The Role of Carbohydrate Intake on the Gut Microbiome: A Weight of Evidence Systematic Review
Abstract
:1. Introduction
1.1. Carbohydrates
- Rapidly digestible starches, which are hydrolyzed after less than 20 min of enzymatic digestion.
- Slowly digestible starches, which are absorbed in the small intestine and hydrolyzed in vitro after 100 min of enzymatic digestion.
- Resistant starches (RS), which are not hydrolyzed after 120 min of enzymatic incubation [3]. RS can occur naturally, such as potato starch [9], but also as a result of food processing (cooking techniques and production process). There are five RS types, depending on why these molecules resist hydrolysis [1]: type 1 RS is physically inaccessible because it is matrixed in food; type 2 RS is not gelatinized, and it is also inaccessible to enzymes; type 3 RS is retrograded after temperature changes, such as heating and cooling on multiple occasions; type 4 RS is a chemically modified starch where new bonds that are not α 1–4 or α 1–6 are present; and type 5 RS is amylose-lipid complex [10].
1.1.1. Carbohydrates as Part of Food
1.1.2. Carbohydrates as Functional Components; Functional Carbohydrates
1.1.3. Carbohydrates as Food Additives
1.2. Gut Microbiota and Microbiome
1.2.1. Gut Microbiota General Composition
1.2.2. Factors That Modify the Microbiome
1.2.3. Carbohydrates as Modulators of the Gut Microbiome
1.3. Statistics Weight of Evidence (WoE) and Information Value (IV)
2. Materials and Methods
2.1. Search Query
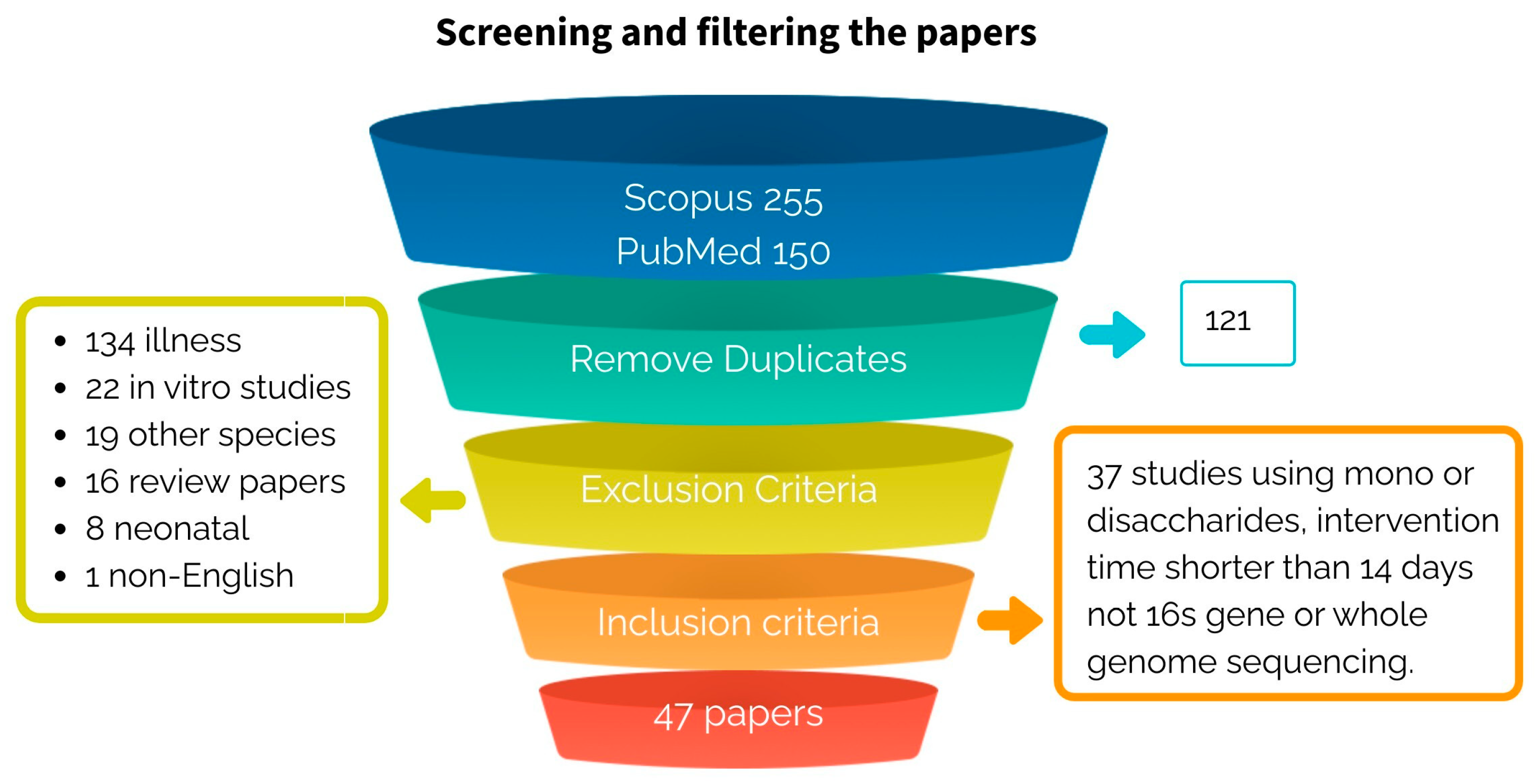
2.2. Screening
2.3. Information Synthesis and Variable Categorization
- 1.
- Carbohydrate use: food additive, functional carbohydrate, or food component
- 2.
- General description of the carbohydrate: Sulfated saccharide, fructan, inulin, oligosaccharide, polysaccharide, starch, gelatinized starch, resistant starch, insoluble fiber, soluble fiber, and antioxidant capacity.Within the response variables, there is only one classification:
- 3.
- Bacterial diversity variables (BDV): Bacterial taxa whose relative abundance showed a significant change compared to the control group after the intervention.
