Storage and Algal Association of Bacteria That Protect Microchloropsis salina from Grazing by Brachionus plicatilis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Algal, Rotifer, and Algal–Bacterial Cultures
2.2. Microalgal Growth Assay, Specific Growth Rate Calculation, and Rotifer Counts
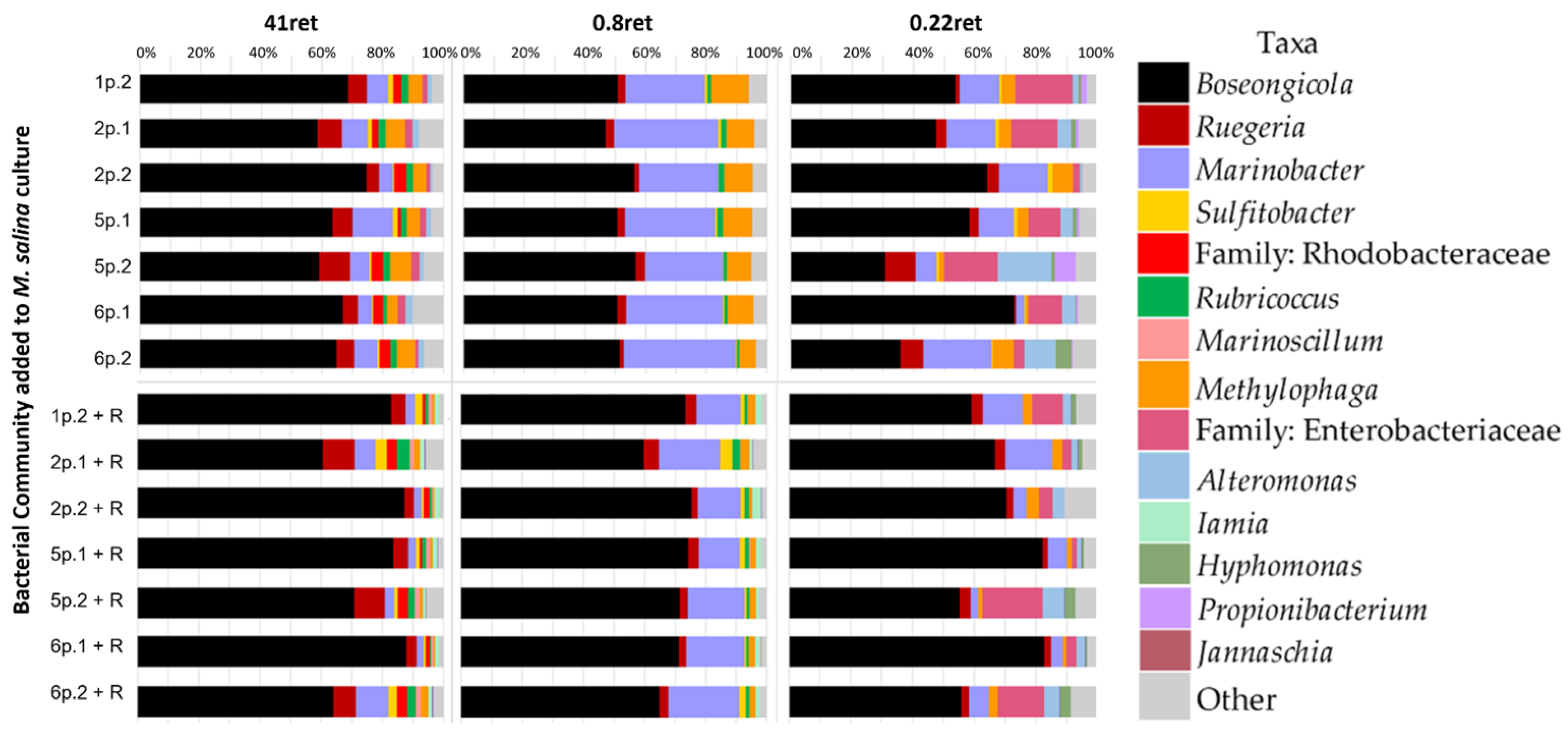
2.3. Fractionation by Sequential Filtration
2.4. DNA Extraction, Library Preparation, SSUrRNA Amplicon Sequencing, and Data Analysis
3. Results
3.1. Microalgae Growth Experiment
3.2. Bacterial Community Composition
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, M.I.; Shin, J.H.; Kim, J.D. The promising future of microalgae: Current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb. Cell Fact. 2018, 17, 36. [Google Scholar] [CrossRef]
- Lane, T.W. Barriers to microalgal mass cultivation. Curr. Opin. Biotechnol. 2020, 73, 323–328. [Google Scholar] [CrossRef]
- Zittelli, G.C.; Biondi, N.; Rodolfi, L.; Tredici, M.R. Photobioreactors for Mass Production of Microalgae. In Handbook of Microalgal Culture; John Wiley & Sons, Ltd.: Oxford, UK, 2013; pp. 225–266. [Google Scholar]
- Kazamia, E.; Riseley, A.S.; Howe, C.J.; Smith, A.G. An Engineered Community Approach for Industrial Cultivation of Microalgae. Ind. Biotechnol. 2014, 10, 184–190. [Google Scholar] [CrossRef] [Green Version]
- Carney, L.T.; Lane, T.W. Parasites in algae mass culture. Front. Microbiol. 2014, 5, 278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.; Jiang, J.; Fa, Y. Overcoming the Biological Contamination in Microalgae and Cyanobacteria Mass Cultivations for Photosynthetic Biofuel Production. Molecules 2020, 25, 5220. [Google Scholar] [CrossRef]
- Lubzens, E. Raising rotifers for use in aquaculture. Hydrobiologia 1987, 147, 245–255. [Google Scholar] [CrossRef]
