Untangling the Effects of Plant Genotype and Soil Conditions on the Assembly of Bacterial and Fungal Communities in the Rhizosphere of the Wild Andean Blueberry (Vaccinium floribundum Kunth)
Abstract
1. Introduction
2. Materials and Methods
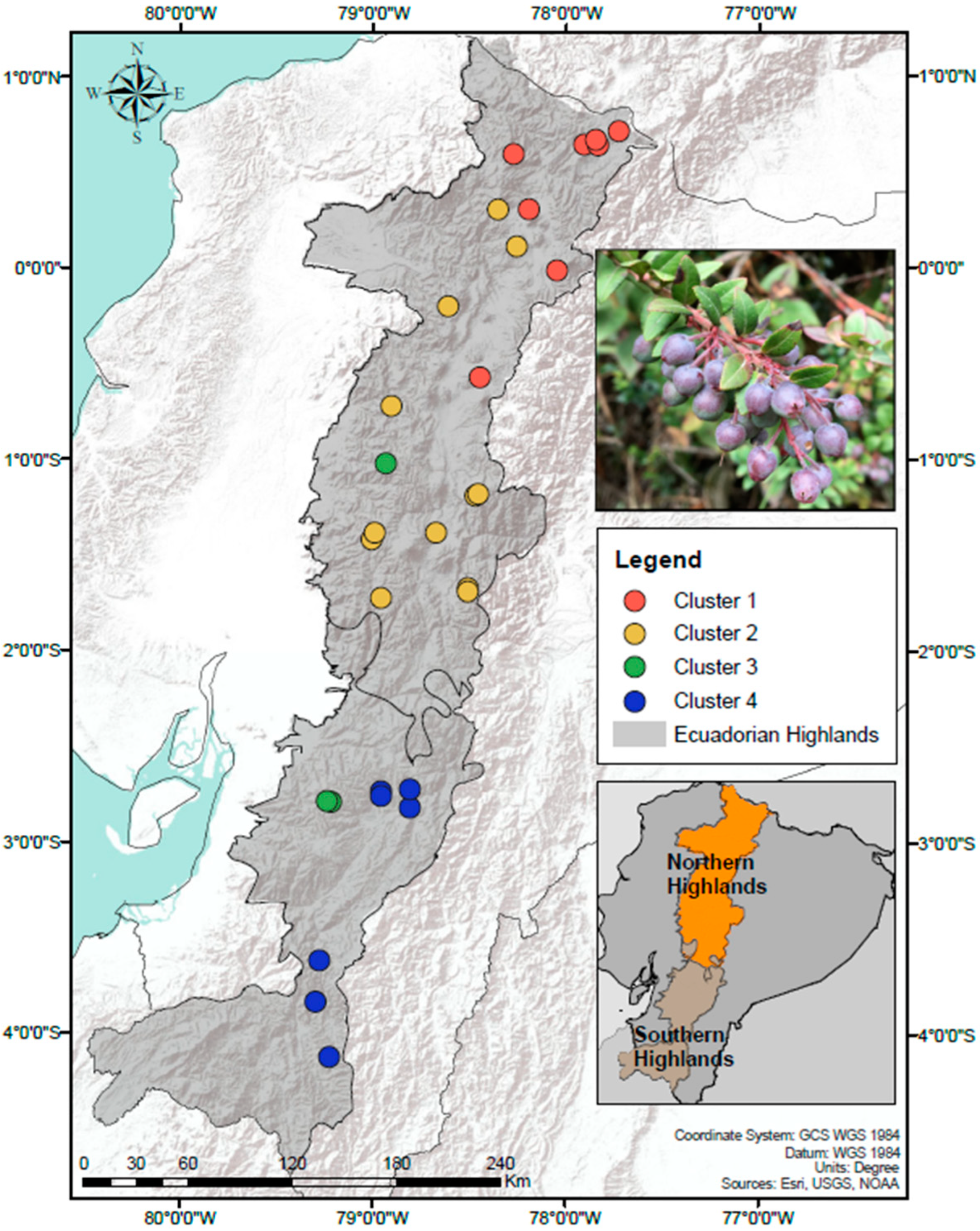
2.1. Sample Collection and Processing
2.2. Amplicon Sequence Data Processing
2.3. Alpha and Beta Diversity Analyses
2.4. Linear Mixed Models for Alpha-Diversity
3. Results
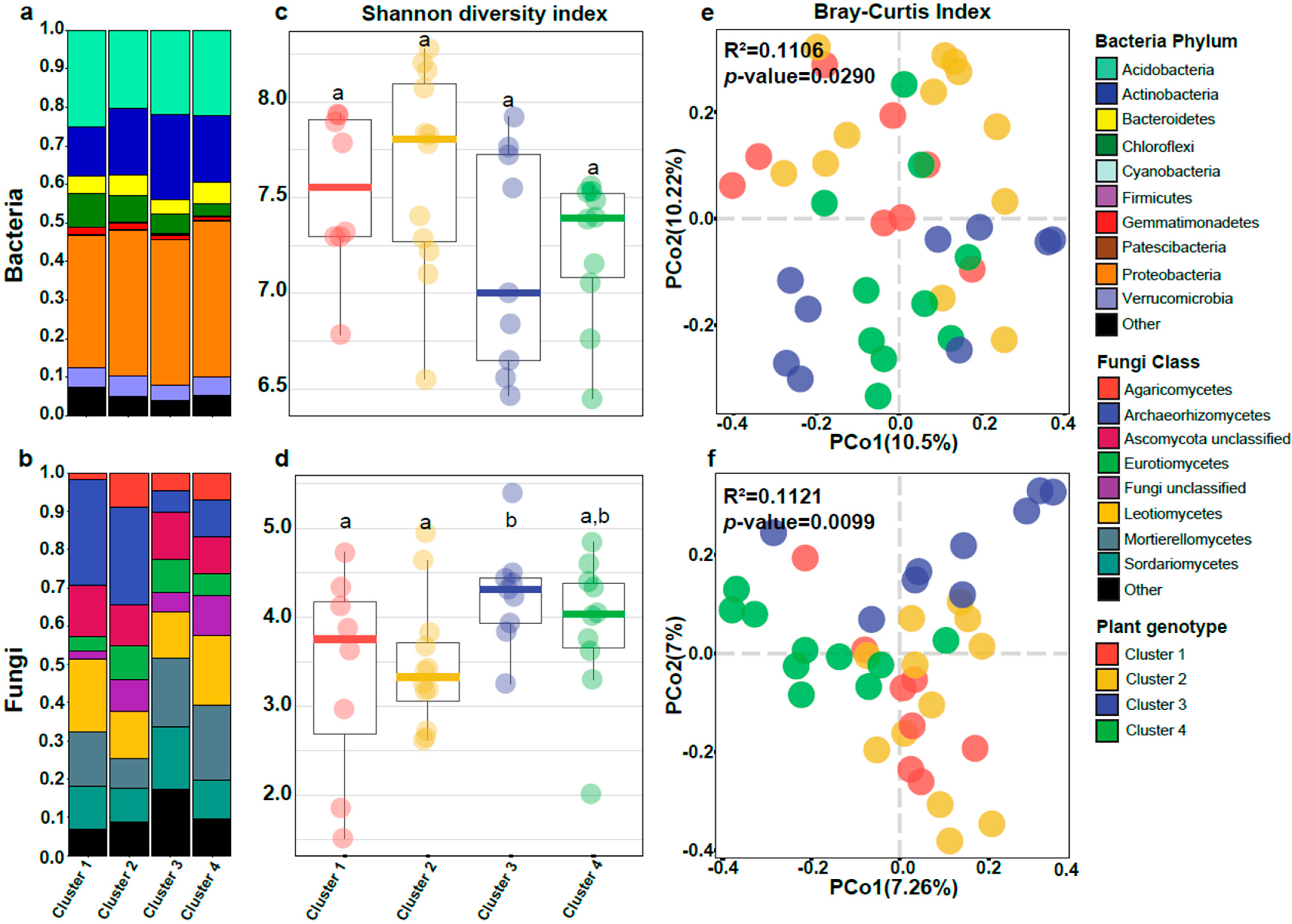
3.1. Bacterial Alpha-Diversity Differed across the Soil Regions of the Ecuadorian Highlands
3.2. Plant Genotype Influences Alpha-Diversity in Fungi
3.3. Soil Region Is the Main Predictor for Alpha-Diversity of Bacterial Communities, While Fungal Diversity Is Driven by the Interaction between Plant Genotype and Altitude
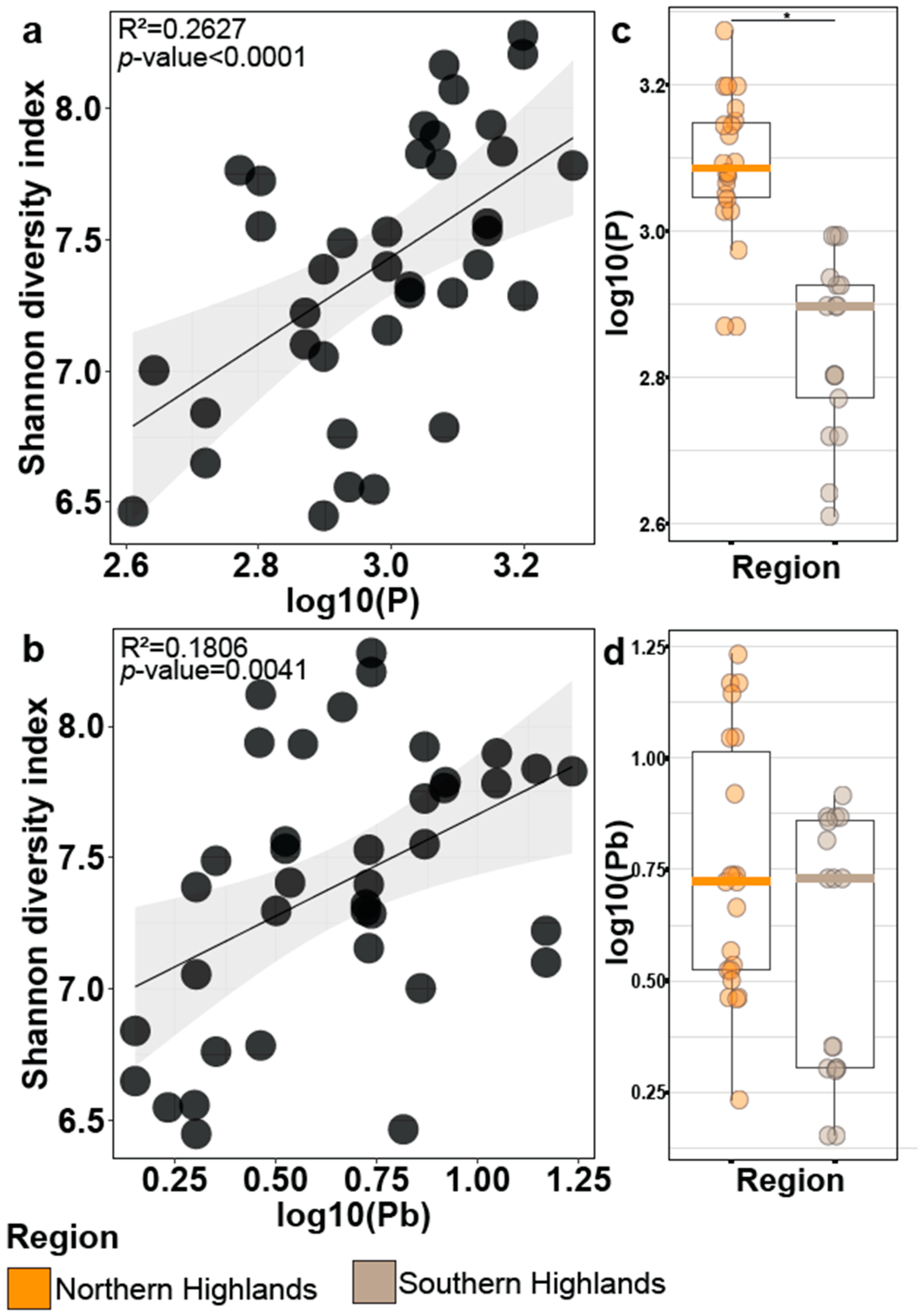
3.4. Bacterial Alpha-Diversity Is Correlated with the Phosphorus and Lead Found in the Páramo Soils
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Fitzpatrick, C.R.; Salas-González, I.; Conway, J.M.; Finkel, O.M.; Gilbert, S.; Russ, D.; Teixeira, P.J.P.L.; Dangl, J.L. The Plant Microbiome: From Ecology to Reductionism and Beyond. Annu. Rev. Microbiol. 2020, 74, 81–100. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Mattupalli, C.; Eversole, K.; Leach, J.E. Enabling sustainable agriculture through understanding and enhancement of microbiomes. New Phytol. 2021, 230, 2129–2147. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef]
- Yurgel, S.N.; Douglas, G.M.; Comeau, A.M.; Mammoliti, M.; Dusault, A.; Percival, D.; Langille, M.G.I. Variation in Bacterial and Eukaryotic Communities Associated with Natural and Managed Wild Blueberry Habitats. Phytobiomes J. 2017, 1, 102–113. [Google Scholar] [CrossRef]
- Klein, M.; Stewart, J.D.; Porter, S.S.; Weedon, J.T.; Kiers, E.T. Evolution of manipulative microbial behaviors in the rhizosphere. Evol. Appl. 2021, 15, 1521–1536. [Google Scholar] [CrossRef]
- Cheng, Z.; Lei, S.; Li, Y.; Huang, W.; Ma, R.; Xiong, J.; Zhang, T.; Jin, L.; Haq, H.U.; Xu, X.; et al. Revealing the Variation and Stability of Bacterial Communities in Tomato Rhizosphere Microbiota. Microorganisms 2020, 8, 170. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.R.; Lundberg, D.S.; del Rio, T.G.; Tringe, S.G.; Dangl, J.L.; Mitchell-Olds, T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 2016, 7, 12151. [Google Scholar] [CrossRef] [PubMed]
