Endogenous CO2 Overpressure Effect on Higher Alcohols Metabolism during Sparkling Wine Production
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganism, Grape Must, and Fermentation Conditions
2.2. Metabolic Analysis
2.2.1. Analysis of Methanol and Higher Alcohols
2.2.2. Analysis of Amino Acids
2.3. Proteomic Analysis
2.4. Statistical Analysis
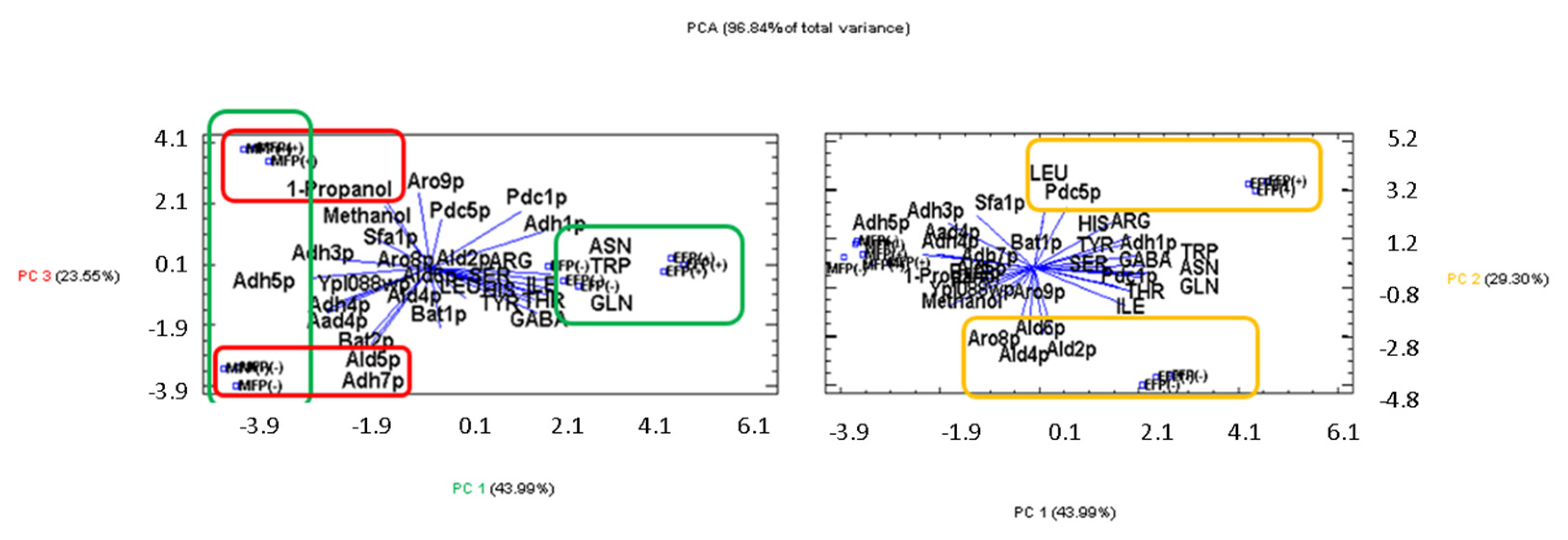
3. Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, F.; Du, B.; Li, J. Aroma compounds in wine. In Grape and Wine Biotechnology; Morata, A., Loira, I., Eds.; Intech Open Journals: Rijeka, Croatia, 2016. [Google Scholar]
- de Castilhos, M.B.; de Queiroga, A.P.; Sabino, L.L.; dos Santos Júnior, J.R.; Santiago-Urbina, J.A.; Nolasco-Cancino, H.; Ruíz-Terán, F.; Del Bianchi, V.L. Flavor Biochemistry of Fermented Alcoholic Beverages. In Natural Flavours, Fragrances, and Perfumes: Chemistry, Production, and Sensory Approach; Wiley: Hoboken, NJ, USA, 2023; pp. 91–114. [Google Scholar]
- Rollero, S.; Mouret, J.R.; Bloem, A.; Sanchez, I.; Ortiz-Julien, A.; Sablayrolles, J.M.; Dequin, S.; Camarasa, C. Quantitative13C-isotope labelling-based analysis to elucidate the influence of environmental parameters on the production of fermentative aromas during wine fermentation. Microb. Biotechnol. 2017, 10, 1649–1662. [Google Scholar] [CrossRef] [PubMed]
- Lambrechts, M.G.; Pretorius, I.S. Yeast and its Importance to Wine Aroma—A Review. S. Afr. J. Enol. Vitic. 2000, 21, 97–129. [Google Scholar] [CrossRef] [Green Version]
- Van Rooyen, P.C.; De Wet, P.; Van Wyk, C.J.; Tromp, A. Chening Blanc wine volatiles and intensity of guava-like flavour. S. Afr. J. Enol. Vitic. 1982, 3, 1–7. [Google Scholar]
- Gonzalez-Ramos, D.; Gonzalez, R. Genetic determinants of the release of mannoproteins of enological interest by Saccharomyces cerevisiae. J. Agric. Food Chem. 2006, 54, 9411–9416. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Rodríguez, A.J.; Polo, M.C.; Carrascosa, A.V. Structural and ultrastructural changes in yeast cells during autolysis in a model wine system and in sparkling wines. Int. J. Food Microbiol. 2001, 71, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Penacho, V.; Valero, E.; Gonzalez, R. Transcription profiling of sparkling wine second fermentation. Int. J. Food Microbiol. 2012, 153, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Molina, A.M.; Guadalupe, V.; Varela, C.; Swiegers, J.H.; Pretorius, I.S.; Agosin, E. Differential synthesis of fermentative aroma compounds of two related commercial wine yeast strains. Food Chem. 2009, 117, 189–195. [Google Scholar] [CrossRef]
- Rankine, B.C. Formation of higher alcohols by wine yeasts, and relationship to taste thresholds. J. Sci. Food Agric. 1967, 18, 583–589. [Google Scholar] [CrossRef]
- Mauricio, J.C.; Moreno, J.; Zea, L.; Ortega, J.M.; Medina, M. The effects of grape must fermentation conditions on volatile alcohols and esters formed by Saccharomyces cerevisiae. J. Sci. Food Agric. 1997, 75, 155–160. [Google Scholar] [CrossRef]