- Huang, Y.; Li, L.; Liu, J.; Lin, W. Botanical pesticides as potential rotifer-control agents in microalgal mass culture. Algal Res. 2014, 4, 62–69. [Google Scholar] [CrossRef]
- Hirayama, K.; Ogawa, S. Fundamental studies on physiology of rotifer for its mass culture-I. Nippon Suisan Gakkai Shi 1972, 38, 1207–1214. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Van Ginkel, S.W.; Pradeep, P.; Igou, T.; Yi, C.; Snell, T.; Chen, Y. The Selective Use of Hypochlorite to Prevent Pond Crashes for Algae-Biofuel Production. Water Environ. Res. 2016, 88, 70–78. [Google Scholar] [CrossRef]
- Fischer, B.B.; Roffler, S.; Eggen, R.I.L. Multiple stressor effects of predation by rotifers and herbicide pollution on different Chlamydomonas strains and potential impacts on population dynamics. Environ. Toxicol. Chem. 2012, 31, 2832–2840. [Google Scholar] [CrossRef]
- Fisher, C.L.; Ward, C.S.; Lane, P.D.; Kimbrel, J.A.; Sale, K.L.; Stuart, R.K.; Mayali, X.; Lane, T.W. Bacterial communities protect the alga Microchloropsis salina from grazing by the rotifer Brachionus plicatilis. Algal Res. 2019, 40, 101500. [Google Scholar] [CrossRef]
- Natrah, F.; Kenmegne, M.M.; Wiyoto, W.; Sorgeloos, P.; Bossier, P.; Defoirdt, T. Effects of micro-algae commonly used in aquaculture on acyl-homoserine lactone quorum sensing. Aquaculture 2011, 317, 53–57. [Google Scholar] [CrossRef]
- Deines, P.; Matz, C.; Jürgens, K. Toxicity of violacein-producing bacteria fed to bacterivorous freshwater plankton. Limnol. Oceanogr. 2009, 54, 1343–1352. [Google Scholar] [CrossRef]
- Ward, C.S.; Rolison, K.; Li, M.; Rozen, S.; Fisher, C.L.; Lane, T.W.; Thelen, M.P.; Stuart, R.K. Janthinobacter additions reduce rotifer grazing of microalga Microchloropsis salina in biotically complex communities. Algal Res. 2021, 58, 102400. [Google Scholar] [CrossRef]