- Fernández, N.V.; Marchelli, P.; Tenreiro, R.; Chaves, S.; Fontenla, S.B. Are the rhizosphere fungal communities of Nothofagus alpina established in two different environments influenced by plant genetic diversity? For. Ecol. Manag. 2020, 473, 118269. [Google Scholar] [CrossRef]
- Fernie, A.R.; Yan, J. De Novo Domestication: An Alternative Route toward New Crops for the Future. Mol. Plant 2019, 12, 615–631. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, M.A.; Menoyo, E.; Lugo, M.A.; Negritto, M.A.; Farías, M.E.; Anton, A.M.; Siñeriz, F. Molecular characterization and in situ detection of bacterial communities associated with rhizosphere soil of high altitude native Poaceae from the Andean Puna region. J. Arid. Environ. 2010, 74, 1177–1185. [Google Scholar] [CrossRef]
- Jorquera, M.A.; Maruyama, F.; Ogram, A.V.; Navarrete, O.U.; Lagos, L.M.; Inostroza, N.G.; Acuña, J.J.; Rilling, J.I.; de La Luz Mora, M. Rhizobacterial Community Structures Associated with Native Plants Grown in Chilean Extreme Environments. Microb. Ecol. 2016, 72, 633–646. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Kiers, E.T. Rewilding plant microbiomes. Science 2022, 378, 599–600. [Google Scholar] [CrossRef]
- Wang, J.; Hu, A.; Meng, F.; Zhao, W.; Yang, Y.; Soininen, J.; Shen, J.; Zhou, J. Embracing mountain microbiome and ecosystem functions under global change. New Phytol. 2022, 234, 1987–2002. [Google Scholar] [CrossRef] [PubMed]
- Luteyn, J.L.; Pedraza-Peñaloza, P. Blueberry Relatives of the New World Tropics (Ericaceae); The New York Botanical Garden: New York, NY, USA, 2012; p. 1. [Google Scholar]
- Coba Santamaría, P.; Coronel, D.; Verdugo, K.; Paredes, M.F.; Yugsi, E.; Huachi, L. Estudio etnobotánico del mortiño (Vaccinium floribundum) como alimento ancestral y potencial alimento funcional. La Granja 2012, 16, 5–13. [Google Scholar] [CrossRef]
- Hofstede, R.; Segarra, P.; Mena, P. Los Páramos del Mundo: Proyecto atlas Mundial de los Páramos; Hofstede, R., Segarra, P., Mena Vásconez, P., Eds.; Global Peatland Initiative/NC-IUCN/EcoCiencia: Quito, Ecuador, 2003; pp. 1–300. [Google Scholar]