2.4. Statistical Analysis and Co-Occurrence of Categorical Variables
3. Results
3.1. Search Strategy
3.2. Screening
3.3. Information Synthesis and Variable Categorization
3.4. Statistical Analysis and Co-Occurrence of Categorical Variables
4. Discussion
4.1. Sulfated Polysaccharides
4.2. Gelatinized Starch
4.3. Fungal Polysaccharides
4.4. Oligosaccharides
4.5. Insoluble Fiber
4.6. Starch
4.7. Soluble Fiber
4.8. Inulin
4.9. Carbohydrates as Food Additives and Natural Food Components
4.10. Carbohydrates as Functional Compounds (Functional Carbohydrates)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leong, S.Y.; Duque, S.M.; Abduh, S.B.M.; Oey, I. Carbohydrates. In Innovative Thermal and Non-Thermal Processing, Bioaccessibility and Bioavailability of Nutrients and Bioactive Compounds; Elsevier: Amsterdam, The Netherlands, 2019; pp. 171–206. [Google Scholar] [CrossRef]
- Toussaint-Samat, M. The History of Cereals. In A History of Food; Wiley: Hoboken, NJ, USA, 2008; pp. 114–180. [Google Scholar] [CrossRef]
- Sajilata, M.; Singhal, R.S.; Kulkarni, P.R. Resistant Starch? A Review. Compr. Rev. Food Sci. Food Saf. 2006, 5, 1–17. [Google Scholar] [CrossRef]
- Qi, X.; Tester, R.F. Fructose, galactose and glucose—In health and disease. Clin. Nutr. ESPEN 2019, 33, 18–28. [Google Scholar] [CrossRef]
- Shoaib, M.; Shehzad, A.; Omar, M.; Rakha, A.; Raza, H.; Sharif, H.R.; Shakeel, A.; Ansari, A.; Niazi, S. Inulin: Properties, health benefits and food applications. Carbohydr. Polym. 2016, 147, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Beukema, M.; Faas, M.M.; de Vos, P. The effects of different dietary fiber pectin structures on the gastrointestinal immune barrier: Impact via gut microbiota and direct effects on immune cells. Exp. Mol. Med. 2020, 52, 1364–1376. [Google Scholar] [CrossRef] [PubMed]
- Sima, P.; Vannucci, L.; Vetvicka, V. β-glucans and cholesterol (Review). Int. J. Mol. Med. 2018, 41, 1799–1808. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.; Liu, Z.; Zhu, C.; Mou, H.; Kong, Q. Nondigestible carbohydrates, butyrate, and butyrate-producing bacteria. Crit. Rev. Food Sci. Nutr. 2019, 59, S130–S152. [Google Scholar] [CrossRef]
- Magallanes-Cruz, P.A.; Flores-Silva, P.C.; Bello-Perez, L.A. Starch Structure Influences Its Digestibility: A Review. J. Food Sci. 2017, 82, 2016–2023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raigond, P.; Ezekiel, R.; Raigond, B. Resistant starch in food: A review. J. Sci. Food Agric. 2015, 95, 1968–1978. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, H.; Li, Z.; Li, Y.; Wang, S.; Zhu, D.; Wen, X.; Li, S. Effects of dietary supplementation of Ulva pertusa and non-starch polysaccharide enzymes on gut microbiota of Siganus canaliculatus. J. Oceanol. Limnol. 2018, 36, 438–449. [Google Scholar] [CrossRef]
- Chi, L.; Khan, I.; Lin, Z.; Zhang, J.; Lee, M.S.; Leong, W.; Hsiao, W.W.; Zheng, Y. Fructo-oligosaccharides from Morinda officinalis remodeled gut microbiota and alleviated depression features in a stress rat model. Phytomedicine 2020, 67, 153157. [Google Scholar] [CrossRef]
- Zeng, P.; Li, J.; Chen, Y.; Zhang, L. The Structures and Biological Functions of Polysaccharides from Traditional Chinese Herbs. In Progress in Molecular Biology and Translational Science; Elsevier B.V.: Amsterdam, The Netherlands, 2019; Volume 163, pp. 423–444. [Google Scholar] [CrossRef]
- Pansai, N.; Chakree, K.; Yupanqui, C.T.; Raungrut, P.; Yanyiam, N.; Wichienchot, S. Gut microbiota modulation and immune boosting properties of prebiotic dragon fruit oligosaccharides. Int. J. Food Sci. Technol. 2020, 55, 55–64. [Google Scholar] [CrossRef]
- Xing, Y.Y.; Xu, Y.Q.; Jin, X.; Shi, L.L.; Guo, S.W.; Yan, S.M.; Shi, B.L. Optimization extraction and characterization of Artemisia ordosica polysaccharide and its beneficial effects on antioxidant function and gut microbiota in rats. RSC Adv. 2020, 10, 26151–26164. [Google Scholar] [CrossRef]
- Wu, T.; Shen, M.; Yu, Q.; Chen, Y.; Chen, X.; Yang, J.; Huang, L.; Guo, X.; Xie, J. Cyclocarya paliurus polysaccharide improves metabolic function of gut microbiota by regulating short-chain fatty acids and gut microbiota composition. Food Res. Int. 2021, 141, 110119. [Google Scholar] [CrossRef]
- Hao, J.; Li, G.; Yu, G. Dietary Polysaccharide from Enteromorpha Clathrata Modulates Gut Microbiota and Promotes the Growth. Mar. Drugs 2018, 16, 167. [Google Scholar] [CrossRef] [Green Version]
- Shang, Q.; Shi, J.; Song, G.; Zhang, M.; Cai, C.; Hao, J.; Li, G.; Yu, G. Structural modulation of gut microbiota by chondroitin sulfate and its oligosaccharide. Int. J. Biol. Macromol. 2016, 89, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Saha, D.; Bhattacharya, S. Hydrocolloids as thickening and gelling agents in food: A critical review. J. Food Sci. Technol. 2010, 47, 587–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Codex Alimentarius. General Standard for Food Additives. 2021. Available online: http://www.fao.org/food/food-safety-quality/scientific-advice/jecfa/jecfa-additives/en/ (accessed on 22 March 2022).
- Chilakapati, J.; Mehendale, H.M. Acceptable Daily Intake (ADI). In Encyclopedia of Toxicology, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 8–9. [Google Scholar] [CrossRef]
- Kapoor, M.P.; Koido, M.; Kawaguchi, M.; Timm, D.; Ozeki, M.; Yamada, M.; Mitsuya, T.; Okubo, T. Lifestyle related changes with partially hydrolyzed guar gum dietary fiber in healthy athlete individuals—A randomized, double-blind, crossover, placebo-controlled gut microbiome clinical study. J. Funct. Foods 2020, 72, 104067. [Google Scholar] [CrossRef]
- Surono, I.S.; Venema, K. Modulation of Gut Microbiota Profile and Short-Chain Fatty Acids of Rats Fed with Taro Flour or Taro Starch. Int. J. Microbiol. 2020, 2020, 8893283. [Google Scholar] [CrossRef]
- Zhang, S.; Hu, H.; He, W.; Muhammad, Z.; Wang, L.; Liu, F.; Pan, S. Regulatory Roles of Pectin Oligosaccharides on Immunoglobulin Production in Healthy Mice Mediated by Gut Microbiota. Mol. Nutr. Food Res. 2019, 63, e1801363. [Google Scholar] [CrossRef]
- Bang, S.-J.; Lee, E.-S.; Song, E.-J.; Nam, Y.-D.; Seo, M.-J.; Kim, H.-J.; Park, C.-S.; Lim, M.Y.; Seo, D.-H. Effect of raw potato starch on the gut microbiome and metabolome in mice. Int. J. Biol. Macromol. 2019, 133, 37–43. [Google Scholar] [CrossRef]
- Newman, M.A.; Petri, R.M.; Grüll, D.; Zebeli, Q.; Metzler-Zebeli, B.U. Transglycosylated Starch Modulates the Gut Microbiome and Expression of Genes Related to Lipid Synthesis in Liver and Adipose Tissue of Pigs. Front. Microbiol. 2018, 9, 224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Role of the gut microbiota in nutrition and health. BMJ 2018, 361, k2179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lederberg, B.J.; McCray, A.T. ‘Ome Sweet’ Omics—A Genealogical Treasury of Words. Scientist 2001, 15, 8. [Google Scholar]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome Definition Re-Visited: Old Concepts and New Challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, Y.; Ai, C.; Song, S.; Chen, X. Caulerpa lentillifera polysaccharides enhance the immunostimulatory activity in immunosuppressed mice in correlation with modulating gut microbiota. Food Funct. 2019, 10, 4315–4329. [Google Scholar] [CrossRef]
- Henke, M.T.; Kenny, D.J.; Cassilly, C.D.; Vlamakis, H.; Xavier, R.J.; Clardy, J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. USA 2019, 116, 12672–12677. [Google Scholar] [CrossRef] [Green Version]
- Laursen, M.F.; Zachariassen, G.; Bahl, M.I.; Bergström, A.; Høst, A.; Michaelsen, K.F.; Licht, T.R. Having older siblings is associated with gut microbiota development during early childhood. BMC Microbiol. 2015, 15, 154. [Google Scholar] [CrossRef]
- Song, M.; Chan, A.T.; Sun, J. Influence of the Gut Microbiome, Diet, and Environment on Risk of Colorectal Cancer. Gastroenterology 2020, 158, 322–340. [Google Scholar] [CrossRef]
