Effect of Brassinolide on Soil Microorganisms in Millet Field Polluted by Tribenuron-Methyl
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Field Trial and Soil Sample Collection
2.3. TBM Residue Analysis
2.4. Detection of Soil Enzyme Activity
2.5. Total DNA Extraction and High-Throughput Sequencing
2.6. Statistical Analysis and Visualization
3. Results and Discussion
3.1. Residues of TBM in Soil
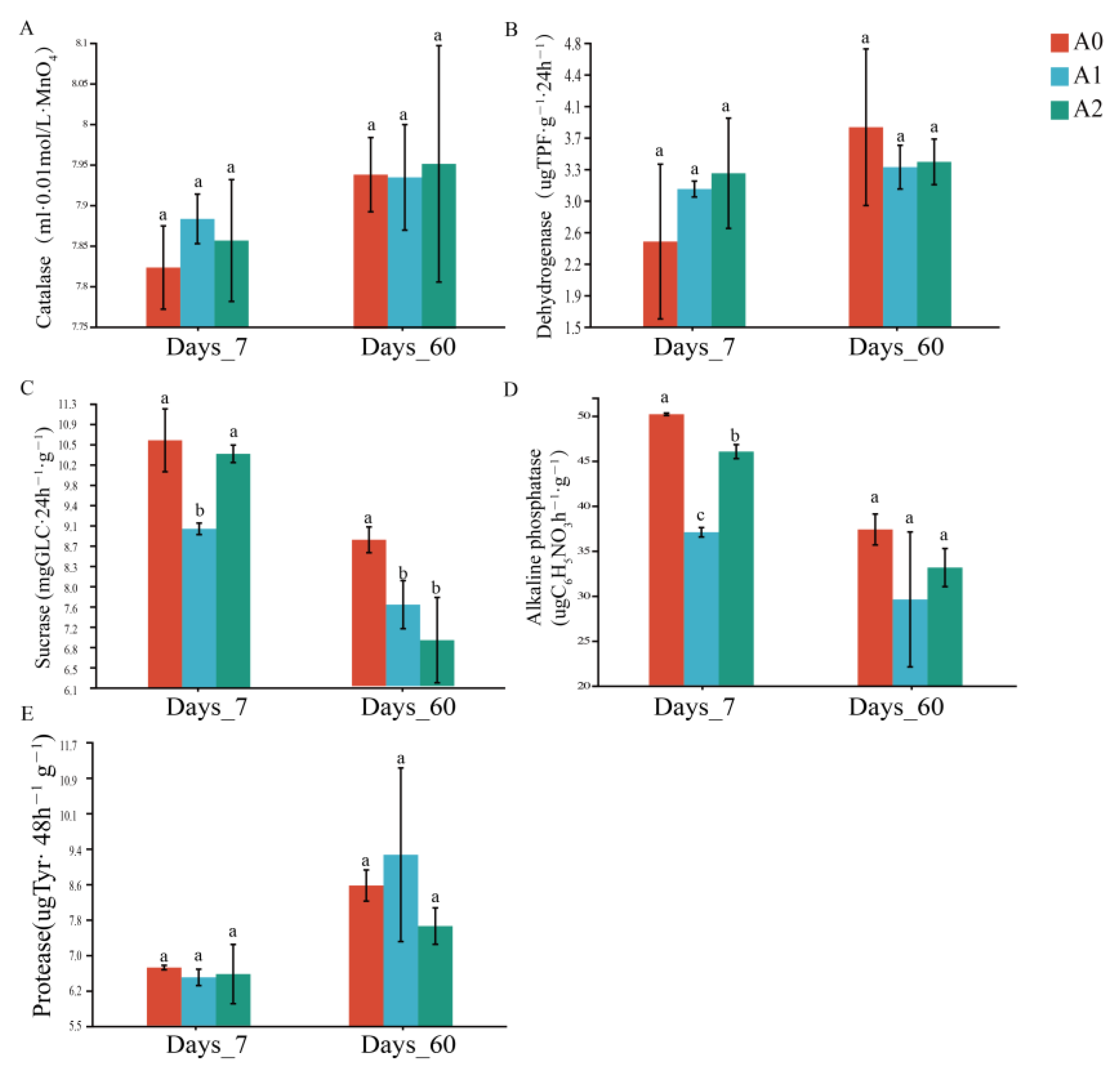
3.2. Effect of BR on Soil Enzyme Activities with TBM
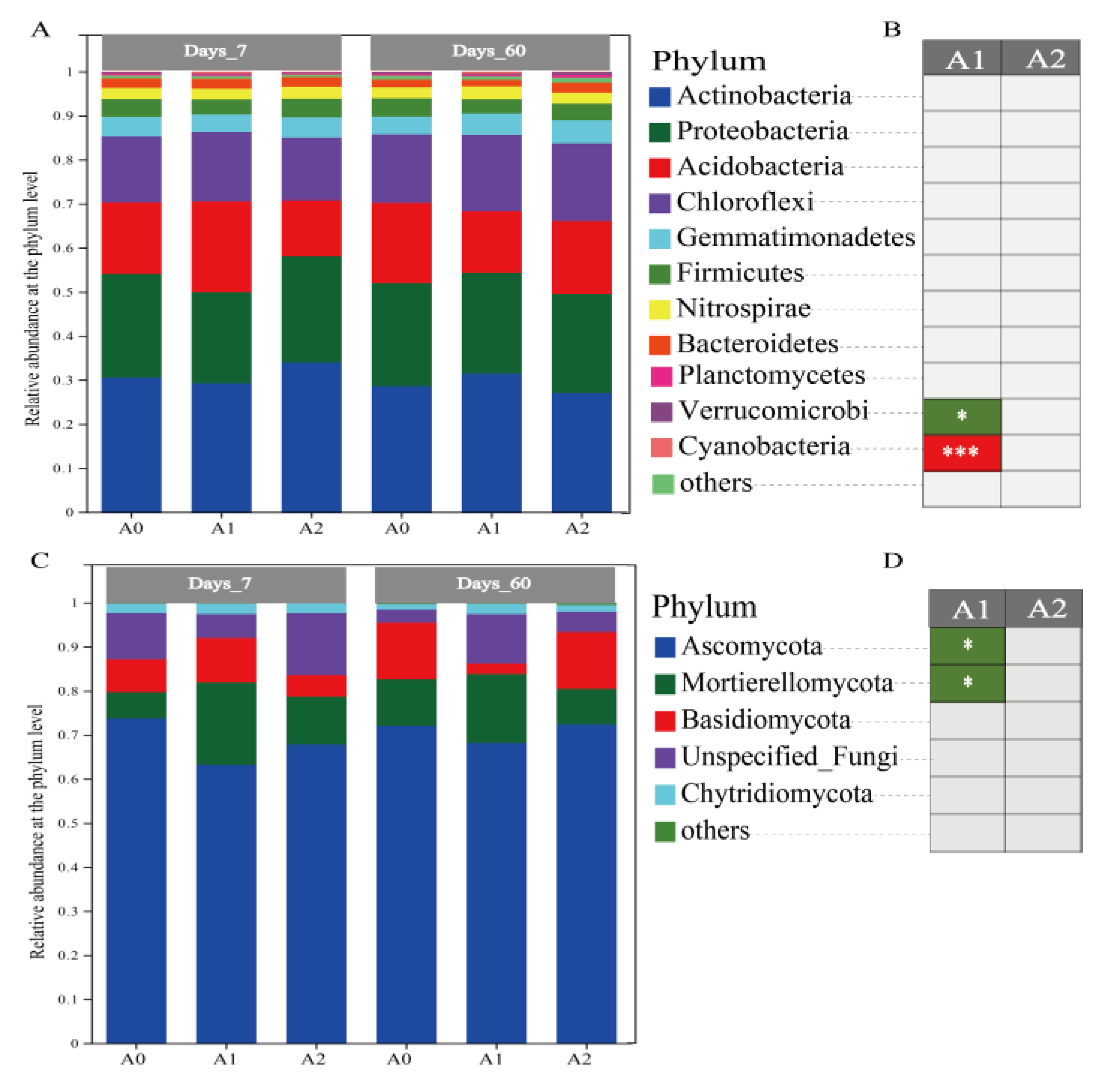
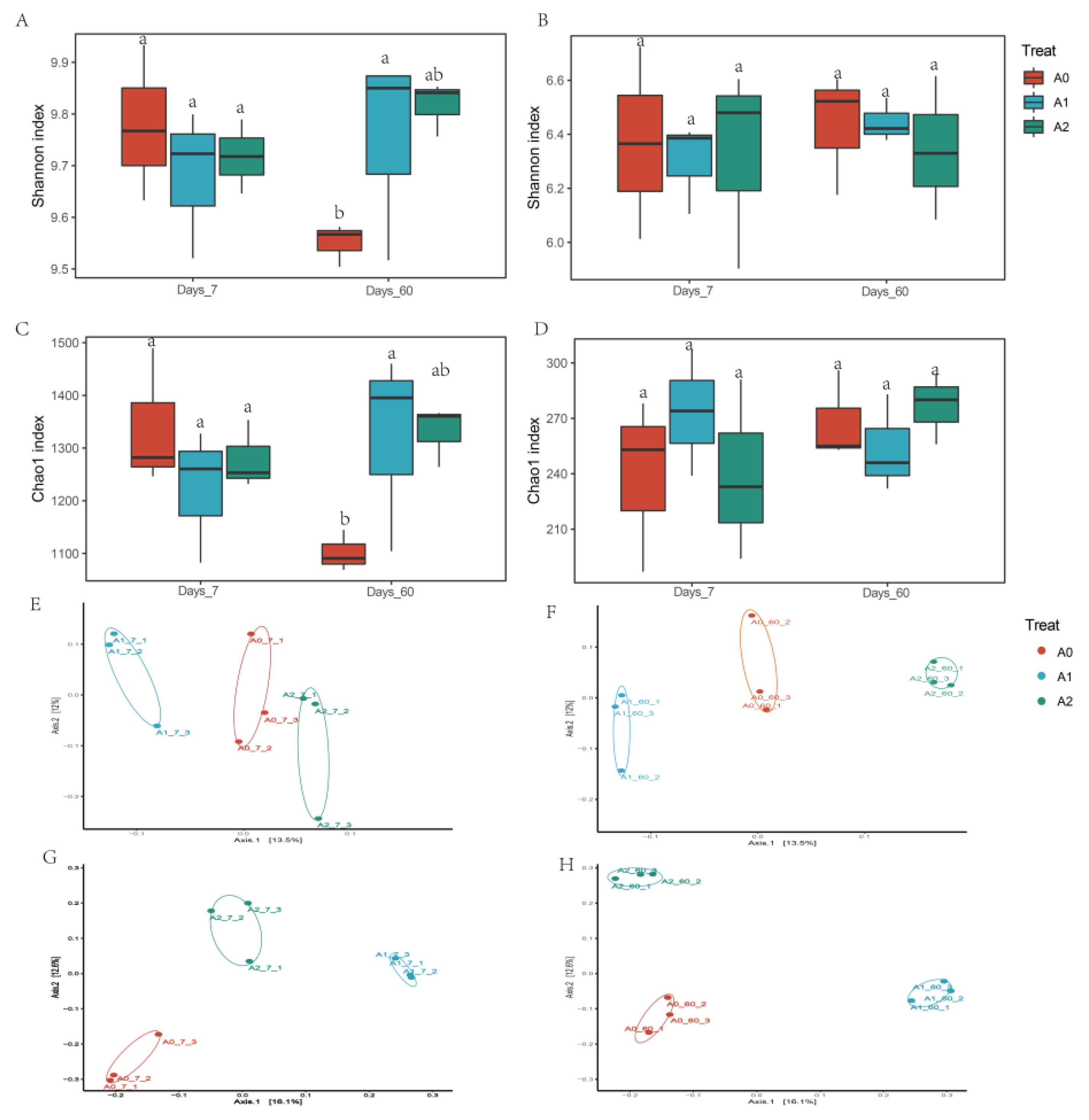
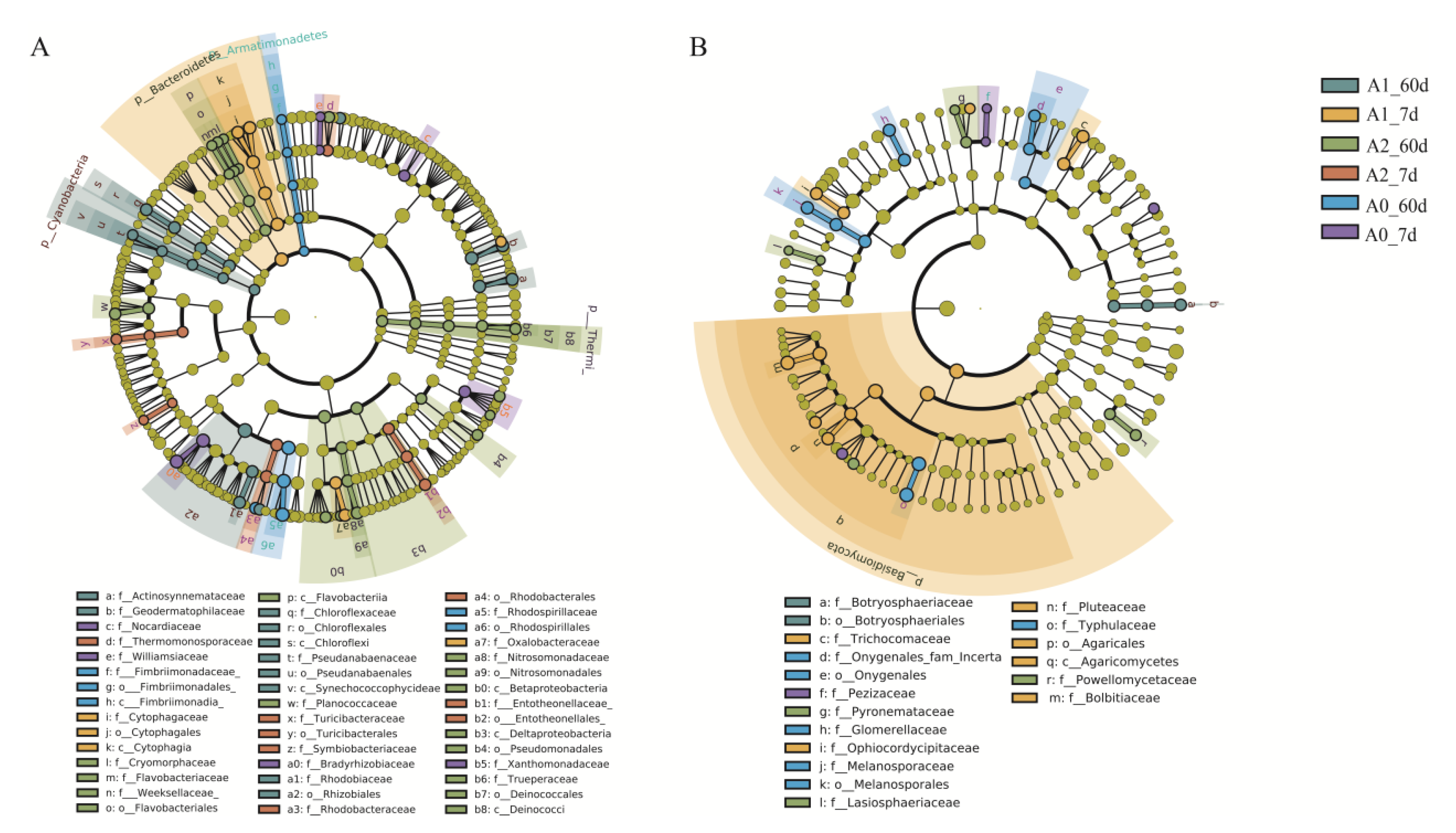
3.3. Effect of BR on Soil Microbiome Community with TBM
3.4. Effect of BR on Soil Function with TBM
3.5. Effect of BR on Millet Yield
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Saleh, A.S.M.; Zhang, Q.; Chen, J.; Shen, Q. Millet Grains: Nutritional Quality, Processing, and Potential Health Benefits. Compr. Rev. Food Sci. Food Saf. 2013, 12, 281–295. [Google Scholar] [CrossRef]
- Devi, P.B.; Vijayabharathi, R.; Sathyabama, S.; Malleshi, N.G.; Priyadarisini, V.B. Health benefits of finger millet (Eleusine coracana L.) polyphenols and dietary fiber: A review. J. Food Sci. Technol.-Mysore 2014, 51, 1021–1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ning, N.; Yuan, X.Y.; Dong, S.Q.; Wen, Y.Y.; Gao, Z.P.; Guo, M.J.; Guo, P.Y. Grain Yield and Quality of Foxtail Millet (Setaria italica L.) in Response to Tribenuron-Methyl. PLoS ONE 2015, 10, e0142557. [Google Scholar] [CrossRef]
