Current and Future Flow Cytometry Applications Contributing to Antimicrobial Resistance Control
Abstract
:1. Introduction
2. The AR Challenge
3. The Landscape of Rapid Methods for Antibiotic Susceptibility Testing
4. Rapid Detection of Bacterial Pathogens and Resistance–Contribution of FCM
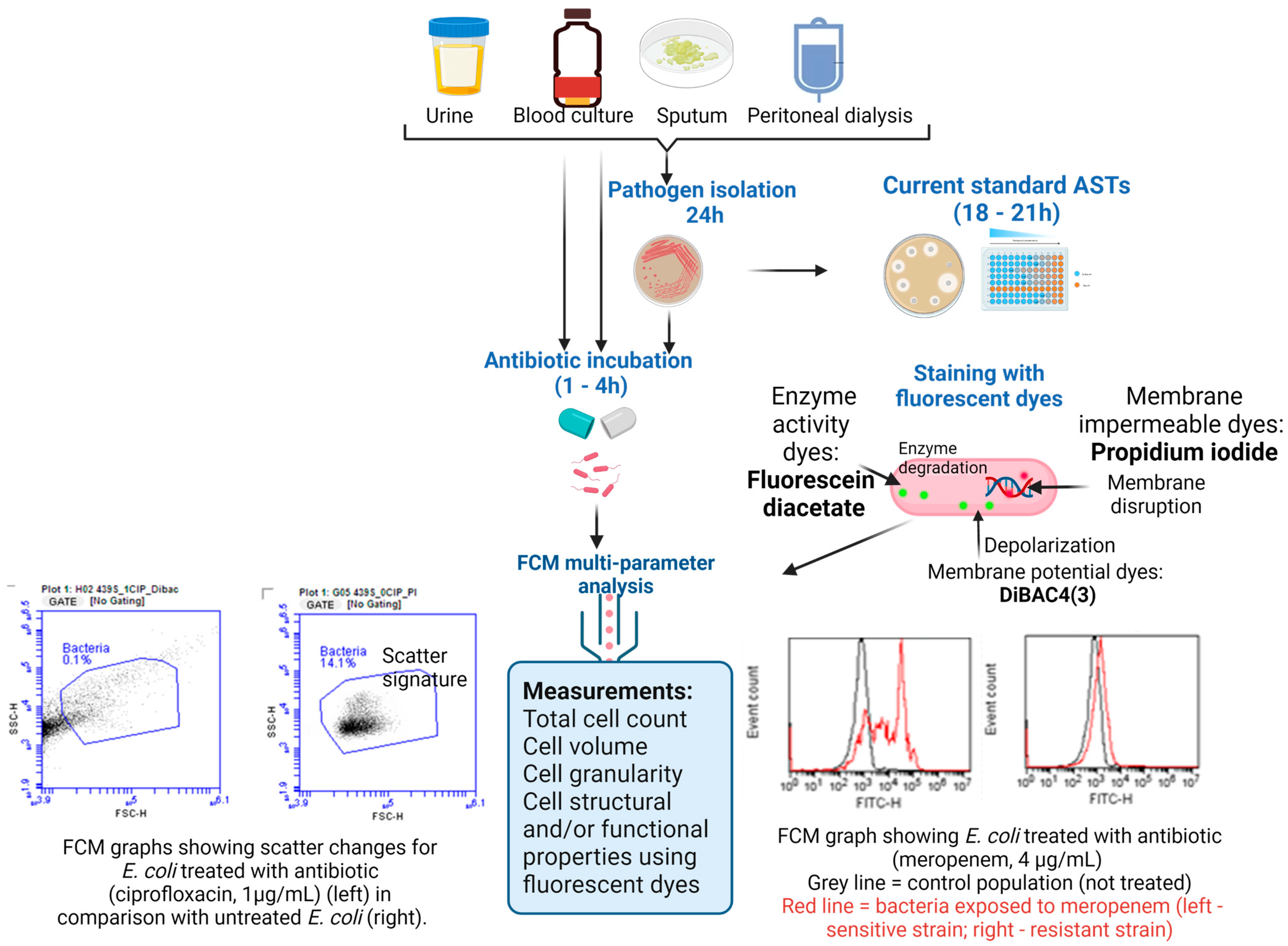
4.1. Direct Pathogen Detection and AST on Clinical Samples Using FCM
4.1.1. Urine Samples
4.1.2. Blood Samples and Hemocultures
4.1.3. Peritoneal Dialysis and Sputum
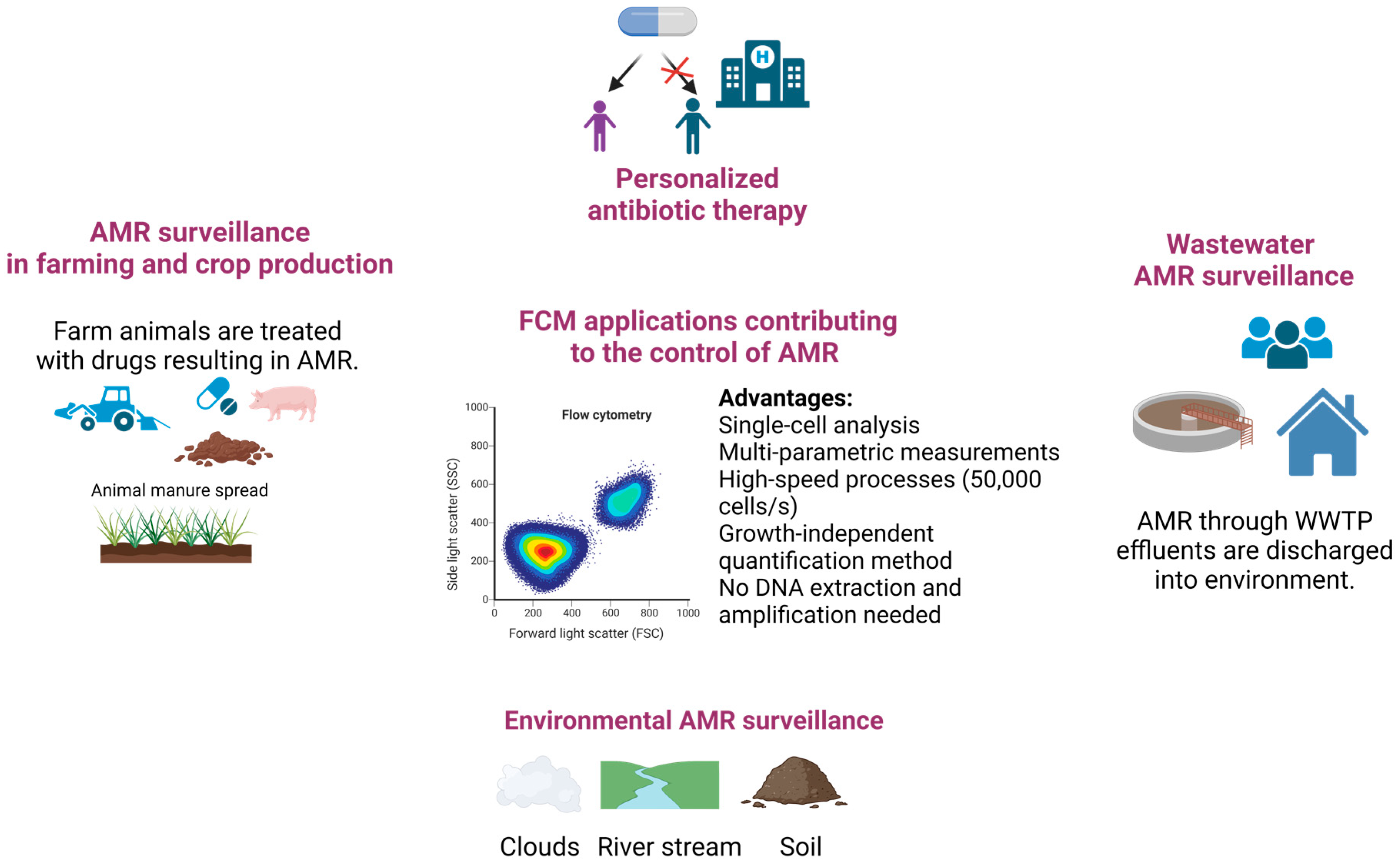
4.2. Detection and Quantification of AMR in the Environment
5. Perspectives for Further Contributions of FCM in Tackling AMR
6. Conclusions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Godeux, A.S.; Lupo, A.; Haenni, M.; Guette-Marquet, S.; Wilharm, G.; Laaberki, M.H.; Charpentier, X. Fluorescence-Based Detection of Natural Transformation in Drug-Resistant Acinetobacter baumannii. J. Bacteriol. 2018, 200, e00181-18. [Google Scholar] [CrossRef]
- Zhang, H.; Song, J.; Zheng, Z.; Li, T.; Shi, N.; Han, Y.; Zhang, L.; Yu, Y.; Fang, H. Fungicide exposure accelerated horizontal transfer of antibiotic resistance genes via plasmid-mediated conjugation. Water Res. 2023, 233, 119789. [Google Scholar] [CrossRef]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. 2016. Available online: https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf (accessed on 1 March 2023).
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.M.; Wertheim, H.F.L.; Sumpradit, N.; Vlieghe, E.; Levy Hara, G.; Gould, I.; Goossens, M.; et al. Antibiotic resistance—The need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef] [PubMed]
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Pant, S.; Gandra, S.; Levin, S.A.; Goossens, H.; Laxminarayan, R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, E3463–E3470. [Google Scholar] [CrossRef]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef]
- Bianco, A.; Papadopoli, R.; Mascaro, V.; Pileggi, C.; Pavia, M. Antibiotic prescriptions to adults with acute respiratory tract infections by Italian general practitioners. Infect. Drug Resist. 2018, 11, 2199–2205. [Google Scholar] [CrossRef] [PubMed]
- Schmiege, D.; Evers, M.; Kistemann, T.; Falkenberg, T. What drives antibiotic use in the community? A systematic review of determinants in the human outpatient sector. Int. J. Hyg. Environ. Health 2020, 226, 113497. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Modarai, M.; Naylor, N.R.; Boyd, S.E.; Atun, R.; Barlow, J.; Holmes, A.H.; Robotham, J.V. Quantifying drivers of antibiotic resistance in humans: A systematic review. Lancet Infect. Dis. 2018, 18, e368–e378. [Google Scholar] [CrossRef] [PubMed]
- Collignon, P.; Beggs, J.; Walsh, T.; Gandra, S.; Laxminarayan, R. Anthropological and socioeconomic factors contributing to global antimicrobial resistance: A univariate and multivariable analysis. Lancet Planet Health 2018, 2, e398–e405. [Google Scholar] [CrossRef] [PubMed]
- Collignon, P.; Beggs, J.J. Socioeconomic Enablers for Contagion: Factors Impelling the Antimicrobial Resistance Epidemic. Antibiotics 2019, 8, 86. [Google Scholar] [CrossRef] [PubMed]
- Thornber, K.; Kirchhelle, C. Hardwiring antimicrobial resistance mitigation into global policy. JAC Antimicrob. Resist. 2022, 4, dlac083. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Céline Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Bassetti, M.; Rello, J.; Blasi, F.; Goossens, H.; Sotgiu, G.; Tavoschi, L.; Zasowski, E.J.; Arber, M.R.; McCool, R.; Patterson, J.V.; et al. Systematic review of the impact of appropriate versus inappropriate initial antibiotic therapy on outcomes of patients with severe bacterial infections. Int. J. Antimicrob. Agents 2020, 56, 106184. [Google Scholar] [CrossRef]
- Kollef, M.H.; Shorr, A.F.; Bassetti, M.; Timsit, J.F.; Micek, S.T.; Michelson, A.P.; Garnacho-Montero, J. Timing of antibiotic therapy in the ICU. Crit. Care 2021, 25, 360. [Google Scholar] [CrossRef]
- Timsit, J.F.; Bassetti, M.; Cremer, O.; Daikos, G.; De Waele, J.; Kallil, A.; Kipnis, E.; Kollef, M.; Laupland, K.; Paiva, J.A.; et al. Rationalizing antimicrobial therapy in the ICU: A narrative review. Intensive Care Med. 2019, 45, 172–189. [Google Scholar] [CrossRef]
