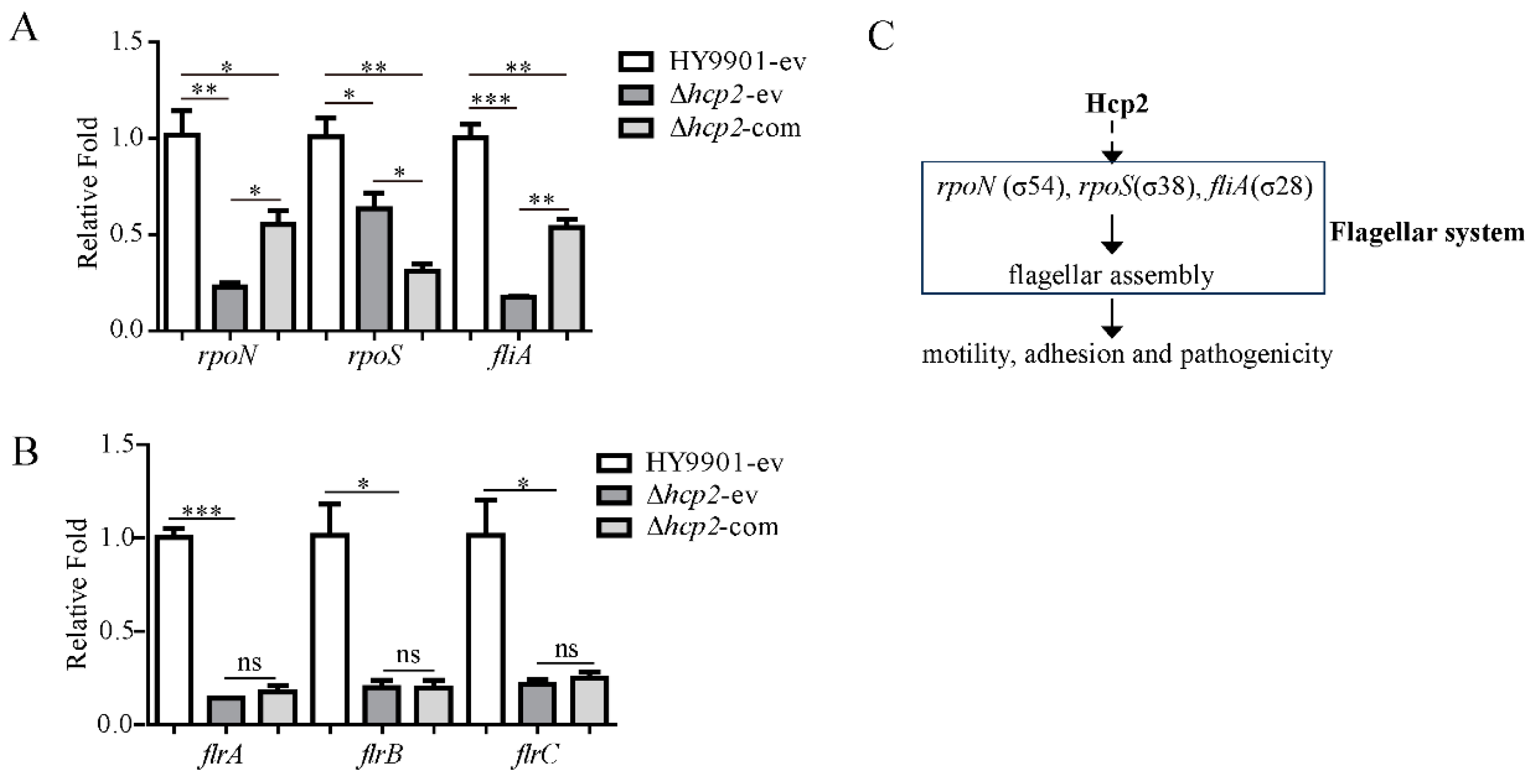
3.6. The Transcriptions of Sigma Factors rpoN and fliA Controlling Flagellin Biosynthesis and Export Were Positively Influenced by hcp2 in Vibrio alginolyticus
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leiman, P.G.; Basler, M.; Ramagopal, U.A.; Bonanno, J.B.; Sauder, J.M.; Pukatzki, S.; Burley, S.K.; Almo, S.C.; Mekalanos, J.J. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. USA 2009, 106, 4154–4159. [Google Scholar] [CrossRef] [PubMed]
- Ho, B.T.; Dong, T.G.; Mekalanos, J.J. A view to a kill: The bacterial type VI secretion system. Cell Host Microbe 2014, 15, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Basler, M.; Ho, B.T.; Mekalanos, J.J. Tit-for-tat: Type VI secretion system counterattack during bacterial cell-cell interactions. Cell 2013, 152, 884–894. [Google Scholar] [CrossRef] [PubMed]
- Coulthurst, S.J. The Type VI secretion system—A widespread and versatile cell targeting system. Res. Microbiol. 2013, 164, 640–654. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhou, X.; Ma, Y.; Zhou, M.; Waldor, M.K.; Zhang, Y.; Wang, Q. Serine/threonine kinase PpkA coordinates the interplay between T6SS2 activation and quorum sensing in the marine pathogen Vibrio alginolyticus. Environ. Microbiol. 2018, 20, 903–919. [Google Scholar] [CrossRef]
- Sheng, L.; Lv, Y.; Liu, Q.; Wang, Q.; Zhang, Y. Connecting type VI secretion, quorum sensing, and c-di-GMP production in fish pathogen Vibrio alginolyticus through phosphatase PppA. Vet. Microbiol. 2013, 162, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Salomon, D.; Klimko, J.A.; Trudgian, D.C.; Kinch, L.N.; Grishin, N.V.; Mirzaei, H.; Orth, K. Type VI Secretion System Toxins Horizontally Shared between Marine Bacteria. PLoS Pathog. 2015, 11, e1005128. [Google Scholar] [CrossRef] [PubMed]
- Durand, E.; Nguyen, V.S.; Zoued, A.; Logger, L.; Péhau-Arnaudet, G.; Aschtgen, M.S.; Spinelli, S.; Desmyter, A.; Bardiaux, B.; Dujeancourt, A.; et al. Biogenesis and structure of a type VI secretion membrane core complex. Nature 2015, 523, 555–560. [Google Scholar] [CrossRef]
- Wang, J.; Brackmann, M.; Castaño-Díez, D.; Kudryashev, M.; Goldie, K.N.; Maier, T.; Stahlberg, H.; Basler, M. Cryo-EM structure of the extended type VI secretion system sheath-tube complex. Nat. Microbiol. 2017, 2, 1507–1512. [Google Scholar] [CrossRef]
- Howard, S.A.; Furniss, R.C.D.; Bonini, D.; Amin, H.; Paracuellos, P.; Zlotkin, D.; Costa, T.R.D.; Levy, A.; Mavridou, D.A.I.; Filloux, A. The Breadth and Molecular Basis of Hcp-Driven Type VI Secretion System Effector Delivery. MBio 2021, 12, e0026221. [Google Scholar] [CrossRef]