- McCourt, J.A.; Pang, S.S.; Guddat, L.W.; Duggleby, R.G. Elucidating the specificity of binding of sulfonylurea herbicides to acetohydroxyacid synthase. Biochemistry 2005, 44, 2330–2338. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Di, Y.; Cai, J.; Chen, Y.; Yuan, S. Target-Site Resistance Mechanisms to Tribenuron-methyl and Cross-resistance Patterns to ALS-inhibiting Herbicides of Catchweed Bedstraw (Galium aparine) with Different ALS Mutations. Weed Sci. 2019, 67, 183–188. [Google Scholar] [CrossRef]
- Yang, Y.J.; Guo, M.J.; Pang, Y.F.; Shao, Q.L.; Wu, Y.Z.; Zhao, H.M.; Ji, A.Q.; Ma, J.H.; Song, X.E.; Sun, C.Q.; et al. Foxtail millet (setaria italica (L.) P. Beauvois) quality response to fertilizer level, herbicide and selenium. Appl. Ecol. Environ. Res. 2021, 19, 4231–4249. [Google Scholar] [CrossRef]
- Duman, F.; Urey, E.; Temizgul, R.; Bozok, F. Biological responses of a non-target aquatic plant (Nasturtium officinale) to the herbicide, tribenuron-methyl. Weed Biol. Manag. 2010, 10, 81–90. [Google Scholar] [CrossRef]
- Hu, M.; Zhang, H.; Kong, L.; Ma, J.; Wang, T.; Lu, X.; Guo, Y.; Zhang, J.; Guan, R.; Chu, P. Comparative proteomic and physiological analyses reveal tribenuron-methyl phytotoxicity and nontarget-site resistance mechanisms in Brassica napus. Plant Cell Environ. 2023, 46, 2255–2272. [Google Scholar] [CrossRef]
- Kayhan, F.E.; Duruel, H.E.E.; Kizilkaya, S.; Dine, S.K.; Kaymak, G.; Akbulut, C.; Ertug, N.D.Y. Toxic effects of herbicide tribenuron-methyl on liver tissue of zebrafish (Danio Rerio). Fresenius Environ. Bull. 2020, 29, 11175–11179. [Google Scholar]