- Vega-Polo, P.; Cobo, M.M.; Argudo, A.; Gutierrez, B.; Rowntree, J.; Torres, M.D.L. Characterizing the genetic diversity of the Andean blueberry (Vaccinium floribundum Kunth.) across the Ecuadorian Highlands. PLoS ONE 2020, 15, e0243420. [Google Scholar] [CrossRef] [PubMed]
- Moreno, J.; Yerovi, F.; Herrera, M.; Yánez, D.; Espinosa, J. Soils from the Highlands. In The Soils of Ecuador; Springer: Berlin/Heidelberg, Germany, 2016; pp. 79–137. [Google Scholar] [CrossRef]
- Winckell, A. Los Grandes Rasgos del Relieve en el Ecuador. In Los Paisajes Naturales del Ecuador; Volúmen 1—Las Condiciones del Medio Natural; IGM: Quito, Ecuador, 1997. [Google Scholar]
- Comeau, A.M.; Douglas, G.M.; Langille, M.G.I. Microbiome Helper: A Custom and Streamlined Workflow for Microbiome Research. Msystems 2017, 2, e00127-16. [Google Scholar] [CrossRef]
- ASTM D 2974; Standard Test Methods for Moisture, Ash, and Organic Matter of Peat and Other Organic Soils. ASTM International: West Conshohocken, PA, USA, 2014.
- US EPA. EPA Method 6010C: Inductively Coupled Plasma-Atomic Emission Spectrometry; EPA Method 6010C; US EPA: Washington, DC, USA, 2007.
- Yourstone, S.M.; Lundberg, D.S.; Dangl, J.L.; Jones, C.D. MT-Toolbox: Improved amplicon sequencing using molecule tags. BMC Bioinform. 2014, 15, 284. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef] [PubMed]
- Salas-Gonzalez, I. Isaisg/Ohchibi: Iskali, version v1.0.0; Zenodo: Genève, Switzerland, 2019. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B. Community Ecology Package, version 2.9. Package Vegan 2013, 2, 1–295. [Google Scholar]
- Lenth, R.; Singmann, H.; Love, J.; Buerkner, P.; Herve, M. Emmeans: Estimated Marginal Measn, Aka Least-Squares Means, R package version 1.1. R Package 2018, 1, 3. [Google Scholar]
- Zhang, Z. Variable selection with stepwise and best subset approaches. Ann. Transl. Med. 2016, 4, 136. [Google Scholar] [CrossRef] [PubMed]
- Zebrowski, C.; Sourdat, M. Los Factores de la Pedogénesis y los Suelos en Ecuador. In Los Paisajes Naturales del Ecuador Tomo I—Las Condiciones Generales del Medio Natural; IGM: Quito, Ecuador, 1997. [Google Scholar]