- Bozdogan, A.; Canbaç, A. The effect of yeast strain, immobilisation and aging time on the amount of free amino acids and amino acids in peptides of sparkling wines obtained from cv. Dimrit Grapes. S. Afr. J. Enol. Vitic. 2012, 33, 257–263. [Google Scholar]
- Amerine, A.M.; Berg, H.V.; Kunkee, R.E.; Ough, C.S.; Singleton, U.L.; Weeb, A.D. The Technology of Winemaking; AVI Technical Books Inc.: Westport, CT, USA, 1980. [Google Scholar]
- Swiegers, J.H.; Pretorius, I.S. Yeast modulation of wine flavor. Adv. Appl. Microbiol. 2005, 57, 131–175. [Google Scholar] [PubMed]
- Dickinson, J.R.; Lanterman, M.M.; Danner, D.J.; Pearson, B.M.; Sanz, P.; Harrison, S.J.; Hewlins, M.J.E. A 13C Nuclear Magnetic Resonance Investigation of the Metabolism of Leucine to Isoamyl Alcohol in Saccharomyces cerevisiae. J. Biol. Chem. 1997, 272, 26871–26878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hazelwood, L.A.; Daran, J.-M.; van Maris, A.J.; Pronk, J.T.; Dickinson, J.R. The Ehrlich Pathway for Fusel Alcohol Production: A Century of Research on Saccharomyces cerevisiae Metabolism. Appl. Environ. Microbiol. 2008, 74, 2259–2266. [Google Scholar] [CrossRef] [Green Version]
- Badura, J.; Medić, M.; van Wyk, N.; Krause, B.; Semmler, H.; Brezina, S.; Pretorius, I.S.; Rauhut, D.; von Wallbrunn, C. Synthesis of Aroma Compounds as a Function of Different Nitrogen Sources in Fermentations Using Non-Saccharomyces Wine Yeasts. Microorganisms 2022, 11, 14. [Google Scholar] [CrossRef]
- Su, Y.; Heras, J.M.; Gamero, A.; Querol, A.; Guillamón, J.M. Impact of nitrogen addition on wine fermentation by Saccharomyces cerevisiae strains with different nitrogen requirements. J. Agric. Food Chem. 2021, 69, 6022–6031. [Google Scholar] [CrossRef]
- Martínez-García, R.; García-Martínez, T.; Puig-Pujol, A.; Mauricio, J.C.; Moreno, J. Changes in sparkling wine aroma during the second fermentation under CO2 pressure in sealed bottle. Food Chem. 2017, 237, 1030–1040. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, R.; Roldán-Romero, Y.; Moreno, J.; Puig-Pujol, A.; Mauricio, J.C.; García-Martínez, T. Use of a flor yeast strain for the second fermentation of sparkling wines: Effect of endogenous CO2 over-pressure on the volatilome. Food Chem. 2019, 308, 125555. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, R.; Mauricio, J.C.; García-Martínez, T.; Peinado, R.A.; Moreno, J. Towards a better understanding of the evolution of odour-active compounds and the aroma perception of sparkling wines during ageing. Food Chem. 2021, 357, 129784. [Google Scholar] [CrossRef]
- Martínez-García, R.; Moreno, J.; Bellincontro, A.; Centioni, L.; Puig-Pujol, A.; Peinado, R.A.; Mauricio, J.C.; García-Martínez, T. Using an electronic nose and volatilome analysis to differentiate sparkling wines obtained under different conditions of temperature, ageing time and yeast formats. Food Chem. 2020, 334, 127574. [Google Scholar] [CrossRef]
