Concealed for a Long Time on the Marches of Empires: Hepatitis B Virus Genotype I
Abstract
:1. Introduction
2. Virus Discovery
3. Genotype I as a True Genotype
4. Genome Architecture
5. Hypothetical Genesis
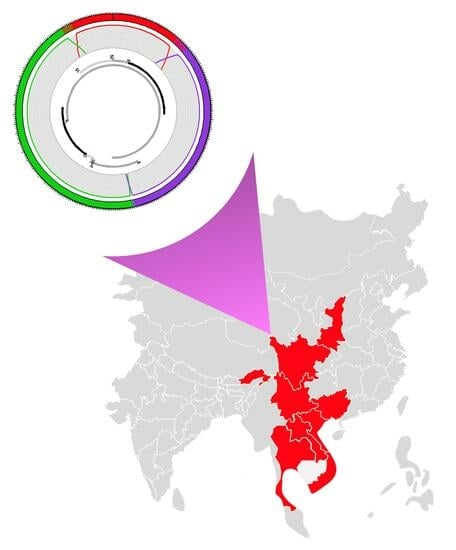
6. Current Geographic Distribution
7. Importance of HBV Genotype I Infections in Its Endemic Area
8. Clinical Consequences of Infections with Genotype I
9. Genetic Variations Observed on HBV Genotype I Genome
10. Infected Populations
11. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Velkov, S.; Ott, J.; Protzer, U.; Michler, T. The global hepatitis B virus genotype distribution approximated from available genotyping data. Genes 2018, 9, 495. [Google Scholar] [CrossRef] [PubMed]
- McNaughton, A.L.; D’Arienzo, V.; Ansari, M.A.; Lumley, S.F.; Littlejohn, M.; Revill, P.; McKeating, J.A.; Matthews, P.C. Insights From Deep Sequencing of the HBV Genome-Unique, Tiny, and Misunderstood. Gastroenterology 2019, 156, 384–399. [Google Scholar] [CrossRef] [PubMed]
- Kramvis, A. Genotypes and genetic variability of hepatitis B virus. Intervirology 2014, 57, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.T.; Trinh, T.N.; Abe, K. New complex recombinant genotype of hepatitis B virus identified in Vietnam. J. Virol. 2008, 82, 5657–5663. [Google Scholar] [CrossRef] [PubMed]
- Olinger, C.; Jutavijittum, P.; Hübschen, J.; Yousukh, A.; Samountry, B.; Thammavaong, T.; Toriyama, K.; Müller, C. Possible new hepatitis B virus genotype, southeast Asia. Emerg. Infect. Dis. 2008, 14, 1777–1780. [Google Scholar] [CrossRef]
- Kurbanov, F.; Tanaka, Y.; Kramvis, A.; Simmonds, P.; Mizokami, M. When should “I” consider a new hepatitis B virus genotype? J. Virol. 2008, 82, 8241–8242. [Google Scholar] [CrossRef]
- Collaborators, G.B.D.H.B. Global, regional, and national burden of hepatitis B, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet Gastroenterol. Hepatol. 2022, 7, 796–829. [Google Scholar] [CrossRef]
- Arankalle, V.; Gandhe, S.; Borkakoty, B.; Walimbe, A.; Biswas, D.; Mahanta, J. A novel HBV recombinant (genotype I) similar to Vietnam/Laos in a primitive tribe in eastern India. J. Viral. Hepat. 2010, 17, 501–510. [Google Scholar] [CrossRef]
- Fang, Z.L.; Hue, S.; Sabin, C.A.; Li, G.J.; Yang, J.Y.; Chen, Q.Y.; Fang, K.X.; Huang, J.; Wang, X.Y.; Harrison, T.J. A complex hepatitis B virus (X/C) recombinant is common in Long An county, Guangxi and may have originated in southern China. J. Gen. Virol. 2011, 92, 402–411. [Google Scholar] [CrossRef]
- Hannoun, C.; Norder, H.; Lindh, M. An aberrant genotype revealed in recombinant hepatitis B virus strains from Vietnam. J. Gen. Virol. 2000, 81, 2267–2272. [Google Scholar] [CrossRef]
- Stuyver, L.; De Gendt, S.; Van Geyt, C.; Zoulim, F.; Fried, M.; Schinazi, R.; Rossau, R. A new genotype of hepatitis B virus: Complete genome and phylogenetic relatedness. J. Gen. Virol. 2000, 81, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Midgley, S. Recombination in the genesis and evolution of hepatitis B virus genotypes. J. Virol. 2005, 79, 15467–15476. [Google Scholar] [CrossRef]