- Beilsmith, K.; Perisin, M.; Bergelson, J. Natural Bacterial Assemblages in Arabidopsis thaliana Tissues Become More Distinguishable and Diverse during Host Development. mBio 2021, 12, e02723-20. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Semchenko, M.; Barry, K.E.; Vries, F.T.; Mommer, L.; Moora, M.; Maciá-Vicente, J.G. Deciphering the role of specialist and generalist plant–microbial interactions as drivers of plant–soil feedback. New Phytol. 2022, 234, 1929–1944. [Google Scholar] [CrossRef]
- Francioli, D.; Schulz, E.; Lentendu, G.; Wubet, T.; Buscot, F.; Reitz, T. Mineral vs. Organic Amendments: Microbial Community Structure, Activity and Abundance of Agriculturally Relevant Microbes Are Driven by Long-Term Fertilization Strategies. Front. Microbiol. 2016, 7, 1446. [Google Scholar] [CrossRef]
- Kidinda, L.K.; Doetterl, S.; Kalbitz, K.; Bukombe, B.; Babin, D.; Mujinya, B.B.; Vogel, C. Relationships between geochemical properties and microbial nutrient acquisition in tropical forest and cropland soils. Appl. Soil Ecol. 2023, 181, 104653. [Google Scholar] [CrossRef]
- Janssen, P.H.; Yates, P.S.; Grinton, B.E.; Taylor, P.M.; Sait, M. Improved Culturability of Soil Bacteria and Isolation in Pure Culture of Novel Members of the Divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 2002, 68, 2391–2396. [Google Scholar] [CrossRef]
- Smit, E.; Leeflang, P.; Gommans, S.; Van Den Broek, J.; van Mil, S.; Wernars, K. Diversity and Seasonal Fluctuations of the Dominant Members of the Bacterial Soil Community in a Wheat Field as Determined by Cultivation and Molecular Methods. Appl. Environ. Microbiol. 2001, 67, 2284–2291. [Google Scholar] [CrossRef]
- Yang, Y.; Chai, Y.; Xie, H.; Zhang, L.; Zhang, Z.; Yang, X.; Hao, S.; Gai, J.; Chen, Y. Responses of soil microbial diversity, network complexity and multifunctionality to three land-use changes. Sci. Total Environ. 2023, 859, 160255. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-H.; Ka, J.-O.; Cho, J.-C. Members of the phylum Acidobacteria are dominant and metabolically active in rhizosphere soil. FEMS Microbiol. Lett. 2008, 285, 263–269. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Mavrodi, O.V.; Hou, J.; Blackmon, C.; Babiker, E.M.; Mavrodi, D. Comparative Analysis of Rhizosphere Microbiomes of Southern Highbush Blueberry (Vaccinium corymbosum L.), Darrow’s Blueberry (V. darrowii Camp), and Rabbiteye Blueberry (V. virgatum Aiton). Front. Microbiol. 2020, 11, 370. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Q.; Wang, F.; Zhang, J.; Chen, Y.; Zhang, C.; Liu, G.; Zhang, H.; Ma, C.; Zhang, J. The Variation in the Rhizosphere Microbiome of Cotton with Soil Type, Genotype and Developmental Stage. Sci. Rep. 2017, 7, 3940. [Google Scholar] [CrossRef]
- Castrillo, G.; Teixeira, P.J.P.L.; Paredes, S.H.; Law, T.F.; De Lorenzo, L.; Feltcher, M.E.; Finkel, O.M.; Breakfield, N.W.; Mieczkowski, P.; Jones, C.D.; et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature 2017, 543, 513–518. [Google Scholar] [CrossRef]
- Finkel, O.M.; Salas-González, I.; Castrillo, G.; Spaepen, S.; Law, T.F.; Teixeira, P.J.; Jones, C.D.; Dangl, J.L. The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response. PLoS Biol. 2019, 17, e3000534. [Google Scholar] [CrossRef]
- Orellana, D.; Machuca, D.; Ibeas, M.A.; Estevez, J.M.; Poupin, M.J. Plant-growth promotion by proteobacterial strains depends on the availability of phosphorus and iron in Arabidopsis thaliana plants. Front. Microbiol. 2022, 13, 4956. [Google Scholar] [CrossRef]
- Hui, N.; Liu, X.X.; Kurola, J.; Mikola, J.; Romantschuk, M. Lead (Pb) contamination alters richness and diversity of the fungal, but not the bacterial community in pine forest soil. Boreal Environment Research. Boreal Environ. Res. 2012, 17, 46–58. [Google Scholar]
- Khan, S.; Hesham, A.E.-L.; Qiao, M.; Rehman, S.; He, J.-Z. Effects of Cd and Pb on soil microbial community structure and activities. Environ. Sci. Pollut. Res. 2009, 17, 288–296. [Google Scholar] [CrossRef]
- Fajardo, C.; Costa, G.; Nande, M.; Botías, P.; García-Cantalejo, J.; Martín, M. Pb, Cd, and Zn soil contamination: Monitoring functional and structural impacts on the microbiome. Appl. Soil Ecol. 2018, 135, 56–64. [Google Scholar] [CrossRef]
- Burns, J.H.; Anacker, B.L.; Strauss, S.Y.; Burke, D.J. Soil microbial community variation correlates most strongly with plant species identity, followed by soil chemistry, spatial location and plant genus. AoB PLANTS 2015, 7, plv0300. [Google Scholar] [CrossRef] [PubMed]