- González-Jiménez, M.D.C.; Moreno-García, J.; García-Martínez, T.; Moreno, J.J.; Puig-Pujol, A.; Capdevilla, F.; Mauricio, J.C. Differential analysis of proteins involved in ester metabolism in two Saccharomyces cerevisiae strains during the second fer-mentation in sparkling wine elaboration. Microorganisms 2020, 8, 403. [Google Scholar] [CrossRef] [Green Version]
- Peinado, R.A.; Moreno, J.A.; Muñoz, D.; Medina, M.; Moreno, J. Gas Chromatographic Quantification of Major Volatile Compounds and Polyols in Wine by Direct Injection. J. Agric. Food Chem. 2004, 52, 6389–6393. [Google Scholar] [CrossRef] [PubMed]
- Vararu, F.; Moreno-García, J.; Zamfir, C.-I.; Cotea, V.V.; Moreno, J. Selection of aroma compounds for the differentiation of wines obtained by fermenting musts with starter cultures of commercial yeast strains. Food Chem. 2016, 197, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Pripis-Nicolau, L.; De Revel, G.; Marchand, S.; Anocibar Beloqui, A.; Bertrand, A. Automated HPLC method for the measurement of free aminoacids including cysteine in must and wines, first application. J. Sci. Food Agric. 2001, 81, 731–738. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. J. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Moreno-García, J.; García-Martínez, T.; Moreno, J.; Millán, M.C.; Mauricio, J.C. A proteomic and metabolomic approach for understanding the role of the flor yeast mitochondria in the velum formation. Int. J. Food Microbiol. 2014, 172, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Moreno-García, J.; Garcia-Martinez, T.; Millán, M.C.; Mauricio, J.C.; Moreno, J. Proteins involved in wine aroma compounds metabolism by a Saccharomyces cerevisiae flor-velum yeast strain grown in two conditions. Food Microbiol. 2015, 51, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ishihama, Y.; Oda, Y.; Tabata, T.; Sato, T.; Nagasu, T.; Rappsilber, J.; Mann, M. Exponentially Modified Protein Abundance Index (emPAI) for Estimation of Absolute Protein Amount in Proteomics by the Number of Sequenced Peptides per Protein. Mol. Cell. Proteom. 2005, 4, 1265–1272. [Google Scholar] [CrossRef] [Green Version]
- Seisonen, S.; Vene, K.; Koppel, K. The current practice in the application of chemometrics for correlation of sensory and gas chromatography data. Food Chem. 2016, 210, 530–540. [Google Scholar] [CrossRef]
- Merín, M.G.; Mendoza, L.M.; Farías, M.E.; Morata de Ambrosini, V.I. Isolation and selection of yeasts from wine grape ecosystem secreting cold-active peptinolytic activity. Int. J. Food Microbiol. 2017, 147, 144–148. [Google Scholar] [CrossRef] [Green Version]
- Wei, X.; Francoise, U.; Qin, M.; Chen, Q.; Li, Y.; Sun, X.; Fang, Y.-L. Effects of different fermentation and storage conditions on methanol content in Chinese spine grape (Vitis davidii Foex) wine. CyTA J. Food 2020, 18, 367–374. [Google Scholar] [CrossRef]