- Kokojka, F.; Bacu, A.; Shahini, S.; Medha, G. Tribenuron-methyl treatment affects glutathione metabolization and other physiological processes in bread wheat. Int. J. Pest Manag. 2023, 69, 248–254. [Google Scholar] [CrossRef]
- Liu, W.; Bai, S.; Zhao, N.; Jia, S.; Li, W.; Zhang, L.; Wang, J. Non-target site-based resistance to tribenuron-methyl and essential involved genes in Myosoton aquaticum (L.). BMC Plant Biol. 2018, 18, 225. [Google Scholar] [CrossRef]
- Chang, Z.; Wang, Y.; Zhao, X.; Wang, Y.; Ma, K.; Yuan, X.; Dong, S. Effects of ‘tribesulfuron-methyl’ on photosynthetic characteristics of different foxtail millet varieties. J. China Agric. Univ. 2021, 26, 35–48. [Google Scholar] [CrossRef]
- Lian, J.-L.; Ren, L.-S.; Zhang, C.; Yu, C.-Y.; Huang, Z.; Xu, A.-X.; Dong, J.-G. How exposure to ALS-inhibiting gametocide tribenuron-methyl induces male sterility in rapeseed. BMC Plant Biol. 2019, 19, 124. [Google Scholar] [CrossRef] [PubMed]
- Mehdizadeh, M.; Alebrahim, M.T.; Roushani, M.; Streibig, J.C. Evaluation of four different crops’ sensitivity to sulfosulfuron and tribenuron methyl soil residues. Acta Agric. Scand. Sect. B-Soil Plant Sci. 2016, 66, 706–713. [Google Scholar] [CrossRef]
- Rachedi, K.; Zermane, F.; Tir, R.; Ayache, F.; Duran, R.; Lauga, B.; Karama, S.; Simon, M.; Boulahrouf, A. Effect of sulfonylurea tribenuron methyl herbicide on soil Actinobacteria growth and characterization of resistant strains. Braz. J. Microbiol. 2018, 49, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Clouse, S.D.; Sasse, J.M. BRASSINOSTEROIDS: Essential Regulators of Plant Growth and Development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 427–451. [Google Scholar] [CrossRef] [Green Version]