- Tassinari, M.; Zannoli, S.; Farabegoli, P.; Pedna, M.F.; Pierro, A.; Mastroianni, A.; Fontan, R.; Luongo, L.; Sarnataro, G.; Menegatti, E.; et al. Rapid diagnosis of bloodstream infections in the critically ill: Evaluation of the broad-range PCR/ESI-MS technology. PLoS ONE 2018, 13, e0197436. [Google Scholar] [CrossRef] [PubMed]
- Waagsbø, B.; Stuve, N.; Afset, J.E.; Klepstad, P.; Mo, S.; Heggelund, L.; Damås, J.K. High levels of discordant antimicrobial therapy in hospital-acquired bloodstream infections is associated with increased mortality in an intensive care, low antimicrobial resistance setting. Infect. Dis. 2022, 54, 738–747. [Google Scholar] [CrossRef] [PubMed]
- Kadri, S.S.; Lai, Y.L.; Warner, S.; Strich, J.R.; Babiker, A.; Ricotta, E.E.; Demirkale, C.Y.; Dekker, J.P.; Palmore, T.N.; Rhee, C.; et al. Forming the National Insititutes of Health Antimicrobial Resistance Outcomes Research Initiative (NIH-ARORI). Inappropriate empirical antibiotic therapy for bloodstream infections based on discordant in-vitro susceptibilities: A retrospective cohort analysis of prevalence, predictors, and mortality risk in US hospitals. Lancet Infect Dis. 2021, 21, 241–251. [Google Scholar] [CrossRef]
- Liu, V.X.; Fielding-Singh, V.; Greene, J.D.; Baker, J.M.; Iwashyna, T.J.; Bhattacharya, J.; Escobar, G.J. The Timing of Early Antibiotics and Hospital Mortality in Sepsis. Am. J. Respir. Crit. Care Med. 2017, 196, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Pulido, M.R.; García-Quintanilla, M.; Martín-Peña, R.; Cisneros, J.M.; McConnell, M.J. Progress on the development of rapid methods for antimicrobial susceptibility testing. J. Antimicrob. Chemother. 2013, 68, 2710–2717. [Google Scholar] [CrossRef]
- Plüddemann, A.; Onakpoya, I.; Harrison, S.; Shinkins, B.; Tompson, A.; Davis, R.; Heneghan, C. Position Paper on Anti-Microbial Resistance Diagnostics; Centre for Evidence-Based Medicine: Oxford, UK, 2015; pp. 1–142. [Google Scholar] [CrossRef]
- Li, Y.; Yang, X.; Zhao, W. Emerging microtechnologies and automated systems for rapid bacterial identification and antibiotic susceptibility testing. SLAS Technol. 2017, 22, 585–608. [Google Scholar] [CrossRef]
- Maurer, F.P.; Christner, M.; Hentschke, M.; Rohde, H. Advances in rapid identification and susceptibility testing of bacteria in the clinical microbiology laboratory: Implications for patient care and antimicrobial stewardship programs. Infect. Dis. Rep. 2017, 9, 6839. [Google Scholar] [CrossRef]
- Syal, K.; Iriya, R.; Yang, Y.; Yu, H.; Wang, S.; Haydel, S.E.; Chen, H.Y.; Tao, N. Antimicrobial susceptibility test with plasmonic imaging and tracking of single bacterial motions on nanometer scale. ACS Nano 2016, 10, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, G.; Lychko, I.; Sobral, R.; Roque, A.C. AIdentification and antibiotic-susceptibility profiling of infectious bacterial agents: A review of current and future trends. Biotechnol. J. 2019, 14, e1700750. [Google Scholar] [CrossRef] [PubMed]
- van Belkum, A.; Burnham, C.A.D.; Rossen, J.W.; Mallard, F.; Rochas, O.; Dunne, W.M., Jr. Innovative and rapid antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. 2020, 18, 299–311. [Google Scholar] [CrossRef]
- Tellapragada, C.; Hasan, B.; Antonelli, A.; Maruri, A.; de Vogel, C.; Gijón, D.; Coppi, M.; Verbon, A.; van Wamel, W.; Rossolini, G.M.; et al. Isothermal microcalorimetry minimal inhibitory concentration testing in extensively drug resistant Gram-negative bacilli: A multicentre study. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2020, 26, 1413.e1–1413.e7. [Google Scholar] [CrossRef] [PubMed]
- Braissant, O.; Bachmann, A.; Bonkat, G. Microcalorimetric assays for measuring cell growth and metabolic activity: Methodology and applications. Methods 2015, 76, 27–34. [Google Scholar] [CrossRef]
- Hannah, S.; Addington, E.; Alcorn, D.; Shu, W.; Hoskisson, P.A.; Corrigan, D.K. Rapid antibiotic susceptibility testing using low-cost, commercially available screen-printed electrodes. Biosens. Bioelectron. 2019, 145, 111696. [Google Scholar] [CrossRef] [PubMed]
- Brosel-Oliu, S.; Abramova, N.; Uria, N.; Bratov, A. Impedimetric transducers based on interdigitated electrode arrays for bacterial detection—A review. Anal. Chim. Acta 2019, 1088, 1–19. [Google Scholar] [CrossRef]