- Foster, J.A.; McVey Neufeld, K.-A. Gut–brain axis: How the microbiome influences anxiety and depression. Trends Neurosci. 2013, 36, 305–312. [Google Scholar] [CrossRef]
- Wisniewski, P.J.; Dowden, R.A.; Campbell, S.C. Role of Dietary Lipids in Modulating Inflammation through the Gut Microbiota. Nutrients 2019, 11, 117. [Google Scholar] [CrossRef] [Green Version]
- Gummesson, A.; Carlsson, L.M.; Storlien, L.H.; Bäckhed, F.; Lundin, P.; Löfgren, L.; Stenlöf, K.; Lam, Y.Y.; Fagerberg, B.; Carlsson, B. Intestinal Permeability Is Associated with Visceral Adiposity in Healthy Women. Obesity 2011, 19, 2280–2282. [Google Scholar] [CrossRef]
- Wang, B.; Yao, M.; Lv, L.; Ling, Z.; Li, L. The Human Microbiota in Health and Disease. Engineering 2017, 3, 71–82. [Google Scholar] [CrossRef]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the Human Intestinal Microbial Flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Li, L.; Ma, S.; Ye, J.; Zhang, H.; Li, Y.; Sair, A.T.; Pan, J.; Liu, X.; Li, X.; et al. High-Dietary Fiber Intake Alleviates Antenatal Obesity-Induced Postpartum Depression: Roles of Gut Microbiota and Microbial Metabolite Short-chain Fatty Acid Involved. J. Agric. Food Chem. 2020, 68, 13697–13710. [Google Scholar] [CrossRef]
- Cheng, W.; Lu, J.; Li, B.; Lin, W.; Zhang, Z.; Wei, X.; Sun, C.; Chi, M.; Bi, W.; Yang, B.; et al. Effect of Functional Oligosaccharides and Ordinary Dietary Fiber on Intestinal Microbiota Diversity. Front. Microbiol. 2017, 8, 1750. [Google Scholar] [CrossRef] [Green Version]
- Payne, A.N.; Chassard, C.; Lacroix, C. Gut microbial adaptation to dietary consumption of fructose, artificial sweeteners and sugar alcohols: Implications for host-microbe interactions contributing to obesity. Obes. Rev. 2012, 13, 799–809. [Google Scholar] [CrossRef]
- Chen, J.; Liu, J.; Yan, C.; Zhang, C.; Pan, W.; Zhang, W.; Lu, Y.; Chen, L.; Chen, Y. Sarcodon aspratus polysaccharides ameliorated obesity-induced metabolic disorders and modulated gut microbiota dysbiosis in mice fed a high-fat diet. Food Funct. 2020, 11, 2588–2602. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Davenport, E.R.; Beaumont, M.; Jackson, M.A.; Knight, R.; Ober, C.; Spector, T.D.; Bell, J.T.; Clark, A.G.; Ley, R.E. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe 2016, 19, 731–743. [Google Scholar] [CrossRef] [Green Version]
- Martin, R.; Makino, H.; Cetinyurek Yavuz, A.; Ben-Amor, K.; Roelofs, M.; Ishikawa, E.; Kubota, H.; Swinkels, S.; Sakai, T.; Oishi, K.; et al. Early-Life Events, Including Mode of Delivery and Type of Feeding, Siblings and Gender, Shape the Developing Gut Microbiota. PLoS ONE 2016, 11, e0158498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korpela, K.; Salonen, A.; Hickman, B.; Kunz, C.; Sprenger, N.; Kukkonen, K.; Savilahti, E.; Kuitunen, M.; de Vos, W.M. Fucosylated oligosaccharides in mother’s milk alleviate the effects of caesarean birth on infant gut microbiota. Sci. Rep. 2018, 8, 13757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, C.; Lane, J.A.; van Sinderen, D.; Hickey, R.M. Human milk oligosaccharides: Shaping the infant gut microbiota and supporting health. J. Funct. Foods 2020, 72, 104074. [Google Scholar] [CrossRef] [PubMed]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Tyakht, A.V.; Kostryukova, E.S.; Popenko, A.S.; Belenikin, M.S.; Pavlenko, A.V.; Larin, A.K.; Karpova, I.Y.; Selezneva, O.V.; Semashko, T.A.; Ospanova, E.A.; et al. Human gut microbiota community structures in urban and rural populations in Russia. Nat. Commun. 2013, 4, 2469. [Google Scholar] [CrossRef] [Green Version]
- Ramirez, J.; Guarner, F.; Fernandez, L.B.; Maruy, A.; Sdepanian, V.L.; Cohen, H. Antibiotics as Major Disruptors of Gut Microbiota. Front. Cell. Infect. Microbiol. 2020, 10, 572912. [Google Scholar] [CrossRef] [PubMed]
- Mailing, L.J.; Allen, J.M.; Buford, T.W.; Fields, C.J.; Woods, J.A. Exercise and the Gut Microbiome: A Review of the Evidence, Potential Mechanisms, and Implications for Human Health. Exerc. Sport Sci. Rev. 2019, 47, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Osadchiy, V.; Martin, C.R.; Mayer, E.A. The gut-brain axis and the microbiome: Mechanisms and clinical implications. Clin. Gastroenterol. Hepatol. 2019, 17, 322–332. [Google Scholar] [CrossRef]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B.; Galla, S.; Chakraborty, S.; Cheng, X.; Yeo, J.; Mell, B.; Zhang, H.; et al. Gut Microbiota in Health and Disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [Green Version]
- Rubin, C.E.; Brandborg, L.L.; Phelps, P.C.; Taylor, H.C. Studies of celiac disease. I. The apparent identical and specific nature of the duodenal and proximal jejunal lesion in celiac disease and idiopathic sprue. Gastroenterology 1968, 54, S800–S802. [Google Scholar]
- Cantarel, B.L.; Lombard, V.; Henrissat, B. Complex Carbohydrate Utilization by the Healthy Human Microbiome. PLoS ONE 2012, 7, e28742. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Bjursell, M.K.; Himrod, J.; Deng, S.; Carmichael, L.K.; Chiang, H.C.; Hooper, L.V.; Gordon, J.I. A Genomic View of the Human- Bacteroides thetaiotaomicron Symbiosis. Science 2003, 299, 2074–2076. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Zhou, P.; Zhang, Y.; Zhang, Z.; Liu, J.; Zhang, H. Short-chain fructo-oligosaccharides alleviates oxidized oil-induced intestinal dysfunction in piglets associated with the modulation of gut microbiota. J. Funct. Foods 2020, 64, 103661. [Google Scholar] [CrossRef]
- Kruschke, J.K. Models of Categorization. In The Cambridge Handbook of Computational Psychology; Cambridge University Press: New York, NY, USA, 2008; pp. 267–301. [Google Scholar]
- Hammer, P.L.; Bonates, T.O. Logical analysis of data—An overview: From combinatorial optimization to medical applications. Ann. Oper. Res. 2006, 148, 203–225. [Google Scholar] [CrossRef]
- Fan, L.; Ding, Y. Research on risk scorecard of sick building syndrome based on machine learning. Build. Environ. 2022, 211, 108710. [Google Scholar] [CrossRef]
- Rosenblatt, E. Calculating Weight of Evidence and Information Value. In Credit Data and Scoring; Elsevier: Amsterdam, The Netherlands, 2020; pp. 99–104. [Google Scholar] [CrossRef]
- Siddiqi, N. Credit Risk Scorecards: Developing and Implementing Intelligent Credit Scoring; Wiley: Danvers, MA, USA, 2006; Volume 1. [Google Scholar]
- Tintó-Moliner, A.; Martin, M. Quantitative weight of evidence method for combining predictions of quantitative structure-activity relationship models. SAR QSAR Environ. Res. 2020, 31, 261–279. [Google Scholar] [CrossRef]
- Rocca, M.; Morford, L.L.; Blanset, D.L.; Halpern, W.G.; Cavagnaro, J.; Bowman, C.J. Applying a weight of evidence approach to the evaluation of developmental toxicity of biopharmaceuticals. Regul. Toxicol. Pharmacol. 2018, 98, 69–79. [Google Scholar] [CrossRef]
- Dekant, W.; Bridges, J. A quantitative weight of evidence methodology for the assessment of reproductive and developmental toxicity and its application for classification and labeling of chemicals. Regul. Toxicol. Pharmacol. 2016, 82, 173–185. [Google Scholar] [CrossRef]
- Cooke, A.; Smith, D.; Booth, A. Beyond PICO: The SPIDER Tool for Qualitative Evidence Synthesis. Qual. Health Res. 2012, 22, 1435–1443. [Google Scholar] [CrossRef]
- Methley, A.M.; Campbell, S.; Chew-Graham, C.; McNally, R.; Cheraghi-Sohi, S. PICO, PICOS and SPIDER: A comparison study of specificity and sensitivity in three search tools for qualitative systematic reviews. BMC Health Serv. Res. 2014, 14, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Siddaway, A.P.; Wood, A.M.; Hedges, L.V. How to Do a Systematic Review: A Best Practice Guide for Conducting and Reporting Narrative Reviews, Meta-Analyses, and Meta-Syntheses. Annu. Rev. Psychol. 2019, 70, 747–770. [Google Scholar] [CrossRef]
- Van Eck, N.J.; Waltman, L. VOS Viewer. 2020. Available online: https://www.vosviewer.com (accessed on 21 June 2020).