- Fujioka, S.; Yokota, T. Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 2003, 54, 137–164. [Google Scholar] [CrossRef]
- Bajguz, A.; Hayat, S. Effects of brassinosteroids on the plant responses to environmental stresses. Plant Physiol. Biochem. 2009, 47, 1–8. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukasinovic, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional Regulators of Plant Growth, Development, and Stress Responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef] [Green Version]
- Nazir, F.; Fariduddin, Q.; Hussain, A.; Khan, T.A. Brassinosteroid and hydrogen peroxide improve photosynthetic machinery, stomatal movement, root morphology and cell viability and reduce Cu-triggered oxidative burst in tomato. Ecotoxicol. Environ. Saf. 2021, 207, 111081. [Google Scholar] [CrossRef]
- Ramirez, V.E.; Poppenberger, B. Modes of Brassinosteroid Activity in Cold Stress Tolerance. Front. Plant Sci. 2020, 11, 583666. [Google Scholar] [CrossRef]
- Wu, Y.; Gao, H.; Zhang, B.; Zhang, H.; Wang, Q.; Liu, X.; Luan, X.; Ma, Y. Effects of 24-Brassinolide on the Fertility, Physiological Characteristics and Cell Ultra-Structure of Soybean Under Saline-Alkali Stress. Sci. Agric. Sin. 2017, 50, 811–821. [Google Scholar] [CrossRef]