- Veses-Garcia, M.; Antypas, H.; Löffler, S.; Brauner, A.; Andersson-Svahn, H.; Richter-Dahlfors, A. Rapid Phenotypic Antibiotic Susceptibility Testing of Uropathogens Using Optical Signal Analysis on the Nanowell Slide. Front. Microbiol. 2018, 9, 1530. [Google Scholar] [CrossRef]
- Diep, T.T.; Needs, S.H.; Bizley, S.; Edwards, A.D. Rapid Bacterial Motility Monitoring Using Inexpensive 3D-Printed OpenFlexure Microscopy Allows Microfluidic Antibiotic Susceptibility Testing. Micromachines 2022, 13, 1974. [Google Scholar] [CrossRef]
- Bolotsky, A.; Ebrahimi, A. Toward Rapid Antibacterial Susceptibility Testing Using Electrochemical Biosensors Based on Organic-Inorganic Catalytic Complexes. Meet. Abstr. 2019, MA2019-02, 2244. [Google Scholar] [CrossRef]
- Besant, J.D.; Sargent, E.H.; Kelley, S.O. Rapid electrochemical phenotypic profiling of antibiotic-resistant bacteria. Lab Chip 2015, 15, 2799–2807. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Chen, L.; Chou, K.C.; Lu, X. Campylobacter jejuni Antimicrobial Resistance Profiles and Mechanisms Determined Using a Raman Spectroscopy-Based Metabolomic Approach. Appl. Environ. Microbiol. 2021, 87, e0038821. [Google Scholar] [CrossRef]
- Velican, A.M.; Măruţescu, L.; Kamerzan, C.; Cristea, V.C.; Banu, O.; Borcan, E.; Chifiriuc, M.C. Rapid Detection and Antibiotic Susceptibility of Uropathogenic Escherichia coli by Flow Cytometry. Microorganisms 2020, 8, 1233. [Google Scholar] [CrossRef]
- Huang, T.H.; Tzeng, Y.L.; Dickson, R.M. FAST: Rapid determinations of antibiotic susceptibility phenotypes using label-free cytometry. Cytometry A 2018, 93, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, E.S.D.; Silva-Dias, A.; Gomes, R.; Martins-Oliveira, I.; Ramos, M.H.; Rodrigues, A.G.; Cantón, R.; Pina-Vaz, C. Evaluation of rapid colistin susceptibility directly from positive blood cultures using a flow cytometry assay. Int. J. Antimicrob. Agents 2019, 54, 820–823. [Google Scholar] [CrossRef]
- Martins-Oliveira, I.; Pérez-Viso, B.; Quintas, S.; Silva-Dias, A.; Gomes, R.; Rodrigues, A.G.; Cantón, R.; Pina-Vaz, C. Evaluation of ultra-rapid susceptibility testing of ceftolozane-tazobactam by a flow cytometry assay directly from positive blood cultures. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2020, 39, 1907–1914. [Google Scholar] [CrossRef] [PubMed]
- Silva-Dias, A.; Pérez-Viso, B.; Martins-Oliveira, I.; Gomes, R.; Rodrigues, A.G.; Cantón, R.; Pina-Vaz, C. Evaluation of FASTinov Ultrarapid Flow Cytometry Antimicrobial Susceptibility Testing Directly from Positive Blood Cultures. J. Clin. Microbiol. 2021, 59, e0054421. [Google Scholar] [CrossRef]
- Mulroney, K.; Kopczyk, M.; Carson, C.; Paton, T.; Inglis, T.; Chakera, A. Same-day confirmation of infection and antimicrobial susceptibility profiling using flow cytometry. EBioMedicine 2022, 82, 104145. [Google Scholar] [CrossRef]
- Kállai, A.; Kelemen, M.; Molnár, N.; Tropotei, A.; Hause, B.; Iványi, Z.; Gál, J.; Ligeti, E.; Kristóf, K.; Lőrincz, Á.M. MICy: A Novel Flow Cytometric Method for Rapid Determination of Minimal Inhibitory Concentration. Microbiol. Spectr. 2021, 9, e0090121. [Google Scholar] [CrossRef]
- Ekelund, O.; Klokkhammer Hetland, M.A.; Høyland Löhr, I.; Schön, T.; Somajo, S. Rapid high-resolution detection of colistin resistance in Gram-negative bacteria using flow cytometry: A comparison with broth microdilution, a commercial screening test and WGS. J. Antimicrob. Chemother. 2021, 76, 3183–3191. [Google Scholar] [CrossRef]
- Sawada, T.; Katayama, M.; Takatani, S.; Ohiro, Y. Early detection of drug-resistant Streptococcus pneumoniae and Haemophilus influenzae by quantitative flow cytometry. Sci. Rep. 2021, 11, 2873. [Google Scholar] [CrossRef] [PubMed]
- Vrioni, G.; Tsiamis, C.; Oikonomidis, G.; Theodoridou, K.; Kapsimali, V.; Tsakris, A. MALDI-TOF mass spectrometry technology for detecting biomarkers of antimicrobial resistance: Current achievements and future perspectives. Ann. Transl. Med. 2018, 6, 240. [Google Scholar] [CrossRef] [PubMed]
- Costa-de-Oliveira, S.; Teixeira-Santos, R.; Silva, A.P.; Pinho, E.; Mergulhão, P.; Silva-Dias, A.; Marques, N.; Martins-Oliveira, I.; Rodrigues, A.G.; Paiva, J.A.; et al. Potential Impact of Flow Cytometry Antimicrobial Susceptibility Testing on the Clinical Management of Gram-Negative Bacteremia Using the FASTinov® Kit. Front. Microbiol. 2017, 8, 2455. [Google Scholar] [CrossRef]