- Berges, J.A.; Franklin, D.J.; Harrison, P.J. Evolution of an artificial seawater medium: Improvements in enriched seawater, artificial water over the last two decades. J. Phycol. 2001, 37, 1138–1145. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Mayers, T.J.; Bramucci, A.R.; Yakimovich, K.M.; Case, R.J. A Bacterial Pathogen Displaying Temperature-Enhanced Virulence of the Microalga Emiliania huxleyi. Front. Microbiol. 2016, 7, 892. [Google Scholar] [CrossRef] [Green Version]
- Sonnenschein, E.C.; Nielsen, K.F.; D’Alvise, P.; Porsby, C.H.; Melchiorsen, J.; Heilmann, J.; Kalatzis, P.G.; López-Pérez, M.; Bunk, B.; Spröer, C.; et al. Global occurrence and heterogeneity of the Roseobacter-clade species Ruegeria mobilis. ISME J. 2016, 11, 569–583. [Google Scholar] [CrossRef] [Green Version]
- Le Chevanton, M.; Garnier, M.; Bougaran, G.; Schreiber, N.; Lukomska, E.; Bérard, J.-B.; Fouilland, E.; Bernard, O.; Cadoret, J.-P. Screening and selection of growth-promoting bacteria for Dunaliella cultures. Algal Res. 2013, 2, 212–222. [Google Scholar] [CrossRef] [Green Version]
- Koch, H.; Germscheid, N.; Freese, H.M.; Noriega-Ortega, B.; Lücking, D.; Berger, M.; Qiu, G.; Marzinelli, E.M.; Campbell, A.H.; Steinberg, C.B.; et al. Genomic, metabolic and phenotypic variability shapes ecological differentiation and intraspecies interactions of Alteromonas macleodii. Sci. Rep. 2020, 10, 809. [Google Scholar] [CrossRef] [Green Version]