- Zhou, Y.; Xia, X.; Yu, G.; Wang, J.; Wu, J.; Wang, M.; Yang, Y.; Shi, K.; Yu, Y.; Chen, Z.; et al. Brassinosteroids play a critical role in the regulation of pesticide metabolism in crop plants. Sci. Rep. 2015, 5, 9018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burns, R.G.; DeForest, J.L.; Marxsen, J.; Sinsabaugh, R.L.; Stromberger, M.E.; Wallenstein, M.D.; Weintraub, M.N.; Zoppini, A. Soil enzymes in a changing environment: Current knowledge and future directions. Soil Biol. Biochem. 2013, 58, 216–234. [Google Scholar] [CrossRef]
- Dong, B.Z.; Qian, W.; Hu, J.Y. Dissipation kinetics and residues of florasulam and tribenuron-methyl in wheat ecosystem. Chemosphere 2015, 120, 486–491. [Google Scholar] [CrossRef]
- Anastassiades, M.; Lehotay, S.J.; Stajnbaher, D.; Schenck, F.J. Fast and easy multiresidue method employing acetonitrile extraction/partitioning and “dispersive solid-phase extraction” for the determination of pesticide residues in produce. J. AOAC Int. 2003, 86, 412–431. [Google Scholar] [CrossRef]
- Gianfreda, L.; Antonietta Rao, M.; Piotrowska, A.; Palumbo, G.; Colombo, C. Soil enzyme activities as affected by anthropogenic alterations: Intensive agricultural practices and organic pollution. Sci. Total Environ. 2005, 341, 265–279. [Google Scholar] [CrossRef]
- Perucci, P.; Casucci, C.; Dumontet, S. An improved method to evaluate the o-diphenol oxidase activity of soil. Soil Biol. Biochem. 2000, 32, 1927–1933. [Google Scholar] [CrossRef]
- Garrido Frenich, A.; Romero-Gonzalez, R.; Luz Gomez-Perez, M.; Martinez Vidal, J.L. Multi-mycotoxin analysis in eggs using a QuEChERS-based extraction procedure and ultra-high-pressure liquid chromatography coupled to triple quadrupole mass spectrometry. J. Chromatogr. A 2011, 1218, 4349–4356. [Google Scholar] [CrossRef]