- Broeren, M.A.; Maas, Y.; Retera, E.; Arents, N.L. Antimicrobial susceptibility testing in 90 min by bacterial cell count monitoring. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2013, 19, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Faria-Ramos, I.; Espinar, M.J.; Rocha, R.; Santos-Antunes, J.; Rodrigues, A.G.; Cantón, R.; Pina-Vaz, C. A novel flow cytometric assay for rapid detection of extended-spectrum beta-lactamases. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2013, 19, E8–E15. [Google Scholar] [CrossRef]
- Saint-Ruf, C.; Crussard, S.; Franceschi, C.; Orenga, S.; Ouattara, J.; Ramjeet, M.; Surre, J.; Matic, I. Antibiotic Susceptibility Testing of the Gram-Negative Bacteria Based on Flow Cytometry. Front. Microbiol. 2016, 7, 1121. [Google Scholar] [CrossRef]
- Inglis, T.J.J.; Paton, T.F.; Kopczyk, M.K.; Mulroney, K.T.; Carson, C.F. Same-day antimicrobial susceptibility test using acoustic-enhanced flow cytometry visualized with supervised machine learning. J. Med. Microbiol. 2020, 69, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Jindal, S.; Thampy, H.; Day, P.J.R.; Kell, D.B. Very rapid flow cytometric assessment of antimicrobial susceptibility during the apparent lag phase of microbial (re)growth. Microbiology 2019, 165, 439–454. [Google Scholar] [CrossRef] [PubMed]
- Filbrun, A.B.; Richardson, J.C.; Khanal, P.C.; Tzeng, Y.L.; Dickson, R.M. Rapid, label-free antibiotic susceptibility determined directly from positive blood culture. Cytometry 2022, 101, 564–576. [Google Scholar] [CrossRef] [PubMed]
- Torres-Sangiao, E.; Lamas Rodriguez, B.; Cea Pájaro, M.; Carracedo Montero, R.; Parajó Pazos, N.; García-Riestra, C. Direct Urine Resistance Detection Using VITEK 2. Antibiotics 2022, 11, 663. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.F.; Li, Y.; Zhang, X.L.; Yu, L.M.; Huang, B.H.; Sun, C.M. A New Method Aimed to Quickly Identify Pathogen and Drug Susceptibility Test Based on Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry Combined with Flow Cytometry. Surg. Infect. 2019, 20, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Kilic, A.; Dogan, E.; Kaya, S.; Oren, S.; Tok, D.; Ardic, N.; Baysallar, M. Rapid identification of Klebsiella pneumoniae by matrix-assisted laser desorption/ionization-time of flight mass spectrometry and detection of meropenem resistance by flow cytometric assay. J. Clin. Lab. Anal. 2016, 30, 1191–1197. [Google Scholar] [CrossRef]
- Edgar, R.H.; Samson, A.P.; Cook, J.; Douglas, M.; Urish, K.; Kellum, J.; Hempel, J.; Viator, J.A. Photoacoustic discrimination of antibiotic-resistant and sensitive Staphylococcus aureus isolates. Lasers Surg. Med. 2022, 54, 418–425. [Google Scholar] [CrossRef]
- Wang, Y.; Hammes, F.; De Roy, K.; Verstraete, W.; Boon, N. Past, present and future applications of flow cytometry in aquatic microbiology. Trends Biotechnol. 2010, 28, 416–424. [Google Scholar] [CrossRef]
- Nebe-von-Caron, G.; Stephens, P.J.; Hewitt, C.J.; Powell, J.R.; Badley, R.A. Analysis of bacterial function by multi-colour fluorescence flow cytometry and single cell sorting. J. Microbiol. Methods 2000, 42, 97–114. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef]
- Boonen, K.J.; Koldewijn, E.L.; Arents, N.L.; Raaymakers, P.A.; Scharnhorst, V. Urine flow cytometry as a primary screening method to exclude urinary tract infections. World J. Urol. 2013, 31, 547–551. [Google Scholar] [CrossRef]
- De Rosa, R.; Grosso, S.; Lorenzi, G.; Bruschetta, G.; Camporese, A. Evaluation of the new sysmex uf-5000 fluorescence flow cytometry analyser for ruling out bacterial urinary tract infection and for prediction of gram negative bacteria in urine cultures. Clin. Chim. Acta Int. J. Clin. Chem. 2018, 484, 171–178. [Google Scholar] [CrossRef]
- Nakamura, A.; Kohno, A.; Noguchi, N.; Kawa, K.; Ohno, Y.; Komatsu, M.; Yamanishi, H. Prediction of Uropathogens by Flow Cytometry and Dip-stick Test Results of Urine Through Multivariable Logistic Regression Analysis. PLoS ONE 2020, 7, e0227257. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Sun, E.; Wang, Y.; Pan, H.; Zhang, Y.; Li, Y.; Zhang, X.; Li, C.; Du, L.; Wang, C. Rapid identification and antimicrobial susceptibility testing for urinary tract pathogens by direct analysis of urine samples using a maldi-tof ms-based combined protocol. Front. Microbiol. 2019, 10, 1182. [Google Scholar] [CrossRef] [PubMed]