- Sana, T.G.; Flaugnatti, N.; Lugo, K.A.; Lam, L.H.; Jacobson, A.; Baylot, V.; Durand, E.; Journet, L.; Cascales, E.; Monack, D.M. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc. Natl. Acad. Sci. USA 2016, 113, E5044–E5051. [Google Scholar] [CrossRef]
- Wang, N.; Liu, J.; Pang, M.; Wu, Y.; Awan, F.; Liles, M.R.; Lu, C.; Liu, Y. Diverse roles of Hcp family proteins in the environmental fitness and pathogenicity of Aeromonas hydrophila Chinese epidemic strain NJ-35. Appl. Microbiol. Biotechnol. 2018, 102, 7083–7095. [Google Scholar] [CrossRef]
- Kalindamar, S.; Abdelhamed, H.; Kordon, A.O.; Pinchuk, L.M.; Karsi, A. Hemolysin Co-regulated Family Proteins Hcp1 and Hcp2 Contribute to Pathogenesis. Front. Vet. Sci. 2021, 8, 681609. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Xiao, X.; Xu, S.; Gao, F.; Wang, J.; Wang, T.; Song, Y.; Pan, J.; Shen, X.; Wang, Y. Roles of RpoS in Yersinia pseudotuberculosis stress survival, motility, biofilm formation and type VI secretion system expression. J. Microbiol. 2015, 53, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Hao, S.; Lan, R.; Wang, G.; Xiao, D.; Sun, H.; Xu, J. The Type VI Secretion System Modulates Flagellar Gene Expression and Secretion in Citrobacter freundii and Contributes to Adhesion and Cytotoxicity to Host Cells. Infect. Immun. 2015, 83, 2596–2604. [Google Scholar] [CrossRef]
- Wang, P.; Dong, J.F.; Li, R.Q.; Li, L.; Zou, Q.H. Roles of the Hcp family proteins in the pathogenicity of Salmonella typhimurium 14,028 s. Virulence 2020, 11, 1716–1726. [Google Scholar] [CrossRef]
- Gong, J.S.; Guan, Y.C.; Zhao, Z.L.; Cai, Y.N.; Shan, X.F. Hcp1 regulates flagella of Aeromonas veronii TH0426 to reduce virulence. Aquaculture 2023, 576, 739899. [Google Scholar]
- Chevance, F.F.V.; Hughes, K.T. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 2008, 6, 455–465. [Google Scholar] [CrossRef]
- Echazarreta, M.A.; Klose, K.E. Vibrio Flagellar Synthesis. Front. Cell. Infect. Microbiol. 2019, 9, 131. [Google Scholar] [CrossRef] [PubMed]
- Danson, A.E.; Jovanovic, M.; Buck, M.; Zhang, X. Mechanisms of σ54-Dependent Transcription Initiation and Regulation. J. Mol. Biol. 2019, 431, 3960–3974. [Google Scholar] [CrossRef]