- Borkakoty, B.J.; Mahanta, J.; Biswas, D. Circulating genotypes of hepatitis B virus in Arunachal Pradesh. Indian J. Med. Res. 2008, 127, 65–70. [Google Scholar]
- Marjenberg, Z.; Wright, C.; Pooley, N.; Cheung, K.W.; Shimakawa, Y.; Vargas-Zambrano, J.C.; Vidor, E. Hepatitis B surface antigen prevalence and the rates of mother-to-child transmission of hepatitis B virus after the introduction of infant vaccination programs in South East Asia and Western Pacific regions: A systematic review. Int. J. Infect. Dis. 2022, 124, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Shi, J.; Hui, L.; Xi, H.; Zhuoma; Quni; Tsedan; Hu, G. The dominant hepatitis B virus genotype identified in Tibet is a C/D hybrid. J. Gen. Virol. 2002, 83, 2773–2777. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Lole, K.; Bollinger, R.; Paranjape, R.; Gadkari, D.; Kulkarni, S.; Novak, N.; Ingersoll, R.; Sheppard, H.; Ray, S. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999, 73, 152–160. [Google Scholar] [CrossRef]
- Schultz, A.; Bulla, I.; Abdou-Chekaraou, M.; Gordien, E.; Morgenstern, B.; Zoulim, F.; Dény, P.; Stanke, M. jpHMM: Recombination analysis in viruses with circular genomes such as the hepatitis B virus. Nucl. Acids Res. 2012, 40, W198–W493. [Google Scholar] [CrossRef]
- Phan, N.M.H.; Faddy, H.; Flower, R.; Spann, K.; Roulis, E. In silico Analysis of Genetic Diversity of Human Hepatitis B Virus in Southeast Asia, Australia and New Zealand. Viruses 2020, 12, 427. [Google Scholar] [CrossRef]
- Li, H.M.; Wang, J.Q.; Wang, R.; Zhao, Q.; Li, L.; Zhang, J.P.; Shen, T. Hepatitis B virus genotypes and genome characteristics in China. World J. Gastroenterol. 2015, 21, 6684–6697. [Google Scholar] [CrossRef] [PubMed]
- Andernach, I.; Jutavijittum, P.; Samountry, B.; Yousukh, A.; Thammavong, T.; Hübschen, J.; Muller, C. A high variability of mixed infections and recent recombinations of hepatitis B virus in Laos. PLoS ONE 2012, 7, e30245. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.; Tran, T.; Nghiem, M.; Rahman, P.; Tran, T.; Dinh, M.; Le, M.; Nguyen, V.; Thwaites, G.; Rahman, M. Molecular characterization of hepatitis B virus in Vietnam. BMC Infect. Dis. 2017, 17, 601. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Yan, X.; Liu, X.; Zhang, B.; Li, L.; Zhang, J.; Wang, J.; Xiao, C. Genotype I of hepatitis B virus was found in east Xishuangbanna, China and molecular dynamics of HBV/I. J. Viral. Hepat. 2015, 22, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Araujo, N.M. Hepatitis B virus intergenotypic recombinants worldwide: An overview. Infect. Genet. Evol. 2015, 36, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Jose-Abrego, A.; Roman, S.; Rebello Pinho, J.R.; de Castro, V.F.D.; Panduro, A. Hepatitis B Virus (HBV) Genotype Mixtures, Viral Load, and Liver Damage in HBV Patients Co-infected With Human Immunodeficiency Virus. Front. Microbiol. 2021, 12, 640889. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Gao, Y.; Sun, X.; Zhu, X.; Li, M. High prevalence of the B2+ C2 subgenotype mixture in patients with chronic hepatitis B in Eastern China. Acta Pharm. Sin. 2012, 33, 1271–1276. [Google Scholar] [CrossRef]