- Leal, H.F.; Azevedo, J.; Silva, G.E.O.; Amorim, A.M.L.; de Roma, L.R.C.; Arraes, A.C.P.; Gouveia, E.L.; Reis, M.G.; Mendes, A.V.; de Oliveira Silva, M.; et al. Bloodstream infections caused by multidrug-resistant gram-negative bacteria: Epidemiological, clinical and microbiological features. BMC Infect. Dis. 2019, 19, 609. [Google Scholar] [CrossRef] [PubMed]
- Mansour, J.D.; Robson, J.A.; Arndt, C.W.; Schulte, T.H. Detection of Escherichia coli in blood using flow cytometry. Cytometry 1985, 6, 186–190. [Google Scholar] [CrossRef]
- Pitt, W.G.; Alizadeh, M.; Husseini, G.A.; McClellan, D.S.; Buchanan, C.M.; Bledsoe, C.G.; Robison, R.A.; Blanco, R.; Roeder, B.L.; Melville, M.; et al. Rapid separation of bacteria from blood-review and outlook. Biotechnol. Prog. 2016, 32, 823–839. [Google Scholar] [CrossRef]
- Hou, H.W.; Bhattacharyya, R.P.; Hung, D.T.; Han, J. Direct detection and drug-resistance profiling of bacteremias using inertial microfluidics. Lab Chip 2015, 15, 2297–2307. [Google Scholar] [CrossRef] [PubMed]
- Tay, A.; Pavesi, A.; Yazdi, S.R.; Lim, C.T.; Warkiani, M.E. Advances in microfluidics in combating infectious diseases. Biotechnol. Adv. 2016, 34, 404–421. [Google Scholar] [CrossRef]
- Gosiewski, T.; Szala, L.; Pietrzyk, A.; Brzychczy-Wloch, M.; Heczko, P.B.; Bulanda, M. Comparison of methods for isolation of bacterial and fungal DNA from human blood. Curr. Microbiol. 2014, 68, 149–155. [Google Scholar] [CrossRef]
- Gosiewski, T.; Jurkiewicz-Badacz, D.; Sroka, A.; Brzychczy-Włoch, M.; Bulanda, M. A novel, nested, multiplex, real-time PCR for detection of bacteria and fungi in blood. BMC Microbiol. 2014, 14, 144. [Google Scholar] [CrossRef]
- Mulroney, K.T.; Hall, J.M.; McGuire, A.L.; Inglis, T.J.J.; Chakera, A. Case Study: Applying Rapid Flow Cytometry Analysis to CAPD Effluent. Perit. Dial. Int. 2018, 38, 376–379. [Google Scholar] [CrossRef]
- Chakera, A.; Mulroney, K.T.; Shak, H.J.; McGuire, A.L.; Eberl, M.; Topley, N. Peritonitis in Peritoneal Dialysis Patients: The Case for Rapid Diagnosis, Targeted Treatment, and Monitoring to Improve Outcomes. EMJ Nephrol. 2018, 6, 56–64. [Google Scholar] [CrossRef]
- Parbhoo, T.; Sampson, S.L.; Mouton, J.M. Recent Developments in the Application of Flow Cytometry to Advance our Understanding of Mycobacterium tuberculosis Physiology and Pathogenesis. Cytometry 2020, 97, 683–693. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, M.R.; Larsen, M.H.; Brown, T.S.; Jain, P.; Munsamy, V.; Wolf, A.; Uccellini, L.; Karim, F.; de Oliveira, T.; Mathema, B.; et al. Early Detection of Emergent Extensively Drug-Resistant Tuberculosis by Flow Cytometry-Based Phenotyping and Whole-Genome Sequencing. Antimicrob. Agents Chemother. 2019, 63, e01834-18. [Google Scholar] [CrossRef] [PubMed]
- Kamariza, M.; Shieh, P.; Ealand, C.S.; Peters, J.S.; Chu, B.; Rodriguez-Rivera, F.P.; Babu Sait, M.R.; Treuren, W.V.; Martinson, N.; Kalscheuer, M.; et al. Rapid detection of Mycobacterium tuberculosis in sputum with a solvatochromic trehalose probe. Sci. Transl. Med. 2018, 10, eaam6310. [Google Scholar] [CrossRef]
- Kamariza, M.; Keyser, S.G.L.; Utz, A.; Knapp, B.D.; Ealand, C.; Ahn, G.; Cambier, C.J.; Chen, T.; Kana, B.; Huang, K.C.; et al. Toward Point-of-Care Detection of Mycobacterium tuberculosis: A Brighter Solvatochromic Probe Detects Mycobacteria within Minutes. JACS Au 2021, 1, 1368–1379. [Google Scholar] [CrossRef]
- Singh, R.; Singh, A.P.; Kumar, S.; Giri, B.S.; Kim, K.H. Antibiotic resistance in major rivers in the world: A systematic review on occurrence, emergence, and management strategies. J. Clean. Prod. 2019, 234, 1484–1505. [Google Scholar] [CrossRef]
- Niegowska, M.; Wögerbauer, M. Improving the risk assessment of antimicrobial resistance (AMR) along the food/feed chain and from environmental reservoirs using qMRA and probabilistic modelling. EFSA J. 2022, 20, e200407. [Google Scholar] [CrossRef]
- Liang, J.; Mao, G.; Yin, X.; Ma, L.; Liu, L.; Bai, Y.; Zhang, T.; Qu, J. Identification and quantification of bacterial genomes carrying antibiotic resistance genes and virulence factor genes for aquatic microbiological risk assessment. Water Res. 2020, 168, 115160. [Google Scholar] [CrossRef]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef]