- Van Eck, N.J.; Waltman, L. Citation-based clustering of publications using CitNetExplorer and VOSviewer. Scientometrics 2017, 111, 1053–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Python Language Reference—Python 3.10.4 Documentation. Available online: https://docs.python.org/3/reference/ (accessed on 23 April 2022).
- Kluyver, T.; Ragan-Kelley, B.; Pérez, F.; Granger, B.; Bussonnier, M.; Frederic, J.; Kelley, K.; Hamrick, J.; Grout, J.; Corlay, S.; et al. Jupyter Notebooks—A Publishing Format for Reproducible Computational Workflows. In Positioning and Power in Academic Publishing: Players, Agents and Agendas; IOS Pres: Amsterdam, The Netherlands, 2016. [Google Scholar] [CrossRef]
- McKinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010. [Google Scholar] [CrossRef] [Green Version]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Gramarly Inc. Gramarly. Available online: https://www.grammarly.com/contact (accessed on 28 February 2023).
- Open, A.I. Chat GPT—A Large Language Model Trained by OpenAI. 2023. Available online: https://openai.com/blog/gpt-3-apps/ (accessed on 28 February 2023).
- Jiang, P.; Zheng, W.; Sun, X.; Jiang, G.; Wu, S.; Xu, Y.; Song, S.; Ai, C. Sulfated polysaccharides from Undaria pinnatifida improved high fat diet-induced metabolic syndrome, gut microbiota dysbiosis and inflammation in BALB/c mice. Int. J. Biol. Macromol. 2021, 167, 1587–1597. [Google Scholar] [CrossRef]
- Wu, T.-Y.; Tsai, S.-J.; Sun, N.-N.; Dai, F.-J.; Yu, P.-H.; Chen, Y.-C.; Chau, C.-F. Enhanced thermal stability of green banana starch by heat-moisture treatment and its ability to reduce body fat accumulation and modulate gut microbiota. Int. J. Biol. Macromol. 2020, 160, 915–924. [Google Scholar] [CrossRef]
- He, N.; Wang, S.; Lv, Z.; Zhao, W.; Li, S. Low molecular weight chitosan oligosaccharides (LMW-COSs) prevent obesity-related metabolic abnormalities in association with the modification of gut microbiota in high-fat diet (HFD)-fed mice. Food Funct. 2020, 11, 9947–9959. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, H.; Yu, B.; Tao, H.; Li, J.; Wu, Z.; Liu, G.; Yuan, C.; Guo, L.; Cui, B. Lycium barbarum polysaccharide attenuates myocardial injury in high-fat diet-fed mice through manipulating the gut microbiome and fecal metabolome. Food Res. Int. 2020, 138, 109778. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Shi, C.-L.; Zhou, X.-J.; Wen, W.; Gao, X.-P.; Wang, L.-Y.; He, B.; Yin, M.; Zhao, J.-Q. Lycium barbarum Polysaccharide Extracted from Lycium barbarum Leaves Ameliorates Asthma in Mice by Reducing Inflammation and Modulating Gut Microbiota. J. Med. Food 2020, 23, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Tuncil, Y.E.; Thakkar, R.D.; Arioglu-Tuncil, S.; Hamaker, B.R.; Lindemann, S.R. Subtle Variations in Dietary-Fiber Fine Structure Differentially Influence the Composition and Metabolic Function of Gut Microbiota. Msphere 2020, 5, e00180-20. [Google Scholar] [CrossRef]
- Marsaux, B.; Abbeele, P.V.D.; Ghyselinck, J.; Prioult, G.; Marzorati, M.; Bogićević, B. Synbiotic Effect of Bifidobacterium lactis CNCM I-3446 and Bovine Milk-Derived Oligosaccharides on Infant Gut Microbiota. Nutrients 2020, 12, 2268. [Google Scholar] [CrossRef]
- Gerasimidis, K.; Bryden, K.; Chen, X.; Papachristou, E.; Verney, A.; Roig, M.; Hansen, R.; Nichols, B.; Papadopoulou, R.; Parrett, A. The impact of food additives, artificial sweeteners and domestic hygiene products on the human gut microbiome and its fibre fermentation capacity. Eur. J. Nutr. 2020, 59, 3213–3230. [Google Scholar] [CrossRef] [Green Version]
- Borewicz, K.; Gu, F.; Saccenti, E.; Arts, I.C.; Penders, J.; Thijs, C.; van Leeuwen, S.S.; Lindner, C.; Nauta, A.; van Leusen, E.; et al. Correlating Infant Fecal Microbiota Composition and Human Milk Oligosaccharide Consumption by Microbiota of 1-Month-Old Breastfed Infants. Mol. Nutr. Food Res. 2019, 63, 1801214. [Google Scholar] [CrossRef] [Green Version]
- Borewicz, K.; Gu, F.; Saccenti, E.; Hechler, C.; Beijers, R.; de Weerth, C.; van Leeuwen, S.S.; Schols, H.A.; Smidt, H. The association between breastmilk oligosaccharides and faecal microbiota in healthy breastfed infants at two, six, and twelve weeks of age. Sci. Rep. 2020, 10, 4270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, H.; Perreau, J.; Powell, J.E.; Han, B.; Zhang, Z.; Kwong, W.K.; Tringe, S.G.; Moran, N.A. Division of labor in honey bee gut microbiota for plant polysaccharide digestion. Proc. Natl. Acad. Sci. USA 2019, 116, 25909–25916. [Google Scholar] [CrossRef]
- Hughes, R.-A.; Ali, R.A.; Mendoza, M.A.; Hassan, H.M.; Koci, M.D. Impact of Dietary Galacto-Oligosaccharide (GOS) on Chicken’s Gut Microbiota, Mucosal Gene Expression, and Salmonella Colonization. Front. Veter-Sci. 2017, 4, 192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, P.; Yang, S.; Hu, C.; Zhao, Z.; Liu, J.; Cheng, Y.; Wang, S.; Chen, Q.; Yu, P.; Zhang, X.; et al. Sargassum fusiforme Polysaccharide Rejuvenat es the Small Intestine in Mice Through Altering its Physiol ogy and Gut Microbiota Composition. Curr. Mol. Med. 2018, 17, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, N.; Kan, J.; Sun, R.; Tang, S.; Wang, Z.; Chen, M.; Liu, J.; Jin, C. Anti-inflammatory activity of alkali-soluble polysaccharides from Arctium lappa L. and its effect on gut microbiota of mice with inflammation. Int. J. Biol. Macromol. 2020, 154, 773–787. [Google Scholar] [CrossRef]
- Pi, Y.; Mu, C.; Gao, K.; Liu, Z.; Peng, Y.; Zhu, W. Increasing the Hindgut Carbohydrate/Protein Ratio by Cecal Infusion of Corn Starch or Casein Hydrolysate Drives Gut Microbiota-Related Bile Acid Metabolism to Stimulate Colonic Barrier Function. Msystems 2020, 5, e00176-20. [Google Scholar] [CrossRef]
- Zhu, W.; Zhou, S.; Liu, J.; McLean, R.J.; Chu, W. Prebiotic, immuno-stimulating and gut microbiota-modulating effects of Lycium barbarum polysaccharide. Biomed. Pharmacother. 2020, 121, 109591. [Google Scholar] [CrossRef]
- Chijiiwa, R.; Hosokawa, M.; Kogawa, M.; Nishikawa, Y.; Ide, K.; Sakanashi, C.; Takahashi, K.; Takeyama, H. Single-Cell Genomics of Uncultured Bacteria Reveals Dietary Fiber Responders in the Mouse Gut Microbiota. Microbiome 2020, 8, 5. [Google Scholar] [CrossRef] [Green Version]