- Yu, C.; Yang, F.; Xue, D.; Wang, X.; Chen, H. The Regulatory Functions of σ54 Factor in Phytopathogenic Bacteria. Int. J. Mol. Sci. 2021, 22, 12692. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhang, S.; Ren, W.; Gong, X.; Long, H.; Zhang, X.; Cai, X.; Huang, A.; Xie, Z. Roles of rpoN in biofilm formation of Vibrio alginolyticus HN08155 at different cell densities. Microbiol. Res. 2021, 247, 126728. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Zhang, Y.; Wang, K.; Li, M.; Jiao, X. Characterization of the RpoN regulon reveals the regulation of motility, T6SS2 and metabolism in Vibrio parahaemolyticus. Front. Microbiol. 2022, 13, 1025960. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Guo, X.; Li, J.; Li, Y.; Sun, H.; Li, A.; Cao, B. RpoN is required for the motility and contributes to the killing ability of Plesiomonas shigelloides. BMC Microbiol. 2022, 22, 299. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Tabassum, N.; Anand, R.; Kim, Y.M. Motility of Vibrio spp.: Regulation and controlling strategies. Appl. Microbiol. Biotechnol. 2020, 104, 8187–8208. [Google Scholar] [CrossRef] [PubMed]
- Patten, C.L.; Kirchhof, M.G.; Schertzberg, M.R.; Morton, R.A.; Schellhorn, H.E. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genom. 2004, 272, 580–591. [Google Scholar] [CrossRef]
- Dong, T.; Yu, R.; Schellhorn, H. Antagonistic regulation of motility and transcriptome expression by RpoN and RpoS in Escherichia coli. Mol. Microbiol. 2011, 79, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Bi, W.; Chen, S.; Zhu, J.; Liu, X. Regulatory function of sigma factors RpoS/RpoN in adaptation and spoilage potential of Shewanella baltica. Food Microbiol. 2021, 97, 103755. [Google Scholar] [CrossRef]
- Cai, S.H.; Wu, Z.H.; Jian, J.C.; Lu, Y.S. Cloning and expression of the gene encoding an extracellular alkaline serine protease from Vibrio alginolyticus strain HY9901, the causative agent of vibriosis in Lutjanus erythopterus (Bloch). J. Fish. Dis. 2007, 30, 493–500. [Google Scholar] [CrossRef]
- Westerfield, M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio); University of Oregon Press: Eugene, OR, USA, 2007. [Google Scholar]
- Luo, P.; He, X.; Liu, Q.; Hu, C. Developing Universal Genetic Tools for Rapid and Efficient Deletion Mutation in Vibrio Species Based on Suicide T-Vectors Carrying a Novel Counterselectable Marker, vmi480. PLoS ONE 2015, 10, e0144465. [Google Scholar] [CrossRef]
- Zhou, Z.; Pang, H.; Ding, Y.; Cai, J.; Huang, Y.; Jian, J.; Wu, Z. VscO, a putative T3SS chaperone escort of Vibrio alginolyticus, contributes to virulence in fish and is a target for vaccine development. Fish Shellfish Immunol. 2013, 35, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Moisi, M.; Jenul, C.; Butler, S.M.; New, A.; Tutz, S.; Reidl, J.; Klose, K.E.; Camilli, A.; Schild, S. A novel regulatory protein involved in motility of Vibrio cholerae. J. Bacteriol. 