- Dennis, E.G.; Keyzers, R.A.; Kalua, C.M.; Maffei, S.M.; Nicholson, E.L.; Boss, P.K. Grape Contribution to Wine Aroma: Production of Hexyl Acetate, Octyl Acetate, and Benzyl Acetate during Yeast Fermentation Is Dependent upon Precursors in the Must. J. Agric. Food Chem. 2012, 60, 2638–2646. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Li, H.; Wang, H.; Zhang, L. Volatile compounds of young Cabernet Sauvignon red wine from Changli County (China). J. Food Compos. Anal. 2008, 21, 689–694. [Google Scholar] [CrossRef]
- Coelho, E.; Coimbra, M.A.; Nogueira, J.M.F.; Rocha, S.M. Quantification approach for assessment of sparkling wine volatiles from different soils, ripening stages, and varieties by stir bar sorptive extraction with liquid desorption. Anal. Chim. Acta 2009, 635, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Loyaux, D.; Roger, S.; Adda, J. The evolution of champagne volatiles during ageing. J. Sci. Food Agric. 1981, 32, 1254–1258. [Google Scholar] [CrossRef]
- Pozo-Bayón, M.; Martín-Álvarez, P.; Moreno-Arribas, M.; Andujar-Ortiz, I.; Pueyo, E. Impact of using Trepat and Monastrell red grape varieties on the volatile and nitrogen composition during the manufacture of rosé Cava sparkling wines. LWT 2010, 43, 1526–1532. [Google Scholar] [CrossRef] [Green Version]
- Sawyer, S.; Longo, R.; Solomon, M.; Nicolotti, L.; Westmore, H.; Merry, A.; Gnoinski, G.; Ylia, A.; Dambergs, R.; Kerslake, F. Autolysis and the duration of ageing on lees independently influence the aroma composition of traditional method sparkling wine. Aust. J. Grape Wine Res. 2021, 28, 146–159. [Google Scholar] [CrossRef]
- Pueyo, E.; Martín-Alvarez, P.J.; Polo, M.C. Relationship Between Foam Characteristics and Chemical Composition in Wines and Cavas (Sparkling Wines). Am. J. Enol. Vitic. 1995, 46, 518–524. [Google Scholar] [CrossRef]
- Procopio, S.; Sprung, P.; Becker, T. Effect of amino acid supply on the transcription of flavour-related genes and aroma compound production during lager yeast fermentation. LWT 2015, 63, 289–297. [Google Scholar] [CrossRef]
- Ljungdahl, P.O.; Daignan-Fornier, B. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 2012, 190, 885–929. [Google Scholar] [CrossRef] [Green Version]
- Belda, I.; Ruiz, J.; Esteban-Fernández, A.; Navascués, E.; Marquina, D.; Santos, A.; Moreno-Arribas, M. Microbial contribution to wine aroma and its intended use for wine quality improvement. Molecules 2017, 22, 189. [Google Scholar] [CrossRef] [Green Version]
- Marsit, S.; Sanchez, I.; Galeote, V.; Dequin, S. Horizontally acquired oligopeptide transporters favour adaptation of Saccharomyces cerevisiae wine yeast to oenological environment. Environ. Microbiol. 2016, 18, 1148–1161. [Google Scholar] [CrossRef] [PubMed]