- Miłobedzka, A.; Ferreira, C.; Vaz-Moreira, I.; Calderón-Franco, D.; Gorecki, A.; Purkrtova, S.; Bartacek, Y.; Dziewit, L.; Singleton, C.M.; Nielsen, P.H.; et al. Monitoring antibiotic resistance genes in wastewater environments: The challenges of filling a gap in the One-Health cycle. J. Hazard. Mater. 2022, 424 Pt C, 127407. [Google Scholar] [CrossRef]
- Qiu, Y.; Zhang, J.; Li, B.; Wen, X.; Liang, P.; Huang, X. A novel microfluidic system enables visualization and analysis of antibiotic resistance gene transfer to activated sludge bacteria in biofilm. Sci. Total Environ. 2018, 642, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, H.; Shi, R.; Lv, J.; Li, B.; Yang, F.; Zheng, X.; Xu, J. Antibiotic resistance genes in different animal manures and their derived organic fertilizer. Environ. Sci. Eur. 2020, 32, 102. [Google Scholar] [CrossRef]
- Jauregi, L.; Epelde, L.; Alkorta, I.; Garbisu, C. Antibiotic Resistance in Agricultural Soil and Crops Associated to the Application of Cow Manure-Derived Amendments from Conventional and Organic Livestock Farms. Front. Vet. Sci. 2021, 8, 633858. [Google Scholar] [CrossRef]
- Keenum, I.; Williams, R.K.; Ray, P.; Garner, E.D.; Knowlton, K.F.; Pruden, A. Combined effects of composting and antibiotic administration on cattle manure–borne antibiotic resistance genes. Microbiome 2021, 9, 81. [Google Scholar] [CrossRef] [PubMed]
- Macedo, G.; Olesen, A.K.; Maccario, L.; Hernandez Leal, L.; vd Maas, P.; Heederik, D.; Mevius, D.; Sørensen, S.J.; Schmitt, H. Horizontal Gene Transfer of an IncP1 Plasmid to Soil Bacterial Community Introduced by Escherichia coli through Manure Amendment in Soil Microcosms. Environ. Sci. Technol. 2022, 56, 11398–11408. [Google Scholar] [CrossRef]
- Scott, L.C.; Aubee, A.; Wilson, M.J.; Esser, S.; Descamps, D.; Lee, N.; Distler, E.; Aw, T.G. Leave No Trace? Ecological and anthropogenic determinants of antibiotic resistant bacteria in a recreational alpine environment. Environ. Res. 2023, 216 Pt 2, 114617. [Google Scholar] [CrossRef]
- Hayward, J.L.; Jackson, A.J.; Yost, C.K.; Truelstrup Hansen, L.; Jamieson, R.C. Fate of antibiotic resistance genes in two Arctic tundra wetlands impacted by municipal. wastewater. Sci. Total Environ. 2018, 642, 1415–1428. [Google Scholar] [CrossRef]
- McCann, C.M.; Christgen, B.; Roberts, J.A.; Su, J.Q.; Arnold, K.E.; Gray, N.D. Understanding drivers of antibiotic resistance genes in high Arctic soil ecosystems. Environ. Int. 2019, 125, 497–504. [Google Scholar] [CrossRef]
- Segawa, T.; Takeuchi, N.; Rivera, A.; Yamada, A.; Yoshimura, Y.; Barcaza, G.; Shinbori, K.; Motoyama, H.; Kohshima, S.; Ushida, K. Distribution of antibiotic resistance genes in glacier environments. Environ. Microbiol. Rep. 2013, 5, 127–134. [Google Scholar] [CrossRef]
- Tan, L.; Li, L.; Ashbolt, N.; Wang, X.; Cui, Y.; Zhu, X.; Xu, Y.; Yang, Y.; Mao, D.; Luo, Y. Arctic antibiotic resistance gene contamination, a result of anthropogenic activities and natural origin. Sci. Total Environ. 2018, 621, 1176–1184. [Google Scholar] [CrossRef]
- Xiang, Q.; Chen, Q.L.; Zhu, D.; An, X.L.; Yang, X.R.; Su, J.Q.; Qiao, M.; Zhu, Y.G. Spatial and temporal distribution of antibiotic resistomes in a peri-urban area is associated significantly with anthropogenic activities. Environ. Pollut. 2018, 235, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Péguilhan, R.; Turgeon, N.; Veillette, N.; Baray, J.L.; Deguillaume, L.; Amato, P.; Duchaine, C. Quantification of antibiotic resistance genes (ARGs) in clouds at a mountain site (puy de Dôme, central France). Sci. Total Environ. 2023, 865, 161264. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.C.; Wilson, M.J.; Esser, S.M.; Lee, N.L.; Wheeler, M.E.; Aubee, A.; Aw, T.G. Assessing visitor use impact on antibiotic resistant bacteria and antibiotic resistance genes in soil and water environments of Rocky Mountain National Park. Sci. Total Environ. 2021, 785, 147122. [Google Scholar] [CrossRef] [PubMed]
- McEwen, S.A.; Collignon, P.J. Antimicrobial resistance: A one health colloquium. Microbiol. Spectr. 2018, 6, 521–547. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Comprehensive Review of the WHO Global Action Plan on Antimicrobial Resistance—Volume 1: Report; WHO Evaluation Office: Geneva, Switzerland, 2021; Available online: https://www.who.int/publications/m/item/comprehensive-review-of-the-who-global-action-plan-on-antimicrobial-resistance (accessed on 1 March 2023).