- Su, L.; Mao, C.; Wang, X.; Li, L.; Tong, H.; Mao, J.; Ji, D.; Lu, T.; Hao, M.; Huang, Z.; et al. The Anti-colitis Effect of Schisandra chinensis Polysaccharide Is Associated with the Regulation of the Composition and Metabolism of Gut Microbiota. Front. Cell. Infect. Microbiol. 2020, 10, 519479. [Google Scholar] [CrossRef]
- Do, M.H.; Lee, H.-B.; Lee, E.; Park, H.-Y. The Effects of Gelatinized Wheat Starch and High Salt Diet on Gut Microbiota and Metabolic Disorder. Nutrients 2020, 12, 301. [Google Scholar] [CrossRef] [Green Version]
- Yan, S.; Shi, R.; Li, L.; Ma, S.; Zhang, H.; Ye, J.; Wang, J.; Pan, J.; Wang, Q.; Jin, X.; et al. Mannan Oligosaccharide Suppresses Lipid Accumulation and Appetite in Western-Diet-Induced Obese Mice Via Reshaping Gut Microbiome and Enhancing Short-Chain Fatty Acids Production. Mol. Nutr. Food Res. 2019, 63, e1900521. [Google Scholar] [CrossRef]
- Kaur, A.; Chen, T.; Green, S.J.; Mutlu, E.; Martin, B.R.; Rumpagaporn, P.; Patterson, J.A.; Keshavarzian, A.; Hamaker, B.R. Physical Inaccessibility of a Resistant Starch Shifts Mouse Gut Microbiota to Butyrogenic Firmicutes. Mol. Nutr. Food Res. 2019, 63, e1801012. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zhu, B.; Sun, Y.; Ai, C.; Wu, S.; Wang, L.; Song, S.; Liu, X. Sulfated polysaccharide from sea cucumber modulates the gut microbiota and its metabolites in normal mice. Int. J. Biol. Macromol. 2018, 120, 502–512. [Google Scholar] [CrossRef]
- Nakata, T.; Kyoui, D.; Takahashi, H.; Kimura, B.; Kuda, T. Inhibitory effects of soybean oligosaccharides and water-soluble soybean fibre on formation of putrefactive compounds from soy protein by gut microbiota. Int. J. Biol. Macromol. 2017, 97, 173–180. [Google Scholar] [CrossRef]
- Chang, S.; Cui, X.; Guo, M.; Tian, Y.; Xu, W.; Huang, K.; Zhang, Y. Insoluble Dietary Fiber from Pear Pomace Can Prevent High-Fat Diet-Induced Obesity in Rats Mainly by Improving the Structure of the Gut Microbiota. J. Microbiol. Biotechnol. 2017, 27, 856–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; Wang, X.; Wang, K.; Deng, Y.; Yang, Y.; Ali, R.; Chen, F.; Wu, Z.; Liao, W.; Mao, L. A novel polysaccharide isolated from Flammulina velutipes, characterization, macrophage immunomodulatory activities and its impact on gut microbiota in rats. J. Anim. Physiol. Anim. Nutr. 2020, 104, 735–748. [Google Scholar] [CrossRef]
- Chen, R.; Liu, B.; Wang, X.; Chen, K.; Zhang, K.; Zhang, L.; Fei, C.; Wang, C.; Liu, Y.; Xue, F.; et al. Effects of polysaccharide from Pueraria lobata on gut microbiota in mice. Int. J. Biol. Macromol. 2020, 158, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Huo, W.; Feng, Z.; Hu, S.; Cui, L.; Qiao, T.; Dai, L.; Qi, P.; Zhang, L.; Liu, Y.; Li, J. Effects of polysaccharides from wild morels on immune response and gut microbiota composition in non-treated and cyclophosphamide-treated mice. Food Funct. 2020, 11, 4291–4303. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, X.; Zhou, S.; Wang, T.T.Y.; Zhou, S.; Yang, K.; Li, Y.; Tian, J.; Wang, J. Dietary fiber isolated from sweet potato residues promotes a healthy gut microbiome profile. Food Funct. 2020, 11, 689–699. [Google Scholar] [CrossRef]
- Peerakietkhajorn, S.; Jeanmard, N.; Chuenpanitkit, P.; K-Da, S.; Bannob, K.; Khuituan, P. Effects of Plant Oligosaccharides Derived from Dragon Fruit on Gut Microbiota in Proximal and Distal Colon of Mice. Sains Malays. 2020, 49, 603–611. [Google Scholar] [CrossRef]
- Chen, Y.; Jin, L.; Li, Y.; Xia, G.; Chen, C.; Zhang, Y. Bamboo-shaving polysaccharide protects against high-diet induced obesity and modulates the gut microbiota of mice. J. Funct. Foods 2018, 49, 20–31. [Google Scholar] [CrossRef]
- Zheng, J.; Cheng, G.; Li, Q.; Jiao, S.; Feng, C.; Zhao, X.; Yin, H.; Du, Y.; Liu, H. Chitin Oligosaccharide Modulates Gut Microbiota and Attenuates High-Fat-Diet-Induced Metabolic Syndrome in Mice. Mar. Drugs 2018, 16, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, C.; Sun, J.; Zhou, B.; Jin, C.; Liu, J.; Kan, J.; Qian, C.; Zhang, N. Effects of polysaccharides from purple sweet potatoes on immune response and gut microbiota composition in normal and cyclophosphamide treated mice. Food Funct. 2018, 9, 937–950. [Google Scholar] [CrossRef]
- Tian, G.; Wu, X.; Chen, D.; Yu, B.; He, J. Adaptation of gut microbiome to different dietary nonstarch polysaccharide fractions in a porcine model. Mol. Nutr. Food Res. 2017, 61, 1700012. [Google Scholar] [CrossRef]
- Higashimura, Y.; Baba, Y.; Inoue, R.; Takagi, T.; Mizushima, K.; Ohnogi, H.; Honda, A.; Matsuzaki, Y.; Naito, Y. Agaro-Oligosaccharides Regulate Gut Microbiota and Adipose Tissue Accumulation in Mice. J. Nutr. Sci. Vitaminol. 2017, 63, 269–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrario, C.; Statello, R.; Carnevali, L.; Mancabelli, L.; Milani, C.; Mangifesta, M.; Duranti, S.; Lugli, G.A.; Jimenez, B.; Lodge, S.; et al. How to Feed the Mammalian Gut Microbiota: Bacterial and Metabolic Modulation by Dietary Fibers. Front. Microbiol. 2017, 8, 1749. [Google Scholar] [CrossRef] [PubMed]
- Ou, J.-Y.; Huang, J.-Q.; Song, Y.; Yao, S.-W.; Peng, X.-C.; Wang, M.-F.; Ou, S.-Y. Feruloylated Oligosaccharides from Maize Bran Modulated the Gut Microbiota in Rats. Plant Foods Hum. Nutr. 2016, 71, 123–128. [Google Scholar] [CrossRef]
- Zhou, S.-S.; Xu, J.; Zhu, H.; Wu, J.; Xu, J.-D.; Yan, R.; Li, X.-Y.; Liu, H.-H.; Duan, S.-M.; Wang, Z.; et al. Gut microbiota-involved mechanisms in enhancing systemic exposure of ginsenosides by coexisting polysaccharides in ginseng decoction. Sci. Rep. 2016, 6, 22474. [Google Scholar] [CrossRef] [Green Version]
- Monteagudo-Mera, A.; Arthur, J.C.; Jobin, C.; Keku, T.; Bruno-Barcena, J.M.; Azcarate-Peril, M.A. High purity galacto-oligosaccharides enhance specific Bifidobacterium species and their metabolic activity in the mouse gut microbiome. Benef. Microbes 2016, 7, 247–264. [Google Scholar] [CrossRef] [Green Version]
- Zhong, L.; Ma, N.; Zheng, H.; Ma, G.; Zhao, L.; Hu, Q. Tuber indicum polysaccharide relieves fatigue by regulating gut microbiota in mice. J. Funct. Foods 2019, 63, 103580. [Google Scholar] [CrossRef]