- Orgiazzi, A.; Bianciotto, V.; Bonfante, P.; Daghino, S.; Ghignone, S.; Lazzari, A.; Lumini, E.; Mello, A.; Napoli, C.; Perotto, S.; et al. 454 Pyrosequencing Analysis of Fungal Assemblages from Geographically Distant, Disparate Soils Reveals Spatial Patterning and a Core Mycobiome. Diversity 2013, 5, 73–98. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef]
- Yao, F.; Yang, S.; Wang, Z.; Wang, X.; Ye, J.; Wang, X.; DeBruyn, J.; Feng, X.; Jiang, Y.; Li, H. Microbial Taxa Distribution Is Associated with Ecological Trophic Cascades along an Elevation Gradient. Front. Microbiol. 2017, 8, 2071. [Google Scholar] [CrossRef] [PubMed]
- Colpaert, J.V.; Van Tichelen, K.K. Decomposition, nitrogen and phosphorus mineralization from beech leaf litter colonized by ectomycorrhizal or litter-decomposing basidiomycetes. New Phytol. 1996, 134, 123–132. [Google Scholar] [CrossRef]
- Janowski, D.; Leski, T. Factors in the Distribution of Mycorrhizal and Soil Fungi. Diversity 2022, 14, 1122. [Google Scholar] [CrossRef]
- Rosling, A.; Cox, F.; Cruz-Martinez, K.; Ihrmark, K.; Grelet, G.-A.; Lindahl, B.D.; Menkis, A.; James, T.Y. Archaeorhizomycetes: Unearthing an Ancient Class of Ubiquitous Soil Fungi. Science 2011, 333, 876–879. [Google Scholar] [CrossRef]
- Dickey, J.R.; Swenie, R.A.; Turner, S.C.; Winfrey, C.C.; Yaffar, D.; Padukone, A.; Beals, K.K.; Sheldon, K.S.; Kivlin, S.N. The Utility of Macroecological Rules for Microbial Biogeography. Front. Ecol. Evol. 2021, 9, 633155. [Google Scholar] [CrossRef]
- Martin, H.; Johanna, M.; Anna, H.; Jürg, E.; Andreas, G.; Meuli, R.G.; Beat, F.; Franco, W. Core and indicative bacterial and fungal taxa define characteristic soil communities of arable land, grassland, and forest. bioRxiv 2021. [Google Scholar] [CrossRef]
- Hoeksema, J.D.; Bever, J.D.; Chakraborty, S.; Chaudhary, V.B.; Gardes, M.; Gehring, C.A.; Hart, M.M.; Housworth, E.A.; Kaonongbua, W.; Klironomos, J.N.; et al. Evolutionary history of plant hosts and fungal symbionts predicts the strength of mycorrhizal mutualism. Commun. Biol. 2018, 1, 116. [Google Scholar] [CrossRef] [PubMed]