- Cebollero, E.; Gonzalez, R. Induction of Autophagy by Second-Fermentation Yeasts during Elaboration of Sparkling Wines. Appl. Environ. Microbiol. 2006, 72, 4121–4127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickinson, J.R.; Salgado, L.E.; Hewlins, J.E. The catabolism of amino acids to long chain and complex alcohols in Saccha-romyces cerevisiae. J. Biol. Chem. 2003, 278, 8028–8034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.; Wen, Y.; Chen, W.; Yan, T.; Tian, X.; Zhou, S. Simultaneously deleting ADH2 and THI3 genes of Saccharomyces cerevisiae for reducing the yield of acetaldehyde and fusel alcohols. FEMS Microbiol. Lett. 2021, 368, fnab094. [Google Scholar] [CrossRef]
- Hazelwood, L.A.; Tai, S.L.; Boer, V.M.; de Winde, J.H.; Pronk, J.T.; Daran, J.M. A new physiological role for Pdr12p in Saccharomyces cerevisiae: Export of aromatic and branched-chain organic acids produced in amino acid catabolism. FEMS Yeast Res. 2006, 6, 937–945. [Google Scholar] [CrossRef] [Green Version]
- Vuralhan, Z.; Morais, M.A.; Tai, S.L.; Piper, M.D.; Pronk, J.T. Identification and characterization of phenylpyruvate decarboxylase genes in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2003, 69, 4534–4541. [Google Scholar] [CrossRef] [Green Version]
Disclaimer/Publisher’s Note: The statements, opinions, and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions, or products referred to in the content. |


| Concentration (mg/L) | |||||
|---|---|---|---|---|---|
| OPT (mg/L) | MFP (+) | EFP (+) | MFP (-) | EFP (-) | |
| Methanol | 668 | 30.85 c ± 1.11 | 26.80 a ± 1.60 | 26.82 ab ± 1.39 | 29.10 bc ± 1.81 |
| 1-Propanol | 830 | 15.01 c ± 1.20 | 12.89 ab ± 0.49 | 11.97 a ± 0.62 | 14.12 bc ± 0.51 |
| Isobutanol | 40 | 21.03 b ± 0.04 | 18.96 a ± 0.96 | 19.69 a ± 0.33 | 19.41 a ± 0.42 |
| Isoamylic Alcohols | 30 | 296.33 b ± 3.29 | 260.97 a ± 10.01 | 288.52 b ± 15.98 | 295.61 b ± 0.25 |
| 2-phenylethanol | 10 | 58.06 b ± 2.99 | 51.11 a ± 3.09 | 52.99 ab ± 2.00 | 52.28 ab ± 1.14 |
| Hexanol | 2.5 | 0.69 a ± 0.72 | 0.68 a ± 0.42 | 0.82 b ± 0.33 | 0.69 a ± 0.40 |
| 2-ethyl-1-hexanol | 8 | 0.91 ns ± 0.27 | 0.72 ns ± 0.17 | 0.60 ns ± 0.17 | 0.49 ns ± 0.13 |
| Concentration (mM) | ||||
|---|---|---|---|---|
| MFP (+) | EFP (+) | MFP (-) | EFP (-) | |
| GABA | 7.1 a ± 0.7 | 25.4 d ± 0.0 | 15.4 b ± 0.1 | 23.7 c ± 0.7 |
| GLN | 5.0 a ± 1.2 | 17.8 c ± 1.0 | 7.2 b ± 0.3 | 15.9 c ± 0.3 |
| TYR | 11.0 a ± 0.4 | 16.6 c ± 0.8 | 13.2 b ± 0.8 | 13.6 b ± 0.7 |
| CYS | 5.0 a ± 0.7 | 11.8 b ± 1.5 | 11.9 b ± 1.4 | 9.5 b ± 1.1 |
| LEU | 5.1 a ± 0.4 | 11.0 c ± 0.5 | 8.0 b ± 1.3 | 4.1 a ± 0.3 |
| ASN | 3.0 a ± 0.2 | 10.0 d ± 0.4 | 3.7 b ± 0.3 | 8.7 c ± 0.8 |
| THR | 6.5 a ± 0.9 | 9.7 c ± 0.4 | 7.8 b ± 0.4 | 10.5 c ± 0.3 |
| GLY | 5.3 a ± 0.1 | 8.9 b ± 0.7 | 5.0 a ± 0.2 | 8.3 b ± 0.4 |
| GLU | 4.8 b ± 0.2 | 7.2 a ± 0.4 | 6.7 b ± 0.5 | 5.6 a ± 0.6 |
| VAL | 3.8 ns ± 0.2 | 6.9 ns ± 0.2 | 3.1 ns ± 2.6 | 3.0 ns ± 0.7 |
| ARG | 3.6 a ± 0.4 | 6.1 b ± 0.1 | 4.2 a ± 0.1 | 3.9 a ± 0.2 |
| SER | 1.2 b ± 0.5 | 5.0 c ± 0.1 | 2.1 a ± 0.1 | 4.8 c ± 0.4 |
| ILE | 2.0 a ± 0.6 | 4.5 b ± 0.3 | 2.6 a ± 0.3 | 7.1 c ± 0.5 |
| ALA | 2.0 a ± 0.6 | 3.5 b ± 0.0 | 3.4 b ± 0.5 | 3.7 b ± 0.2 |