- Reimer, R.A.; Soto-Vaca, A.; Nicolucci, A.C.; Mayengbam, S.; Park, H.; Madsen, K.L.; Menon, R.; Vaughan, E.E. Effect of chicory inulin-type fructan–containing snack bars on the human gut microbiota in low dietary fiber consumers in a randomized crossover trial. Am. J. Clin. Nutr. 2020, 111, 1286–1296. [Google Scholar] [CrossRef]
- Bush, J.R.; Alfa, M.J. Increasing levels of Parasutterella in the gut microbiome correlate with improving low-density lipoprotein levels in healthy adults consuming resistant potato starch during a randomised trial. BMC Nutr. 2020, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Alfa, M.J.; Strang, D.; Tappia, P.S.; Graham, M.; Van Domselaar, G.; Forbes, J.D.; Laminman, V.; Olson, N.; DeGagne, P.; Bray, D.; et al. A randomized trial to determine the impact of a digestion resistant starch composition on the gut microbiome in older and mid-age adults. Clin. Nutr. 2018, 37, 797–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vital, M.; Howe, A.; Bergeron, N.; Krauss, R.M.; Jansson, J.K.; Tiedje, J.M. Metagenomic Insights into the Degradation of Resistant Starch by Human Gut Microbiota. Appl. Environ. Microbiol. 2018, 84, e01562-18. [Google Scholar] [CrossRef] [Green Version]
- Kiewiet, M.B.G.; Elderman, M.E.; El Aidy, S.; Burgerhof, J.G.M.; Visser, H.; Vaughan, E.E.; Faas, M.M.; de Vos, P. Flexibility of Gut Microbiota in Ageing Individuals during Dietary Fiber Long-Chain Inulin Intake. Mol. Nutr. Food Res. 2021, 65, e2000390. [Google Scholar] [CrossRef] [PubMed]
- Hughes, R.L.; Horn, W.H.; Finnegan, P.; Newman, J.W.; Marco, M.L.; Keim, N.L.; Kable, M.E. Resistant Starch Type 2 from Wheat Reduces Postprandial Glycemic Response with Concurrent Alterations in Gut Microbiota Composition. Nutrients 2021, 13, 645. [Google Scholar] [CrossRef]
- Zhang, L.; Ouyang, Y.; Li, H.; Shen, L.; Ni, Y.; Fang, Q.; Wu, G.; Qian, L.; Xiao, Y.; Zhang, J.; et al. Metabolic phenotypes and the gut microbiota in response to dietary resistant starch type 2 in normal-weight subjects: A randomized crossover trial. Sci. Rep. 2019, 9, 4736. [Google Scholar] [CrossRef] [Green Version]
- Healey, G.; Murphy, R.; Butts, C.; Brough, L.; Whelan, K.; Coad, J. Habitual dietary fibre intake influences gut microbiota response to an inulin-type fructan prebiotic: A randomised, double-blind, placebo-controlled, cross-over, human intervention study. Br. J. Nutr. 2018, 119, 176–189. [Google Scholar] [CrossRef]
- Maier, T.V.; Lucio, M.; Lee, L.H.; VerBerkmoes, N.C.; Brislawn, C.J.; Bernhardt, J.; Lamendella, R.; McDermott, J.E.; Bergeron, N.; Heinzmann, S.S.; et al. Impact of Dietary Resistant Starch on the Human Gut Microbiome, Metaproteome, and Metabolome. mBio 2017, 8, e01343-17. [Google Scholar] [CrossRef] [Green Version]
- Tanes, C.; Bittinger, K.; Gao, Y.; Friedman, E.S.; Nessel, L.; Paladhi, U.R.; Chau, L.; Panfen, E.; Fischbach, M.A.; Braun, J.; et al. Role of dietary fiber in the recovery of the human gut microbiome and its metabolome. Cell Host Microbe 2021, 29, 394–407.e5. [Google Scholar] [CrossRef]
- Bendiks, Z.A.; Knudsen, K.E.; Keenan, M.J.; Marco, M.L. Conserved and variable responses of the gut microbiome to resistant starch type 2. Nutr. Res. 2020, 77, 12–28. [Google Scholar] [CrossRef]
- Setny, P.; Baron, R.; McCammon, J.A. How Can Hydrophobic Association Be Enthalpy Driven? J. Chem. Theory Comput. 2010, 6, 2866–2871. [Google Scholar] [CrossRef]
- Wu, T.; Shen, M.; Guo, X.; Huang, L.; Yang, J.; Yu, Q.; Chen, Y.; Xie, J. Cyclocarya paliurus polysaccharide alleviates liver inflammation in mice via beneficial regulation of gut microbiota and TLR4/MAPK signaling pathways. Int. J. Biol. Macromol. 2020, 160, 164–174. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, H.; Zheng, J.; Jiang, N.; Sun, G.; Bao, X.; Lin, A.; Liu, H. Chitosan oligosaccharides attenuate loperamide-induced constipation through regulation of gut microbiota in mice. Carbohydr. Polym. 2020, 253, 117218. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Dong, L.; Jia, X.; Liu, L.; Chi, J.; Huang, F.; Ma, Q.; Zhang, M.; Zhang, R. Bound Phenolics Ensure the Antihyperglycemic Effect of Rice Bran Dietary Fiber in db/db Mice via Activating the Insulin Signaling Pathway in Skeletal Muscle and Altering Gut Microbiota. J. Agric. Food Chem. 2020, 68, 4387–4398. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Liu, M.; Zhang, P.; Fan, S.; Huang, J.; Yu, S.; Zhang, C.; Li, H. Fucoidan and galactooligosaccharides ameliorate high-fat diet–induced dyslipidemia in rats by modulating the gut microbiota and bile acid metabolism. Nutrition 2019, 65, 50–59. [Google Scholar] [CrossRef]
- Gotteland, M.; Riveros, K.; Gasaly, N.; Carcamo, C.; Magne, F.; Liabeuf, G.; Beattie, A.; Rosenfeld, S. The Pros and Cons of Using Algal Polysaccharides as Prebiotics. Front. Nutr. 2020, 7, 163. [Google Scholar] [CrossRef] [PubMed]
- Baffert, C.; Kpebe, A.; Avilan, L.; Brugna, M. Hydrogenases and H2 metabolism in sulfate-reducing bacteria of the Desulfovibrio genus. Adv. Microb. Physiol. 2019, 74, 143–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez, I.; Stegen, J.C.; Maldonado-Gómez, M.X.; Eren, A.M.; Siba, P.M.; Greenhill, A.R.; Walter, J. The Gut Microbiota of Rural Papua New Guineans: Composition, Diversity Patterns, and Ecological Processes. Cell Rep. 2015, 11, 527–538. [Google Scholar] [CrossRef] [Green Version]
- De Filippis, F.; Pellegrini, N.; Vannini, L.; Jeffery, I.B.; La Storia, A.; Laghi, L.; Serrazanetti, D.I.; Di Cagno, R.; Ferrocino, I.; Lazzi, C.; et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut Microbiota 2016, 65, 1812–1821. [Google Scholar] [CrossRef]
- Pedersen, H.K.; Gudmundsdottir, V.; Nielsen, H.B.; Hyotylainen, T.; Nielsen, T.; Jensen, B.A.H.; Forslund, K.; Hildebrand, F.; Prifti, E.; Falony, G.; et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016, 535, 376–381. [Google Scholar] [CrossRef]