2009, 191, 7027–7038. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Xu, T.; Fossheim, L.E.; Zhang, X.H. FliC, a flagellin protein, is essential for the growth and virulence of fish pathogen Edwardsiella tarda. PLoS ONE 2012, 7, e45070. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.G.; Varcoe, L.T.; Attridge, S.R.; Manning, P.A. Vibrio cholerae Hcp, a secreted protein coregulated with HlyA. Infect. Immun. 1996, 64, 283–289. [Google Scholar] [CrossRef]
- Silverman, J.M.; Agnello, D.M.; Zheng, H.; Andrews, B.T.; Li, M.; Catalano, C.E.; Gonen, T.; Mougous, J.D. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol. Cell 2013, 51, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Bouteiller, M.; Dupont, C.; Bourigault, Y.; Latour, X.; Barbey, C.; Konto-Ghiorghi, Y.; Merieau, A. Flagella: Generalities and Specificities. Int. J. Mol. Sci. 2021, 22, 3337. [Google Scholar] [CrossRef] [PubMed]
- Kamber, T.; Pothier, J.F.; Pelludat, C.; Rezzonico, F.; Duffy, B.; Smits, T.H.M. Role of the type VI secretion systems during disease interactions of Erwinia amylovora with its plant host. BMC Genom. 2017, 18, 628. [Google Scholar] [CrossRef]
- Bouteiller, M.; Gallique, M.; Bourigault, Y.; Kosta, A.; Hardouin, J.; Massier, S.; Konto-Ghiorghi, Y.; Barbey, C.; Latour, X.; Chane, A.; et al. Crosstalk between the Type VI Secretion System and the Expression of Class IV Flagellar Genes in the MFE01 Strain. Microorganisms 2020, 8, 622. [Google Scholar] [CrossRef]
- Dong, T.G.; Mekalanos, J.J. Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res. 2012, 40, 7766–7775. [Google Scholar] [CrossRef]
- Thormann, K.M.; Beta, C.; Kühn, M.J. Wrapped Up: The Motility of Polarly Flagellated Bacteria. Annu. Rev. Microbiol. 2022, 76, 349–367. [Google Scholar] [CrossRef]
- Liang, H.; Xia, L.; Wu, Z.; Jian, J.; Lu, Y. Expression, characterization and immunogenicity of flagellin FlaC from Vibrio alginolyticus strain HY9901. Fish Shellfish Immunol. 2010, 29, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.Y.; Wu, Z.H.; Jian, J.C.; Liu, Z.H. Construction of a fusion flagellin complex and evaluation of the protective immunity of it in red snapper (Lutjanus sanguineus). Lett. Appl. Microbiol. 2012, 55, 115–121. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Wu, F.; Huang, Y.C.; Jian, J.C.; Cai, S.H. Mechanisms Underlying the Virulence and Intestinal Colonization of the Vibrio alginolyticus HY9901 DctP Protein with Proteomic Analysis. Appl. Biochem. Microbiol. 2023, 59, 646–658. [Google Scholar] [CrossRef]