- Strullu-Derrien, C.; Selosse, M.A.; Kenrick, P.; Martin, F.M. The origin and evolution of mycorrhizal symbioses: From palaeomycology to phylogenomics. New Phytol. 2018, 220, 1012–1030. [Google Scholar] [CrossRef] [PubMed]
- Bergelson, J.; Mittelstrass, J.; Horton, M.W. Characterizing both bacteria and fungi improves understanding of the Arabidopsis root microbiome. Sci. Rep. 2019, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Gehring, C.A.; Sthultz, C.M.; Flores-Rentería, L.; Whipple, A.V.; Whitham, T.G. Tree genetics defines fungal partner communities that may confer drought tolerance. Proc. Natl. Acad. Sci. USA 2017, 114, 11169–11174. [Google Scholar] [CrossRef] [PubMed]
- Frąc, M.; Hannula, E.S.; Bełka, M.; Salles, J.F.; Jedryczka, M. Soil mycobiome in sustainable agriculture. Front. Microbiol. 2022, 13, 1033824. [Google Scholar] [CrossRef]
- Bayranvand, M.; Akbarinia, M.; Salehi Jouzani, G.; Gharechahi, J.; Kooch, Y.; Baldrian, P. Composition of soil bacterial and fungal communities in relation to vegetation composition and soil characteristics along an altitudinal gradient. FEMS Microbiol. Ecol. 2020, 97, fiaa201. [Google Scholar] [CrossRef]
- D’Alò, F.; Baldrian, P.; Odriozola, I.; Morais, D.; Větrovský, T.; Zucconi, L.; Ripa, C.; Cannone, N.; Malfasi, F.; Onofri, S. Composition and functioning of the soil microbiome in the highest altitudes of the Italian Alps and potential effects of climate change. FEMS Microbiol. Ecol. 2022, 98, fiac025. [Google Scholar] [CrossRef]
- Tang, M.; Li, L.; Wang, X.; You, J.; Li, J.; Chen, X. Elevational is the main factor controlling the soil microbial community structure in alpine tundra of the Changbai Mountain. Sci. Rep. 2020, 10, 12442. [Google Scholar] [CrossRef]
- Dassen, S.; Cortois, R.; Martens, H.; de Hollander, M.; Kowalchuk, G.A.; van der Putten, W.H.; De Deyn, G.B. Differential responses of soil bacteria, fungi, archaea and protists to plant species richness and plant functional group identity. Mol. Ecol. 2017, 26, 4085–4098. [Google Scholar] [CrossRef]
- de Vries, F.T.; Griffiths, R.I.; Bailey, M.; Craig, H.; Girlanda, M.; Gweon, H.S.; Hallin, S.; Kaisermann, A.; Keith, A.M.; Kretzschmar, M.; et al. Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 2018, 9, 3033. [Google Scholar] [CrossRef] [PubMed]

| Shannon Diversity Index | ||
|---|---|---|