- Chen, G.; Cao, P.; Liu, R. A multi-residue method for fast determination of pesticides in tea by ultra performance liquid chromatography-electrospray tandem mass spectrometry combined with modified QuEChERS sample preparation procedure. Food Chem. 2011, 125, 1406–1411. [Google Scholar] [CrossRef]
- Cao, J.; Lv, Y.; Qi, Y.; Qin, S.; Wang, X.; Li, J. Dissipation, terminal residue and dietary risk assessment of flonicamid in cabbage. Int. J. Environ. Anal. Chem. 2022. [Google Scholar] [CrossRef]
- Zhao, J.; Tan, Z.; Wen, Y.; Fan, S.; Liu, C. Dissipation of fluazinam in citrus groves and a risk assessment for its dietary intake. J. Sci. Food Agric. 2020, 100, 2052–2056. [Google Scholar] [CrossRef]
- Neemisha; Sharma, S. Soil enzymes and their role in nutrient cycling. In Structure and Functions of Pedosphere; Springer: Berlin/Heidelberg, Germany, 2022; pp. 173–188. [Google Scholar]
- Mazorra, L.M.; Nunez, M.; Hechavarria, M.; Coll, F.; Sánchez-Blanco, M.J. Influence of brassinosteroids on antioxidant enzymes activity in tomato under different temperatures. Biol. Plant 2002, 45, 593–596. [Google Scholar] [CrossRef]
- Dick, R.P. Soil enzyme activities as indicators of soil quality. Defin. Soil Qual. A Sustain. Environ. 1994, 35, 107–124. [Google Scholar]
- Zhou, X.; Shu, R.; Huang, H.; Yang, S.; Long, Y.; Liu, W.; Yin, X. Effects of different remediate agents on tobacco plants caused phytotoxicity by quinclorac and its soil microorganisms and enzyme activities. J. South. Agric. 2018, 49, 2210–2217. [Google Scholar]