- Kato, H.; Orito, E.; Gish, R.G.; Bzowej, N.; Newsom, M.; Sugauchi, F.; Suzuki, S.; Ueda, R.; Miyakawa, Y.; Mizokami, M. Hepatitis B e antigen in sera from individuals infected with hepatitis B virus of genotype G. Hepatology 2002, 35, 922–929. [Google Scholar] [CrossRef]
- Araujo, N.; Osiowy, A. Hepatitis B Virus Genotype G: The Odd Cousin of the Family. Front. Microb. 2022, 13, 1208. [Google Scholar] [CrossRef]
- Osiowy, C.; Gordon, D.; Borlang, J.; Giles, E.; Villeneuve, J.P. Hepatitis B virus genotype G epidemiology and co-infection with genotype A in Canada. J. Gen. Virol. 2008, 89, 3009–3015. [Google Scholar] [CrossRef]
- Sakamoto, T.; Tanaka, Y.; Simonetti, J.; Osiowy, C.; Borresen, M.L.; Koch, A.; Kurbanov, F.; Sugiyama, M.; Minuk, G.Y.; McMahon, B.J.; et al. Classification of hepatitis B virus genotype B into 2 major types based on characterization of a novel subgenotype in Arctic indigenous populations. J. Infect. Dis. 2007, 196, 1487–1492. [Google Scholar] [CrossRef] [PubMed]
- Revill, P.; Tu, T.; Netter, H.; Yuen, L.; Locarnini, S.; Littlejohn, M. The evolution and clinical impact of hepatitis B virus genome diversity. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 618–634. [Google Scholar] [CrossRef] [PubMed]
- Locarnini, S.; Littlejohn, M.; Yuen, L. Origins and Evolution of the Primate Hepatitis B Virus. Front. Microb. 2021, 12, 653684. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Ran, J.; Feng, Y.M.; Miao, J.; Zhao, Y.; Jia, Y.; Li, Z.; Yue, W.; Xia, X. Genetic diversity of hepatitis B virus in Yunnan, China: Identification of novel subgenotype C17, an intergenotypic B/I recombinant, and B/C recombinants. J. Gen. Virol. 2020, 101, 972–981. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.H.; Chen, Q.Y.; Jiang, Z.H.; Wang, X.Y.; Zhang, W.J.; He, X.; Harrison, T.J.; Jackson, J.B.; Wu, L.; Fang, Z.L. A novel subgenotype I3 of hepatitis B virus in Guangxi, China: A 15-year follow-up study. Virus Genes 2023, 59, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Yuan, Q.; Ge, S.X.; Wang, H.Y.; Zhang, Y.L.; Chen, Q.R.; Zhang, J.; Chen, P.J.; Xia, N.S. Molecular and phylogenetic analyses suggest an additional hepatitis B virus genotype “I”. PLoS ONE 2010, 5, e9297. [Google Scholar] [CrossRef] [PubMed]
- Holzmayer, V.; Hance, R.; Defechereux, P.; Grant, R.; Kuhns, M.; Cloherty, G.; Rodgers, M. Identification of hepatitis B virus genotype I in Thailand. J. Med. Virol. 2019, 91, 717–721. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.C.; Li, X.; Li, L.; Li, X.F.; Zhang, L.; Wei, J. Evolution of full-length genomes of HBV quasispecies in sera of patients with a coexistence of HBsAg and anti-HBs antibodies. Sci. Rep. 2017, 7, 661. [Google Scholar] [CrossRef] [PubMed]