- Scher, J.U.; Sczesnak, A.; Longman, R.S.; Segata, N.; Ubeda, C.; Bielski, C.; Rostron, T.; Cerundolo, V.; Pamer, E.G.; Abramson, S.B.; et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2013, 2, e01202. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.M. The immune response toPrevotellabacteria in chronic inflammatory disease. Immunology 2017, 151, 363–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dillon, S.M.; Lee, E.J.; Kotter, C.V.; Austin, G.L.; Gianella, S.; Siewe, B.; Smith, D.M.; Landay, A.L.; McManus, M.C.; Robertson, C.E.; et al. Gut dendritic cell activation links an altered colonic microbiome to mucosal and systemic T-cell activation in untreated HIV-1 infection. Mucosal Immunol. 2016, 9, 24–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, N.; Vogensen, F.K.; Van Den Berg, F.W.J.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sørensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut Microbiota in Human Adults with Type 2 Diabetes Differs from Non-Diabetic Adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, M.; Pang, X.; Zhao, Y.; Wang, L.; Zhao, L. Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J. 2012, 6, 1848–1857. [Google Scholar] [CrossRef]
- Wang, J.; Tang, H.; Zhang, C.; Zhao, Y.; Derrien, M.; Rocher, E.; van-Hylckama Vlieg, J.E.T.; Strissel, K.; Zhao, L.; Obin, M.; et al. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 2015, 9, 1–15. [Google Scholar] [CrossRef]
- Everard, A.; Lazarevic, V.; Gaïa, N.; Johansson, M.; Ståhlman, M.; Bäckhed, F.; Delzenne, N.M.; Schrenzel, J.; Francois, P.; Cani, P.D. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J. 2014, 8, 2116–2130. [Google Scholar] [CrossRef]
- Everard, A.; Lazarevic, V.; Derrien, M.; Girard, M.; Muccioli, G.G.; Neyrinck, A.M.; Possemiers, S.; Van Holle, A.; François, P.; de Vos, W.M.; et al. Responses of Gut Microbiota and Glucose and Lipid Metabolism to Prebiotics in Genetic Obese and Diet-Induced Leptin-Resistant Mice. Diabetes 2011, 60, 2775–2786. [Google Scholar] [CrossRef] [Green Version]
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R.; et al. Gut Microbiota from Twins Discordant for Obesity Modulate Metabolism in Mice. Science 2013, 341, 1241214. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Shen, D.; Fang, Z.; Jie, Z.; Qiu, X.; Zhang, C.; Chen, Y.; Ji, L. Human Gut Microbiota Changes Reveal the Progression of Glucose Intolerance. PLoS ONE 2013, 8, e71108. [Google Scholar] [CrossRef] [PubMed]
- Tanida, M.; Shen, J.; Maeda, K.; Horii, Y.; Yamano, T.; Fukushima, Y.; Nagai, K. High-fat diet-induced obesity is attenuated by probiotic strain Lactobacillus paracasei ST11 (NCC2461) in rats. Obes. Res. Clin. Pract. 2008, 2, 159–169. [Google Scholar] [CrossRef]
- Naito, E.; Yoshida, Y.; Makino, K.; Kounoshi, Y.; Kunihiro, S.; Takahashi, R.; Matsuzaki, T.; Miyazaki, K.; Ishikawa, F. Beneficial effect of oral administration of Lactobacillus casei strain Shirota on insulin resistance in diet-induced obesity mice. J. Appl. Microbiol. 2011, 110, 650–657. [Google Scholar] [CrossRef]
- Chiu, Y.-H.; Tsai, J.-J.; Lin, S.-L.; Lin, M.-Y. Lactobacillus casei MYL01 modulates the proinflammatory state induced by ethanol in an in vitro model. J. Dairy Sci. 2014, 97, 2009–2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mäkeläinen, H.; Saarinen, M.; Stowell, J.; Rautonen, N.; Ouwehand, A. Xylo-oligosaccharides and lactitol promote the growth of Bifidobacterium lactis and Lactobacillus species in pure cultures. Benef. Microbes 2010, 1, 139–148. [Google Scholar] [CrossRef]
- Elsen, L.V.D.; Tims, S.; Jones, A.; Stewart, A.; Stahl, B.; Garssen, J.; Knol, J.; Forbes-Blom, E.; Land, B.V. Prebiotic oligosaccharides in early life alter gut microbiome development in male mice while supporting influenza vaccination responses. Benef. Microbes 2019, 10, 279–291. [Google Scholar] [CrossRef]
- Paganini, D.; Uyoga, M.A.; Kortman, G.A.M.; Cercamondi, C.I.; Moretti, D.; Barth-Jaeggi, T.; Schwab, C.; Boekhorst, J.; Timmerman, H.M.; Lacroix, C.; et al. Prebiotic galacto-oligosaccharides mitigate the adverse effects of iron fortification on the gut microbiome: A randomised controlled study in Kenyan infants. Gut 2017, 66, 1956–1967. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Li, H.; Zhang, X.; Jiang, M.; Luo, C.; Lu, Z.; Xu, Z.-H.; Shi, J.-S. Prebiotic Mannan-Oligosaccharides Augment the Hypoglycemic Effects of Metformin in Correlation with Modulating Gut Microbiota. J. Agric. Food Chem. 2018, 66, 5821–5831. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Chen, J.; Miao, S.; Deng, K.; Liu, J.; Zeng, S.; Zheng, B.; Lu, X. Lotus seed oligosaccharides at various dosages with prebiotic activity regulate gut microbiota and relieve constipation in mice. Food Chem. Toxicol. 2019, 134, 110838. [Google Scholar] [CrossRef]
- Binda, C.; Lopetuso, L.R.; Rizzatti, G.; Gibiino, G.; Cennamo, V.; Gasbarrini, A. Actinobacteria: A relevant minority for the maintenance of gut homeostasis. Dig. Liver Dis. 2018, 50, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, E.D.; Zheng, H.; Joglekar, P.; Higginbottom, S.K.; Firbank, S.J.; Bolam, D.N.; Sonnenburg, J.L. Specificity of Polysaccharide Use in Intestinal Bacteroides Species Determines Diet-Induced Microbiota Alterations. Cell 2010, 141, 1241–1252. [Google Scholar] [CrossRef] [Green Version]
- Moens, F.; Weckx, S.; De Vuyst, L. Bifidobacterial inulin-type fructan degradation capacity determines cross-feeding interactions between bifidobacteria and Faecalibacterium prausnitzii. Int. J. Food Microbiol. 2016, 231, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Mao, B.; Gu, J.; Wu, J.; Cui, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W.W. Blautia—A new functional genus with potential probiotic properties? Gut Microbes 2021, 13, 1875796. [Google Scholar] [CrossRef] [PubMed]
- Zafar, H.; Saier, M.H. Gut Bacteroides species in health and disease. Gut Microbes 2021, 13, 1848158. [Google Scholar] [CrossRef] [PubMed]


| SPIDER Tool Search Description | ||
|---|---|---|
| S | Sample | Fecal or gut samples from humans or animals that could provide insights into the human gut microbiome. |
| P I | Phenomenon of Interest | This study aims to investigate changes in the microbiome composition following dietary interventions with different oligosaccharides and polysaccharides, which can be found naturally in food, added as food additives, or be functional carbohydrates found in nature. |