- Andrade, F.F.; Gomes, R.; Martins-Oliveira, I.; Dias, A.; Rodrigues, A.G.; Pina-Vaz, C.A. Rapid Flow Cytometric Antimicrobial Susceptibility Assay (FASTvet) for Veterinary Use—Preliminary Data. Front. Microbiol. 2020, 7, 1944. [Google Scholar] [CrossRef] [PubMed]
- Göröcs, Z.; Tamamitsu, M.; Bianco, V.; Wolf, P.; Roy, S.; Shindo, K.; Yanny, K.; Wu, Y.; Koydemir, H.C.; Rivenson, Y.; et al. A deep learning-enabled portable imaging flow cytometer for cost-effective, high-throughput, and label-free analysis of natural water samples. Light Sci. Appl. 2018, 7, 66. [Google Scholar] [CrossRef]
- Wang, K.L.; Zhang, J.X.; Min, D.; Lv, J.L.; Liu, D.F.; Yu, H.Q. Detection and Quantification of Antimicrobial-Resistant Cells in Aquatic Environments by Bioorthogonal Noncanonical Amino Acid Tagging. Environ. Sci. Technol. 2022, 56, 15685–15694. [Google Scholar] [CrossRef]
- Williams, A.J.; Cooper, W.M.; Ramsaroop, S.; Alusta, P.; Buzatu, D.A.; Wilkes, J.G. Rapid Flow Cytometry Detection of a Single Viable Escherichia coli O157:H7 Cell in Raw Spinach Using a Simplified Sample Preparation Technique. Front. Microbiol. 2017, 8, 1493. [Google Scholar] [CrossRef]
- Wang, M.; Bai, Z.; Liu, S.; Liu, Y.; Wang, Z.; Zhou, G.; Gong, X.; Jiang, Y.; Sui, Z. Accurate quantification of total bacteria in raw milk by flow cytometry using membrane potential as a key viability parameter. LWT 2023, 173, 114315. [Google Scholar] [CrossRef]
- Li, S.; Liu, Z.; Süring, C.; Chen, L.; Müller, S.; Zeng, P. The Impact of the Antibiotic Fosfomycin on Wastewater Communities Measured by Flow Cytometry. Front. Microbiol. 2022, 12, 737831. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Flach, C.F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Li, G.; Wang, M.; Chen, C.; Xu, Z.; An, T. Photoelectrocatalytic inactivation mechanism of E. coli DH5α (TET) and synergistic degradation of corresponding antibiotics in water. Water Res. 2022, 215, 118240. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Tian, C.; Zhao, M.; Yu, P.; Zheng, T.; Li, Z.; Wang, H. Removal of tetracycline-resistant Escherichia coli and its genes through ultrasound treatment combined with ultraviolet light emitting diodes. Environ. Res. 2021, 197, 111007. [Google Scholar] [CrossRef] [PubMed]
- Briaud, P.; Camus, L.; Bastien, S.; Doléans-Jordheim, A.; Vandenesch, F.; Moreau, K. Coexistence with Pseudomonas aeruginosa alters Staphylococcus aureus transcriptome, antibiotic resistance and internalization into epithelial cells. Sci. Rep. 2019, 9, 16564. [Google Scholar] [CrossRef]
- Magalhães, A.P.; Lopes, S.P.; Pereira, M.O. Insights into cystic fibrosis polymicrobial consortia: The role of species interactions in biofilm development, phenotype, and response to in-use antibiotics. Front. Microbiol. 2016, 7, 2146. [Google Scholar] [CrossRef]
- O’Brien, T.J.; Figueroa, W.; Welch, M. Decreased efficacy of antimicrobial agents in a polymicrobial environment. ISME J. 2022, 16, 1694–1704. [Google Scholar] [CrossRef]
- Liu, Z.; Cichocki, N.; Hübschmann, T.; Süring, C.; Ofiţeru, I.D.; Sloan, W.T.; Grimm, V.; Müller, S. Neutral mechanisms and niche differentiation in steady-state insular microbial communities revealed by single cell analysis. Environ. Microbiol. 2019, 21, 164–181. [Google Scholar] [CrossRef]
- Massicotte, R.; Mafu, A.A.; Ahmad, D.; Deshaies, F.; Pichette, G.; Belhumeur, P. Comparison between Flow Cytometry and Traditional Culture Methods for Efficacy Assessment of Six Disinfectant Agents against Nosocomial Bacterial Species. Front. Microbiol. 2017, 8, 112. [Google Scholar] [CrossRef]
- Mohiuddin, S.G.; Kavousi, P.; Orman, M.A. Flow-cytometry analysis reveals persister resuscitation characteristics. BMC Microbiol. 2020, 20, 202. [Google Scholar] [CrossRef]
- Zwerling, A.; Dowdy, D.; von Delft, A.; Taylor, H.; Merritt, M.W. Incorporating social justice and stigma in cost-effectiveness analysis: Drug-resistant tuberculosis treatment. Int. J. Tuber. Lung Dis. 2017, 21, 69–74. [Google Scholar] [CrossRef]
- Bongiorno, D.; Musso, N.; Lazzaro, L.M.; Mongelli, G.; Stefani, S.; Campanile, F. Detection of methicillin-resistant Staphylococcus aureus persistence in osteoblasts using imaging flow cytometry. Microbiol. Open 2020, 9, e1017. [Google Scholar] [CrossRef] [PubMed]

| Methods | Sample Analyzed (n = Number of Samples Tested) | Tested Bacteria (AMR Phenotype) | Principle of Detection | Tested Antibiotics | Time to Results | References |
|---|---|---|---|---|---|---|