- Appelt, S.; Heuner, K. The Flagellar Regulon of Legionella-A Review. Front. Cell. Infect. Microbiol. 2017, 7, 454. [Google Scholar] [CrossRef]
- Liu, E.; Ye, J.; Song, S.; Wang, K.; Zhang, Y.; Zhang, H. Impact of co-deficiency of RpoN and RpoS on stress tolerance, virulence and gene regulation in Edwardsiella tarda. J. Basic Microbiol. 2014, 54, 678–687. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Zhang, X.; Zhang, Y.; Zhu, M.; Yang, P.; Yuan, J.; Xie, Y.; Zhou, T.; Wang, W.; Chen, S.; et al. RpoN-Dependent Direct Regulation of Quorum Sensing and the Type VI Secretion System in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2018, 200, e00205–e00218. [Google Scholar] [CrossRef] [PubMed]
- Homma, M.; Kobayakawa, T.; Hao, Y.; Nishikino, T.; Kojima, S. Function and Structure of FlaK, a Master Regulator of the Polar Flagellar Genes in Marine Vibrio. J. Bacteriol. 2022, 204, e0032022. [Google Scholar] [CrossRef]
- Luo, G.; Huang, L.; Su, Y.; Qin, Y.; Xu, X.; Zhao, L.; Yan, Q. flrA, flrB and flrC regulate adhesion by controlling the expression of critical virulence genes in Vibrio alginolyticus. Emerg. Microbes Infect. 2016, 5, e85. [Google Scholar] [CrossRef]
- Wu, C.M.; Huang, H.H.; Li, L.H.; Lin, Y.T.; Yang, T.C. Molecular Characterization of Three Tandemly Located Flagellin Genes of Stenotrophomonas maltophilia. Int. J. Mol. Sci. 2022, 23, 3863. [Google Scholar] [CrossRef]
- Groisman, E.A. Feedback Control of Two-Component Regulatory Systems. Annu. Rev. Microbiol. 2016, 70, 103–124. [Google Scholar] [CrossRef]
- Manera, K.; Caro, F.; Li, H.; Pei, T.T.; Hersch, S.J.; Mekalanos, J.J.; Dong, T.G. Sensing of intracellular Hcp levels controls T6SS expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 2021, 118, e2104813118. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, S.; Tang, J.; Wang, B.; Cai, J.; Jian, J. Roles of Hcp2, a Hallmark of T6SS2 in Motility, Adhesive Capacity, and Pathogenicity of Vibrio alginolyticus. Microorganisms 2023, 11, 2893. https://doi.org/10.3390/microorganisms11122893
Wu S, Tang J, Wang B, Cai J, Jian J. Roles of Hcp2, a Hallmark of T6SS2 in Motility, Adhesive Capacity, and Pathogenicity of Vibrio alginolyticus. Microorganisms. 2023; 11(12):2893. https://doi.org/10.3390/microorganisms11122893
Chicago/Turabian StyleWu, Shuilong, Jufen Tang, Bei Wang, Jia Cai, and Jichang Jian. 2023. "Roles of Hcp2, a Hallmark of T6SS2 in Motility, Adhesive Capacity, and Pathogenicity of Vibrio alginolyticus" Microorganisms 11, no. 12: 2893. https://doi.org/10.3390/microorganisms11122893