| D | Design | Inclusion Criteria:
|
| E | Evaluation | Gut or fecal microbiome composition, as well as some biomarkers and/or histological changes. |
| R | Research type | Quantitative. |
| Type | Variable Name | Number of Appearances | References |
|---|---|---|---|
| 1 | Carbohydrates as food additive | 12 | [22,23,24,25,26,96,105,107,109,110,116,121] |
| 1 | Carbohydrates as functional components (Functional carbohydrates) | 34 | [14,15,16,17,18,22,23,24,25,88,89,91,93,95,97,98,100,101,102,103,104,106,107,110,111,112,113,115,116,117,118,119,122,125] |
| 1 | Carbohydrates naturally occurring in food (Food components) | 31 | [14,17,18,23,24,25,89,91,93,94,96,98,99,100,101,102,103,104,105,107,108,111,113,114,116,118,119,120,121,124,126] |
| 2 | Fungal polysaccharides | 3 | [100,102,114] |
| 3 | Polysaccharides with antioxidant capacity | 10 | [15,17,88,89,91,95,101,102,114,127] |
| 3 | Sulfated polysaccharides | 3 | [18,88,97] |
| 3 | Fructans | 4 | [104,115,119,122] |
| 3 | Inulin | 4 | [92,115,119,122] |
| 3 | Oligosaccharides | 11 | [14,18,24,95,98,104,106,109,111,113,122] |
| 3 | Polysaccharides | 18 | [15,16,17,88,89,91,93,97,100,101,102,105,107,108,112,114,116,120] |
| 3 | Starch | 11 | [23,25,26,90,94,96,116,117,118,120,121] |
| 3 | Gelatinized starch | 3 | [23,90,94] |
| 3 | Resistant starch | 11 | [25,26,90,96,110,116,117,118,120,121,123] |
| 3 | Insoluble fiber | 11 | [89,90,92,93,97,99,103,108,110,111,124] |
| 3 | Soluble fiber | 16 | [22,24,89,90,92,93,97,98,103,108,109,110,112,119,122,124] |
| Microorganism Taxa | Decrease | Increase |
|---|---|---|
| Actinobacteria | 1 [101] | 3 [22,111,128] |
| Actinobacteria_Bifidobacterium | n/a | 13 [14,17,23,24,90,95,104,113,115,117,119,120,122] |
| Bacteroidetes | 2 [96,118] | 8 [15,24,94,99,100,103,107,110] |
| Bacteroidetes odoribacter | 2 [18,99] | n/a |
| Bacteroidetes_Bacteroidales_oscillospira | n/a | 2 [16,107] |
| Bacteroidetes_Oscillospira_Ruminococcus | n/a | 4 [15,109,120,123] |
| Bacteroidetes_Alistipes | n/a | 2 [15,99] |
| Bacteroidetes _bacteroides | n/a | 7 [90,92,97,109,111,112,113] |
| Bacteroidetes_prevotellaceae | n/a | 3 [18,91,123] |
| Bacteroidetes_Prevotellaceae_prevotella | n/a | 4 [16,23,90,98] |
| Firmicutes | 9 [24,94,99,100,107,108,110,111,124] | 4 [91,94,96,118] |
| Firmicutes_Enterococcus | 3 [24,97,104] | n/a |
| Firmicutes_lactobacillus | 3 [14,15,88] | 10 [17,18,24,91,95,104,105,111,112,113] |
| Firmicutes_Clostridia | n/a | 2 [16,96] |
| Firmicutes_Clostridium | 2 [111,113] | 2 [116,124] |
| Firmicutes_Lachnospiraceae_blautia | 3 [93,116,121] | n/a |
| Firmicutes_Ruminococcaceae_Faecalibacterium | n/a | 2 [120,122] |
| Firmicutes_Coprococcus | 2 [121,122] | n/a |
| Firmicutes_Lachnospira | 2 [115,121] | n/a |
| Firmicutes_Roseburia | n/a | 3 [15,97,120] |
| Firmicutes_Turicibacter | n/a | 2 [25,97] |
| Proteobacteria | 2 [94,114] | 2 [91,108] |
| Proteobacteria_Sutterella | n/a | 2 [23,25] |
| Proteobacteria_Desulfovibrio | n/a | 2 [18,109] |
| Tenericutes | 3 [25,101,122] | n/a |
| Verrucomicrobia_Akkermansia | n/a | 3 [25,91,103] |
| Verrucomicrobia _Akkermansia_muciniphila | n/a | 2 [17,123] |
| Predictive Variable | Response Variable BDV | Information Value (IV) | Number of Bins | References |
|---|---|---|---|---|
| Sulfated polysaccharides | Lactibacillus reduction | 1.808 | 1 | [88] |
| Gelatinized starch | Prevotella increase | 1.781 | 2 | [23,90] |
| Fructan | Faecalibacterium increase | 1.435 | 1 | [122] |
| Sulfated polysaccharides | Desulfovibrio increase | 1.398 | 1 | [18] |
| Fungal polysaccharides | Proteobacteria decrease | 1.398 | 1 | [114] |
| Gelatinized starch | Proteobacteria decrease | 1.398 | 1 | [94] |
| Sulfated polysaccharides | Turicibacter increase | 1.398 | 1 | [97] |
| Sulfated polysaccharides | Odoribacter increase | 1.397 | 1 | [18] |
| Fructan | Coprococcus increase | 1.144 | 1 | [122] |
| Inulin | Faecalibacterium increase | 1.144 | 1 | [122] |
| Oligosaccharides | Lactobacillus increase | 1.051 | 6 | [18,24,95,104,111,113] |
| Insoluble fiber | Actinobacteria increase | 0.95 | 2 | [111,129] |
| Starch | Blautia decrease | 0.948 | 2 | [24,116] |
| Resistant starch | Blautia decrease | 0.948 | 2 | [24,116] |
| Oligosaccharides | Enterococcus reduction | 0.947 | 2 | [24,104] |
| Resistant Starch | Firmicutes increase | 0.947 | 2 | [96,118] |
| Starch | Firmicutes increase | 0.947 | 2 | [96,118] |
| Carbohydrates used as food additives | Blautia decrease | 0.842 | 2 | [116,121] |
| Carbohydrates naturally occurring in food | Bacteroides increase | 0.828 | 2 | [111,113] |
| Insoluble fiber | Bacteroides increase | 0.828 | 4 | [90,92,97,111] |
| Insoluble fiber | Firmicutes reduction | 0.754 | 5 | [99,108,110,111,124] |
| Soluble fiber | Bacteroides increase | 0.729 | 5 | [90,92,97,109,112] |
| Sulfated polysaccharides | Roseburia increase | 0.677 | 1 | [97] |
| Sulfated polysaccharides | Enterococcus reduction | 0.676 | 1 | [97] |
| Inulin | Bifidobacterium increase | 0.538 | 3 | [115,119,122] |
| Fructans | Enterococcus reduction | 0.509 | 1 | [104] |
| Soluble fiber | Actinobacteria increase | 0.500 | 2 | [22,89] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mora-Flores, L.P.; Moreno-Terrazas Casildo, R.; Fuentes-Cabrera, J.; Pérez-Vicente, H.A.; de Anda-Jáuregui, G.; Neri-Torres, E.E. The Role of Carbohydrate Intake on the Gut Microbiome: A Weight of Evidence Systematic Review. Microorganisms 2023, 11, 1728. https://doi.org/10.3390/microorganisms11071728
Mora-Flores LP, Moreno-Terrazas Casildo R, Fuentes-Cabrera J, Pérez-Vicente HA, de Anda-Jáuregui G, Neri-Torres EE. The Role of Carbohydrate Intake on the Gut Microbiome: A Weight of Evidence Systematic Review. Microorganisms. 2023; 11(7):1728. https://doi.org/10.3390/microorganisms11071728
Chicago/Turabian StyleMora-Flores, Lorena P., Rubén Moreno-Terrazas Casildo, José Fuentes-Cabrera, Hugo Alexer Pérez-Vicente, Guillermo de Anda-Jáuregui, and Elier Ekberg Neri-Torres. 2023. "The Role of Carbohydrate Intake on the Gut Microbiome: A Weight of Evidence Systematic Review" Microorganisms 11, no. 7: 1728. https://doi.org/10.3390/microorganisms11071728