- Sitbounlang, P.; Marchio, A.; Bertani, S.; Sychaleun, S.; Dejean, A.; Paboriboune, P.; Deharo, E.; Pineau, P. in preparation. 2023. [Google Scholar]
- Osiowy, C.; Kaita, K.; Solar, K.; Mendoza, K. Molecular characterization of hepatitis B virus and a 9-year clinical profile in a patient infected with genotype I. J. Med. Virol. 2010, 82, 942–948. [Google Scholar] [CrossRef]
- Colson, P.; Roquelaure, B.; Tamalet, C. Detection of a newly identified hepatitis B virus genotype in southeastern France. J. Clin. Virol. 2009, 45, 165–167. [Google Scholar] [CrossRef]
- Kyaw, Y.Y.; Lwin, A.A.; Aye, K.S.; Thu, H.M.; Htun, M.M.; Soe, H.O.; Aye, K.T.; Thant, K.Z.; Hwang, H.J.; Cheong, J. Distribution of hepatitis B virus genotypes in the general population of Myanmar via nationwide study. BMC Infect. Dis. 2020, 20, 552. [Google Scholar] [CrossRef] [PubMed]
- Puri, P.; Srivastava, S. Lower chronic hepatitis B in South Asia despite all odds: Bucking the trend of other infectious diseases. Trop. Gastroenterol. 2012, 33, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Lesmana, L.; Leung, N.; Mahachai, V.; Phiet, P.; Suh, D.; Yao, G.; Zhuang, H. Hepatitis B: Overview of the burden of disease in the Asia-Pacific region. Liver Int. 2006, 26, 3–10. [Google Scholar] [CrossRef]
- Ismail, A.M.; Puhazhenthi, K.S.; Sivakumar, J.; Eapen, C.E.; Kannangai, R.; Abraham, P. Molecular epidemiology and genetic characterization of hepatitis B virus in the Indian subcontinent. Int. J. Infect. Dis. 2014, 20, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Li, J.R.; Zhao, Z.X.; Xu, L.; Yu, W.; Kang, W.Y.; Li, Q.F. Molecular epidemiology of hepatitis B virus genotypes and subgenotypes in ethnic minority populations, Yunnan province, China. Epidemiol. Infect. 2021, 150, e11. [Google Scholar] [CrossRef]
- Fan, J.; Li, X.; Tang, Y.; Lai, X.; Li, X.; Zhang, L.; Wei, J. Molecular Characterization of Coexistence of HBsAg and Anti-HBs in a Patient Infected with HBV Genotype I. Hepat. Mon. 2019, 19, e81740. [Google Scholar] [CrossRef]
- Li, G.J.; Hue, S.; Harrison, T.J.; Yang, J.Y.; Chen, Q.Y.; Wang, X.Y.; Fang, Z.L. Hepatitis B virus candidate subgenotype I1 varies in distribution throughout Guangxi, China and may have originated in Long An county, Guangxi. J. Med. Virol. 2013, 85, 799–807. [Google Scholar] [CrossRef]
- Liu, C.; Calin, G.; Meloon, B.; Gamliel, N.; Sevignani, C.; Ferracin, M.; Dumitru, C.; Shimizu, M.; Zupo, S.; Dono, M.; et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. USA 2004, 101, 9740–9744. [Google Scholar] [CrossRef]
- Fu, Y.; Wu, S.; Hu, Y.; Chen, T.; Zeng, Y.; Liu, C.; Ou, Q. Mutational characterization of HBV reverse transcriptase gene and the genotype-phenotype correlation of antiviral resistance among Chinese chronic hepatitis B patients. Emerg. Microb. Infect. 2020, 9, 2381–2393. [Google Scholar] [CrossRef]
- Levrero, M.; Zucman-Rossi, J. Mechanisms of HBV-induced hepatocellular carcinoma. J. Hepatol. 2016, 64, S84–S101. [Google Scholar] [CrossRef]