- Wang, G.; Li, X.; Xi, X.; Cong, W.-F. Crop diversification reinforces soil microbiome functions and soil health. Plant Soil 2022, 476, 375–383. [Google Scholar] [CrossRef]
- Bergmann, G.T.; Bates, S.T.; Eilers, K.G.; Lauber, C.L.; Caporaso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol. Biochem. 2011, 43, 1450–1455. [Google Scholar] [CrossRef] [Green Version]
- Fuerst, J.A. Phylum Verrucomicrobia. In Encyclopedia of Microbiology, 4th ed.; Schmidt, T.M., Ed.; Academic Press: Oxford, UK, 2019; pp. 551–563. [Google Scholar]
- Wang, X.; Bian, Q.; Jiang, Y.; Zhu, L.; Chen, Y.; Liang, Y.; Sun, B. Organic amendments drive shifts in microbial community structure and keystone taxa which increase C mineralization across aggregate size classes. Soil Biol. Biochem. 2021, 153, 108062. [Google Scholar] [CrossRef]
- Chang, F.; He, S.; Dang, C. Assisted selection of biomarkers by linear discriminant analysis effect size (LEfSe) in microbiome data. JoVE (J. Vis. Exp.) 2022, 183, e61715. [Google Scholar] [CrossRef]
- Li, F.; Chen, L.; Zhao, Z.-H.; Li, Y.; Yu, H.-Y.; Wang, Y.; Zhang, J.-B.; Han, Y.-L. The changes of chemical molecular components in soil organic matter are associated with fungus Mortierella capitata K. Soil Tillage Res. 2023, 227, 105598. [Google Scholar] [CrossRef]
- Yadav, S.K.; Soni, R.; Rajput, A.S. Role of microbes in organic farming for sustainable agro-ecosystem. In Microorganisms for Green Revolution: Volume 2: Microbes for Sustainable Agro-Ecosystem; Springer: Singapore, 2018; pp. 241–252. [Google Scholar]
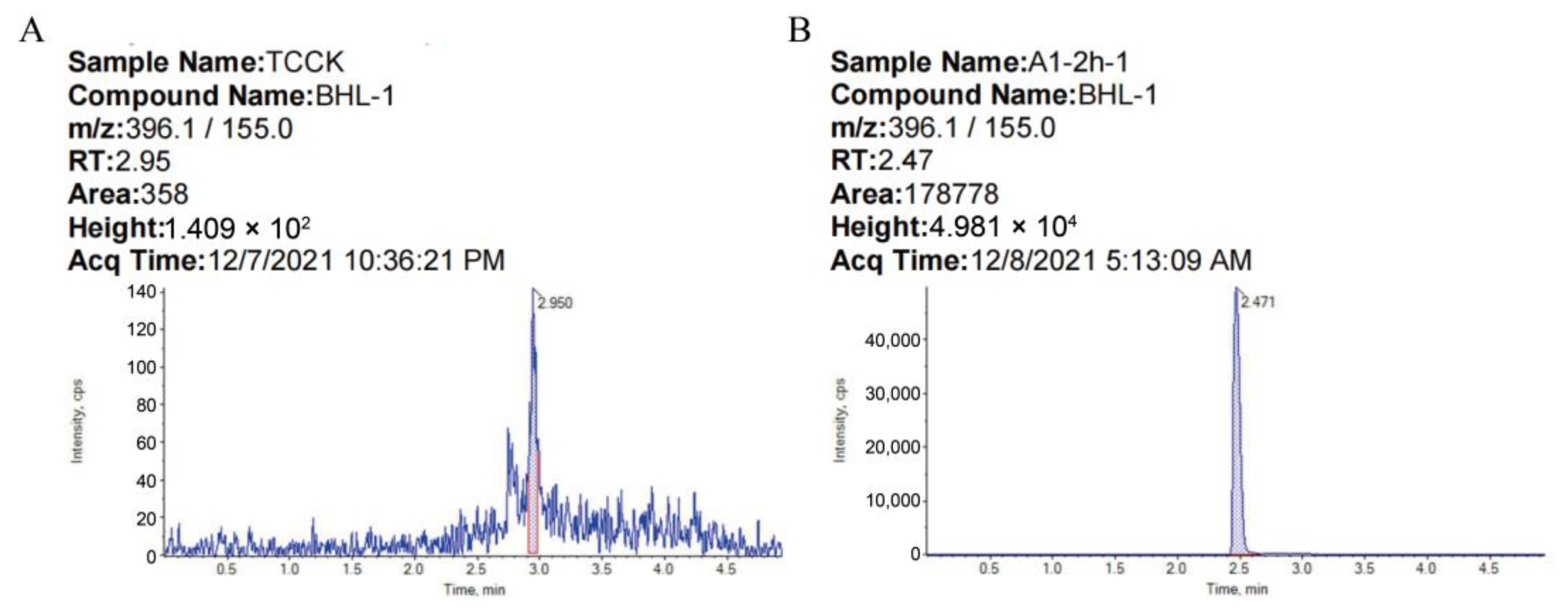