| Bacteria | Fungi | |
| R2 = 0.3623 (0.0058) | R2 = 0.4694 (0.0155) | |
| Region | 7.857 (0.0088) | 3.581 (0.0681) |
| Plant genotype | 0.096 (0.9618) | 2.814 (0.0561) |
| Altitude range | 0.316 (0.7314) | 0.195 (0.8242) |
| LDepth | 1.875 (0.1811) | 2.159 (0.1521) |
| Plant genotype × Altitude range | 2.181 (0.1501) | 9.652 (0.0036) |
| Shannon Diversity Index | |
|---|---|
| R2 = 0.4137 (0.0005) | |
| log10P | 2.213 (0.0341) |
| log10Pb | 2.105 (0.0432) |
| Edaphic factor PC1 | 0.176 (0.8614) |
| Edaphic factor PC2 | 1.322 (0.1957) |
| Edaphic factor PC3 | 0.237 (0.8144) |
| only log10P + log10Pb | R2 = 0.4065 (3.15 × 10−5) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramirez-Villacis, D.X.; Pinos-Leon, A.; Vega-Polo, P.; Salas-González, I.; Jones, C.D.; Torres, M.d.L. Untangling the Effects of Plant Genotype and Soil Conditions on the Assembly of Bacterial and Fungal Communities in the Rhizosphere of the Wild Andean Blueberry (Vaccinium floribundum Kunth). Microorganisms 2023, 11, 399. https://doi.org/10.3390/microorganisms11020399
Ramirez-Villacis DX, Pinos-Leon A, Vega-Polo P, Salas-González I, Jones CD, Torres MdL. Untangling the Effects of Plant Genotype and Soil Conditions on the Assembly of Bacterial and Fungal Communities in the Rhizosphere of the Wild Andean Blueberry (Vaccinium floribundum Kunth). Microorganisms. 2023; 11(2):399. https://doi.org/10.3390/microorganisms11020399
Chicago/Turabian StyleRamirez-Villacis, Dario X., Andrea Pinos-Leon, Pamela Vega-Polo, Isai Salas-González, Corbin D. Jones, and Maria de Lourdes Torres. 2023. "Untangling the Effects of Plant Genotype and Soil Conditions on the Assembly of Bacterial and Fungal Communities in the Rhizosphere of the Wild Andean Blueberry (Vaccinium floribundum Kunth)" Microorganisms 11, no. 2: 399. https://doi.org/10.3390/microorganisms11020399
APA StyleRamirez-Villacis, D. X., Pinos-Leon, A., Vega-Polo, P., Salas-González, I., Jones, C. D., & Torres, M. d. L. (2023). Untangling the Effects of Plant Genotype and Soil Conditions on the Assembly of Bacterial and Fungal Communities in the Rhizosphere of the Wild Andean Blueberry (Vaccinium floribundum Kunth). Microorganisms, 11(2), 399. https://doi.org/10.3390/microorganisms11020399