- Amin, S.A.; Parker, M.S. Armbrust EV Interactions between Diatoms and Bacteria. Microbiol. Mol. Biol. Rev. 2012, 76, 667–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amin, S.A.; Green, D.H.; Hart, M.C.; Küpper, F.C.; Sunda, W.G.; Carrano, C.J. Photolysis of iron–siderophore chelates promotes bacterial–algal mutualism. Proc. Natl. Acad. Sci. USA 2019, 106, 17071–17076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
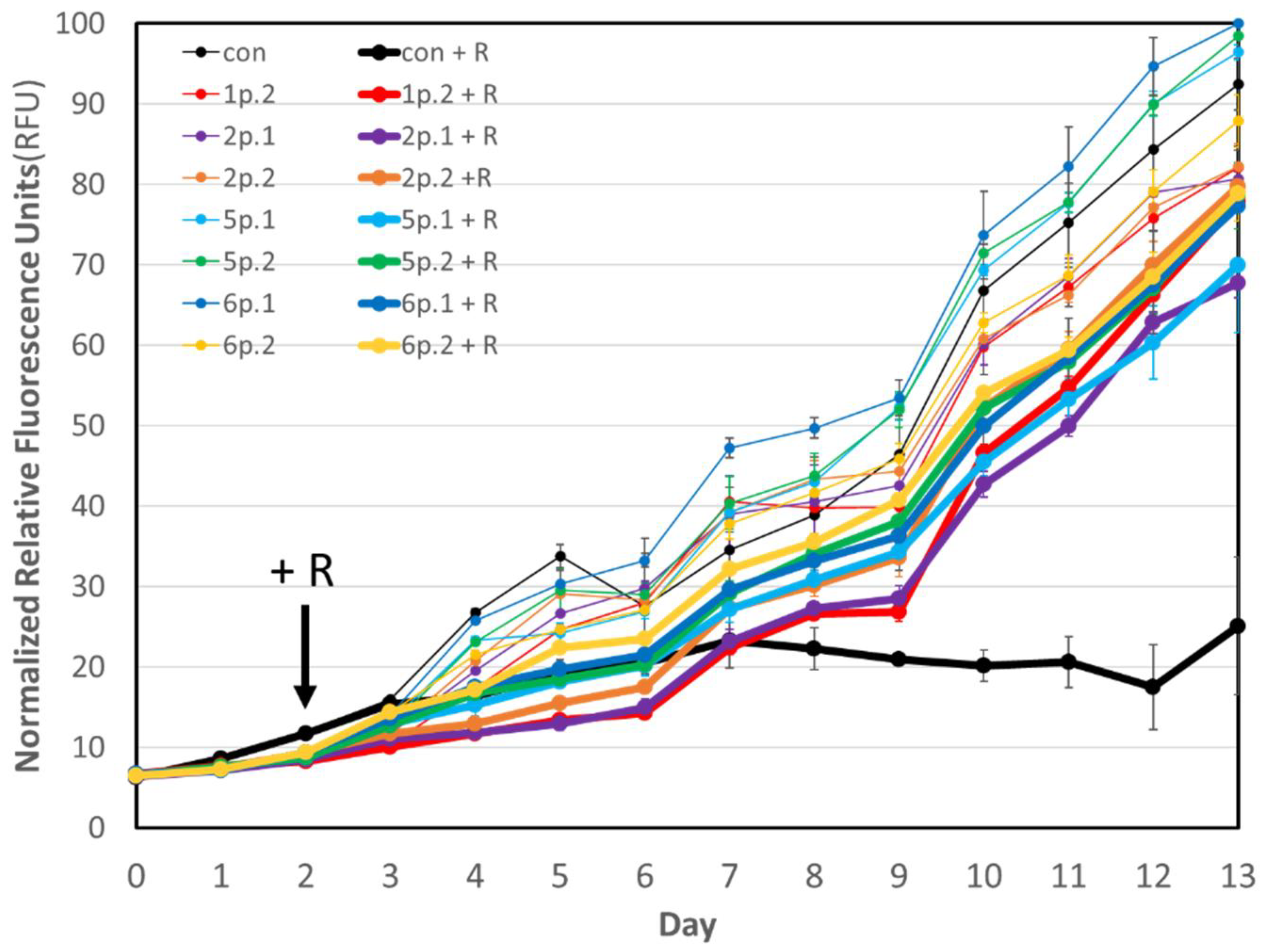
| Normalized Average Specific Growth Rate (Day−1) | Normalized Average Specific Growth Rate (Day−1) | Average Motile Rotifers mL−1 | ||||
|---|---|---|---|---|---|---|
| Day 6 | Day 11 | Day 13 | ||||
| CON | 100 | CON + R | 13.5 | 25 | 75 | 37.5 |
| 1p.2 | 108.6 | 1p.2 + R | 123.2 | 2.5 | 1.7 | 1.7 |
| 2p.1 | 117.0 | 2p.1 + R | 105.8 | 10 | 5 | 0 |
| 2p.2 | 109.6 | 2p.2 + R | 106.9 | 11.7 | 1.7 | 0.8 |
| 5p.1 | 115.2 | 5p.1 + R | 133.6 | 22.5 | 5 | 8.3 |
| 5p.2 | 106.1 | 5p.2 + R | 127.0 | 7.5 | 3.3 | 1.7 |
| 6p.1 | 115.0 | 6p.1 + R | 112.8 | 15.8 | 9.2 | 0.8 |
| 6p.2 | 108.9 | 6p.2 + R | 105.1 | 4.2 | 8.3 | 5.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fisher, C.L.; Fong, M.V.; Lane, P.D.; Carlson, S.; Lane, T.W. Storage and Algal Association of Bacteria That Protect Microchloropsis salina from Grazing by Brachionus plicatilis. Microorganisms 2023, 11, 786. https://doi.org/10.3390/microorganisms11030786
Fisher CL, Fong MV, Lane PD, Carlson S, Lane TW. Storage and Algal Association of Bacteria That Protect Microchloropsis salina from Grazing by Brachionus plicatilis. Microorganisms. 2023; 11(3):786. https://doi.org/10.3390/microorganisms11030786
Chicago/Turabian StyleFisher, Carolyn L., Michelle V. Fong, Pamela D. Lane, Skylar Carlson, and Todd W. Lane. 2023. "Storage and Algal Association of Bacteria That Protect Microchloropsis salina from Grazing by Brachionus plicatilis" Microorganisms 11, no. 3: 786. https://doi.org/10.3390/microorganisms11030786