| HIS | 1.7 a ± 0.1 | 3.3 c ± 0.1 | 2.1 b ± 0.1 | 2.3 b ± 0.2 |
| ASP | 0.4 a ± 0.4 | 1.9 b ± 0.2 | 0.6 a ± 0.1 | 1.8 b ± 0.1 |
| TRP | 0.8 a ± 0.0 | 1.8 c ± 0.1 | 0.9 a ± 0.2 | 1.5 b ± 0.1 |
| MET | 0.7 ns ± 0.0 | 0.7 ns ± 0.0 | 0.8 ns ± 0.3 | 0.5 ns ± 0.1 |
| Protein Name | Protein Content (mol%) | |||
|---|---|---|---|---|
| MFP (+) | EFP (+) | MFP (-) | EFP (-) | |
| 1 AAD14P | n.d. a | n.d. a | 0.0355 b ± 0.0004 | n.d. a |
| 1 AAD4P | 0.0149 b ± 0.0001 | n.d. a | 0.0503 c ± 0.0005 | n.d. a |
| 1 ADH1P | 0.424 a ± 0.004 | 0.841 c ± 0.008 | 0.200 b ± 0.002 | 0.539 d ± 0.005 |
| 1 ADH3P | 0.122 c ± 0.001 | 0.078 a ± 0.001 | 0.121 c ± 0.001 | 0.0356 b ± 0.0004 |
| 1 ADH4P | 0.0195 b ± 0.0002 | n.d. a | 0.0675 c ± 0.0007 | n.d. a |
| 1 ADH5P | 0.0404 b ± 0.0004 | n.d. a | 0.0578 c ± 0.0004 | n.d. a |
| 1 ADH6P | n.d. a | n.d. a | 0.0567 b ± 0.0006 | n.d. a |
| 1 ADH7P | n.d. a | n.d. a | 0.120 b ± 0.001 | 0.0187 c ± 0.0002 |
| 2 ALD2P | 0.0102 c ± 0.0001 | n.d. a | 0.0091 b ± 0.0001 | 0.0399 d ± 0.0004 |
| 2 ALD4P | 0.0365 b ± 0.0004 | 0.0120 a ± 0.0001 | 0.0519 c ± 0.0005 | 0.109 d ± 0.001 |
| 2 ALD5P | n.d. a | n.d. a | 0.0280 c ± 0.0003 | 0.0110 b ± 0.0001 |
| 2 ALD6P | 0.0295 d ± 0.0003 | n.d. a | 0.0768 c ± 0.0008 | 0.370 b ± 0.004 |
| 3 ARO10P | n.d. a | n.d. a | 0.00389 b ± 0.00004 | n.d. a |
| 4 ARO8P | 0.0317 ab ± 0.0003 | n.d. a | 0.111 b ± 0.001 | 0.211 c ± 0.002 |
| 4 ARO9P | 0.0280 d ± 0.0003 | n.d. a | 0.0061 b ± 0.0001 | 0.0176 c ± 0.0002 |
| 4 BAT1P | n.d. a | 0.0930 b ± 0.0009 | 0.103 c ± 0.001 | n.d. a |
| 4 BAT2P | n.d. a | n.d. a | 0.0496 c ± 0.0005 | 0.0187 b ± 0.0002 |
| 3 PDC1P | 0.397 b ± 0.004 | 0.430 c ± 0.004 | 0.246 a ± 0.003 | 0.453 d ± 0.005 |
| 3 PDC5P | 0.377 c ± 0.004 | 0.408 d ± 0.004 | 0.253 b ± 0.003 | 0.197 a ± 0.0002 |
| 3 PDC6P | n.d. a | n.d. a | 0.0488 b ± 0.0005 | n.d. a |
| 1 SFA1P | 0.0913 c ± 0.0009 | 0.0800 b ± 0.0008 | 0.0793 b ± 0.0008 | 0.0556 a ± 0.0006 |
| 1 YPL088WP | 0.0234 b ± 0.0002 | n.d. a | 0.0443 d ± 0.0004 | 0.0298 c ± 0.0003 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Jiménez, M.d.C.; Mauricio, J.C.; Moreno-García, J.; Puig-Pujol, A.; Moreno, J.; García-Martínez, T. Endogenous CO2 Overpressure Effect on Higher Alcohols Metabolism during Sparkling Wine Production. Microorganisms 2023, 11, 1630. https://doi.org/10.3390/microorganisms11071630
González-Jiménez MdC, Mauricio JC, Moreno-García J, Puig-Pujol A, Moreno J, García-Martínez T. Endogenous CO2 Overpressure Effect on Higher Alcohols Metabolism during Sparkling Wine Production. Microorganisms. 2023; 11(7):1630. https://doi.org/10.3390/microorganisms11071630
Chicago/Turabian StyleGonzález-Jiménez, María del Carmen, Juan Carlos Mauricio, Jaime Moreno-García, Anna Puig-Pujol, Juan Moreno, and Teresa García-Martínez. 2023. "Endogenous CO2 Overpressure Effect on Higher Alcohols Metabolism during Sparkling Wine Production" Microorganisms 11, no. 7: 1630. https://doi.org/10.3390/microorganisms11071630