- Sung, F.Y.; Lan, C.Y.; Huang, C.J.; Lin, C.L.; Liu, C.J.; Chen, P.J.; Lin, S.M.; Yu, M.W. Progressive accumulation of mutations in the hepatitis B virus genome and its impact on time to diagnosis of hepatocellular carcinoma. Hepatology 2016, 64, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Lee, S.; Kim, B. Naturally occurring hepatitis B virus mutations leading to endoplasmic reticulum stress and their contribution to the progression of hepatocellular carcinoma. Int. J. Mol. Sci. 2019, 20, 597. [Google Scholar] [CrossRef]
- Tong, W.; He, J.; Sun, L.; He, S.; Qi, Q. Hepatitis B virus with a proposed genotype I was found in Sichuan Province, China. J. Med. Virol. 2012, 84, 866–870. [Google Scholar] [CrossRef] [PubMed]
- Araujo, N.M.; Teles, S.A.; Spitz, N. Comprehensive Analysis of Clinically Significant Hepatitis B Virus Mutations in Relation to Genotype, Subgenotype and Geographic Region. Front. Microbiol. 2020, 11, 616023. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.; Scornavacca, C. Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. System. Biol. 2012, 61, 1061–1067. [Google Scholar] [CrossRef]
- Puri, P. Tackling the Hepatitis B Disease Burden in India. J. Clin. Exp. Hepatol. 2014, 4, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Hartmann, J.; Liu, J.; Huang, P.S.C.G. Geographic patterns of Zhuang (Tai) kinship terms in Guangxi and border areas: A GIS analysis of language and culture change. Soc. Cult. Geo. 2007, 8, 575–596. [Google Scholar] [CrossRef]
- Su, H.; Liu, Y.; Xu, Z.; Cheng, S.; Ye, H.; Xu, Q.; Liu, Q.; Tan, S.; Xu, D.; Liu, Y. A novel complex A/C/G intergenotypic recombinant of hepatitis B virus isolated in southern China. PLoS ONE 2014, 9, e84005. [Google Scholar] [CrossRef]
- McElwee, P. The Politics of Indigenous Environmental Knowledge in Vietnam. Hum. Ecol. 2022, 50, 214–258. [Google Scholar] [CrossRef]
- Pholsena, V. L’Evolution du paysage ethnique. In Laos, un Pays en Mutation; Schlemmer, G., Ed.; Belin: Paris, France, 2011; pp. 2010–2089. [Google Scholar]
- Chuon, C.; Takahashi, K.; Matsuo, J.; katayama, K.; Yamamoto, C.; Hok, S.; Nagashima, S.; Akbar, S.; Tanaka, J. High possibility of hepatocarcinogenesis in HBV genotype C1 infected Cambodians is indicated by 340 HBV C1 full-genomes analysis from GenBank. Sci. Rep. 2019, 9, 12186. [Google Scholar] [CrossRef]
- Michaud, J. Seeing the Forest for the Trees: Scale, Magnitude, and Range in the Southeast Massif. In Historical dictionary of the peoples of the Southeast Asian massif; Michaud, J., Swain, M., Barkataki-Ruscheweyh, M., Eds.; Rowman & Littlefield: Lanham, MA, USA, 2016; pp. 1–40. [Google Scholar]
- van Schendel, W. Geographies of Knowing, Geographies of Ignorance: Jumping Scale in Southeast Asia. Soc. Space 2002, 20, 647–668. [Google Scholar] [CrossRef]
- Scott, J. The Art of Not Being Governed: An Anarchist History of Upland Southeast Asia; NUS Press: Singapore, 2010; p. 464. [Google Scholar]
- Shi, S. Ethnic flows in the Tibetan-Yi corridor throughout history. Int. J. Anthropol. Ethnol. 2018, 2, 2–22. [Google Scholar] [CrossRef]