| FCM coupled with MALDI TOF MS and VITEK 2 | Urine samples (n = 211) | Escherichia coli | Cell-counting (cut-off 150 bacteria/mL) | Ampicillin, Amoxicillin/Clavulanic acid, Cefuroxime, Cefoxitin, Cefotaxime, Ceftazidime, Cefepime, Imipenem, Ertapenem, Gentamycin, Tobramycin, Nalidixic Acid, Ciprofloxacin, Fosfomycin, Nitrofurantoin, Cotrimoxazole | 7 h | [54] |
| FCM | Urine samples (n = 107) E. coli strains (n = 19) | E. coli, Klebsiella pneumoniae Proteus mirabilis, Pseudomonas aeruginosa | Fluorescent dyes: DiBAC4(3) | Ceftriaxone, ciprofloxacin, nitrofurantoin, trimethoprim–sulfamethoxazole | 4 h | [37] |
| FCM Fastinov | Positive blood cultures (n = 447) | Gram-positive and Gram-negative | Fluorescent dyes | Gram-negative: ampicillin, amoxicillin-clavulanic acid, cefotaxime, ceftazidime, ceftolozane-tazobactam, piperacillin–tazobactam, meropenem, imipenem, gentamicin, amikacin, ciprofloxacin, and colistin Gram-positive: ampicillin, penicillin, imipenem, vancomycin, linezolid, cefoxitin, and gentamicin | <2 h | [41] |
| FCM Fastinov | Spiked blood cultures (n = 204) | Enterobacterales, Pseudomonas spp., Acinetobacter baumannii | Fluorescent dyes | Colistin | <2 h | [39] |
| FCM Fastinov | Spiked blood cultures (n = 162) | E. coli, K. pneumoniae, Enterobacter ssp, Serratia marcescens, Providencia spp., Morganella morgani, Proteus spp. | Fluorescent membrane potential dye | Ceftolozane–tazobactam | <2 h | [40] |
| FCM | Blood spiked | E. coli, K. pneumoniae, A. nosocomialis | No | 8 h | [38] | |
| FCM and MALDI-TOF MS | Positive blood cultures (n = 238) | Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Enterobacter aerogenes, Acinetobacter baumannii, Klebsiella oxytoca, Proteus mirabilis, Enterobacter cloacae, Citrobacter freundii, Staphylococcus aureus, Staphylococcus saparophytics, Staphylococcus hominis, Enterococcus faecalis, Staphylococcus epidermidis, Staphylococcus simulans, Enterococcus faecium, Candida albicans, Candida tropicalis, Candida pseudotropicalis, Candida parapsilosis | FDA PI | Ampicillin, vancomycin, cefotaxime, oxacillin, methicillin, ceftazidime amikacin, cefotaxime, ciprofloxacin | 3 h | [55] |
| FCM | Blood culture samples | E. coli, P. aeruginosa, S. aureus | Antibiotic-induced changes in count rate | Ceftazidime, meropenem, tobramycin, oxacillin | 5 h | [53] |
| Acoustic-enhanced FCM | Peritoneal dialysis effluent specimens | Escherichia coli, Pseudomonas aeruginosa, Staphyloccocus aureus, Staphylococcus epidermidis, Klebsiella pneumoniae | Live/DEAD™ Fixable Violet viability stain | Piperacillin–tazobactam, benzyl-penicillin, oxacillin, cefoxitin, vancomycin, teicoplanin, gentamicin, trimethoprim–sulfamethoxazole, daptomycin, erythromycin, clindamycin, amoxicillin, linezolid, ceftriaxone, ciprofloxacin, trimethoprim, cefepime, tigecycline, amikacin, aztreonam, amoxicillin-clavulanic acid, piperacillin–tazobactam, meropenem | 4 h | [42] |
| MALDI-TOF and FCM | Clinical strains (n = 174) | K. pneumoniae (carbapenem resistant) | Fluorescent dyes: propidium iodide and thiazole orange | Meropenem | 2 h | [56] |
| FCM | Clinical strains (n = 174) | E. coli and K. pneumoniae | YoPro-1 | Colistin | 3 h | [44] |
| Photoacoustic FCM | Clinical strains | S. aureus | Bacteriophage labeled with Direct Red 81 | Daptomycin | 4 h | [57] |
| FCM | Clinical and reference strains | S. pneumoniae H. influenzae | SYTO9 and PI | Penicillin G, cefotaxime | 10 min | [45] |
| FCM | Reference strains (n = 6) | Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Staphylococcus aureus, Streptococcus pyogenes, Enterococcus faecalis | Fluorescent dyes: acridine orange | Vancomycin, ciprofloxacin, levofloxacin, ceftriaxon, cefepime, amplicilin, piperacillin–tazobactam, trimethoprim–sulfamethoxazole, cefazolin, colistin, imipenem, gentamycin | 4 h | [43] |
| Current Rapid ASTs | FCM Assays |
|---|---|
Growth-dependent quantification methods
| Growth-independent quantification method
|
Genotypic AST methods
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marutescu, L.G. Current and Future Flow Cytometry Applications Contributing to Antimicrobial Resistance Control. Microorganisms 2023, 11, 1300. https://doi.org/10.3390/microorganisms11051300
Marutescu LG. Current and Future Flow Cytometry Applications Contributing to Antimicrobial Resistance Control. Microorganisms. 2023; 11(5):1300. https://doi.org/10.3390/microorganisms11051300
Chicago/Turabian StyleMarutescu, Luminita Gabriela. 2023. "Current and Future Flow Cytometry Applications Contributing to Antimicrobial Resistance Control" Microorganisms 11, no. 5: 1300. https://doi.org/10.3390/microorganisms11051300