| Structural Formula | Retention Time (min) | Parent Ion (m/z) | Product Ion (m/z) | Collision Energy (eV) |
|---|---|---|---|---|
 | 2.5 | 396.1 | 155.0 | 21 |
| 181.0 | 27 |
| Treat | First-Order Kinetic Equation | R2 | Half-Life (Days) |
|---|---|---|---|
| A0 | Ct = 0.0125e−0.033x | 0.4490 | 21.0 |
| A1 | Ct = 0.0132e−0.039x | 0.5323 | 17.8 |
| A2 | Ct = 0.0121e−0.034x | 0.4499 | 20.4 |
| Treatments | Panicle Length (cm) | Panicle Width (mm) | Panicle Weigth (g) | Thousand-Grain Weight (g) | Yield (kg hm−2) |
|---|---|---|---|---|---|
| A0 | 25.06 ± 1.65 a | 27.64 ± 4.76 a | 24.2 ± 6.37 a | 3.17 ± 0.02 b | 4447 ± 27.3 a |
| A1 | 22.12 ± 2.75 b | 24.08 ± 4 a | 22.17 ± 6.26 ab | 3.17 ± 0.04 b | 4552 ± 28.0 a |
| A2 | 22.03 ± 2.39 b | 26.3 ± 4.28 a | 16.39 ± 4.88 b | 3.24 ± 0.02 a | 5054 ± 25.4 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, X.; Cao, J.; Guo, S.; Wang, H.; Dong, Q.; Guo, P.; Yuan, X. Effect of Brassinolide on Soil Microorganisms in Millet Field Polluted by Tribenuron-Methyl. Microorganisms 2023, 11, 1829. https://doi.org/10.3390/microorganisms11071829
Song X, Cao J, Guo S, Wang H, Dong Q, Guo P, Yuan X. Effect of Brassinolide on Soil Microorganisms in Millet Field Polluted by Tribenuron-Methyl. Microorganisms. 2023; 11(7):1829. https://doi.org/10.3390/microorganisms11071829
Chicago/Turabian StyleSong, Xi’e, Junli Cao, Shuai Guo, Hao Wang, Qianhui Dong, Pingyi Guo, and Xiangyang Yuan. 2023. "Effect of Brassinolide on Soil Microorganisms in Millet Field Polluted by Tribenuron-Methyl" Microorganisms 11, no. 7: 1829. https://doi.org/10.3390/microorganisms11071829