- Ning, C.; Wang, C.C.; Gao, S.; Yang, Y.; Zhang, X.; Wu, X.; Zhang, F.; Nie, Z.; Tang, Y.; Robbeets, M.; et al. Ancient Genomes Reveal Yamnaya-Related Ancestry and a Potential Source of Indo-European Speakers in Iron Age Tianshan. Curr. Biol. 2019, 29, 2526–2532. [Google Scholar] [CrossRef] [PubMed]
- McColl, H.; Racimo, F.; Vinner, L.; Demeter, F.; Gakuhari, T.; Moreno-Mayar, J.V.; van Driem, G.; Gram Wilken, U.; Seguin-Orlando, A.; de la Fuente Castro, C.; et al. The prehistoric peopling of Southeast Asia. Science 2018, 361, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.A.; Fan, X.; Sun, B.; Chen, C.; Lang, J.; Ko, Y.C.; Tsang, C.H.; Chiu, H.; Wang, T.; Bao, Q.; et al. Ancient DNA indicates human population shifts and admixture in northern and southern China. Science 2020, 369, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Wall, J.; Yang, M.; Jay, F.; Kim, S.; Durand, E.; Stevison, L.; Gignoux, C.; Woerner, A.; Hammer, M.; Slatkin, M. Higher levels of Neanderthal ancestry in East Asians than in Europeans. Genetics 2013, 194, 199–209. [Google Scholar] [CrossRef]
- Massilani, D.; Skov, L.; Hajdinjak, M.; Gunchinsuren, B.; Tseveendorj, D.; Yi, S.; Lee, J.; Nagel, S.; Nickel, B.; Deviese, T.; et al. Denisovan ancestry and population history of early East Asians. Science 2020, 370, 579–583. [Google Scholar] [CrossRef]






| Genes | Position on HBV Genome | Genotype A | Genotype C | Genotype G |
|---|---|---|---|---|
| Polymerase | 2307-1623 | 25% | 25% | 43% |
| PreS1 | 2848-3204 | 56% | 28% | |
| PreS2 | 3205-154 | 100% | ||
| HBs | 155-835 | 35% | 63% | |
| PreC-C (HBe-HBc) | 1814-2454 | 100% | ||
| HBx | 1374-1838 | 70% | 27% |
| Gene or Regulatory Element | Nucleotide or Amino-Acid | Viet Nam (6) I1 | Viet Nam (2) I2 | Laos (4) I1 | Laos (12) I2 | China (88) I1 | India (8) I2 | Thailand (1) I1 |
|---|---|---|---|---|---|---|---|---|
| Pol | rtA181T/V | 0 | 0 | 0 | 0 | 2 (T) | 0 | 0 |
| Pol | rtM204I/V/L | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Pol | rtL229V/M/F | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pol | rtN236T/I | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| pre-S2 | 3174-76, M1V/T/I | 0 | 0 | 0 | 0 | 1 (I)/1 (R) | 1 (T) | 0 |
| pre-S2 | T54C/TT53-54CC/T53C, F22S/P/L | 0 | 0 | 0 | 0 | 1 (S)/4 (P)/9 (L) | 1 | 0 |
| HBs | C312T, S53L | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| HBs | G542A, G130R | 0 | 0 | 0 | 0 | 14 (S) | 0 | 0 |
| HBs | G553A, M133T | 0 | 0 | 0 | 0 | 4 (I) | 0 | 0 |
| HBs | G587A, G145R | 1 | 0 | 0 | 1 | 0 | 3 | 0 |
| HBs | G700A, W182Stop | 0 | 0 | 0 | 0 | 6 | 0 | 0 |
| HBs | T704G, W184A | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| HBs | 763-765, P203Q | 0 | 0 | 0 | 1 (Q) | 1(S) | 0 | 0 |
| HBs | T766A, S204R | 0 | 0 | 0 | 1 (N) | 2 (G) | 7 (N)/1 (R) | 0 |
| HBs | T784G, S210R | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| HBs | T801A, L216Stop | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| HBs | T813G/T814G, F220L/C | 1 (L) | 0 | 0 | 0 | 40 (C) | 0 | 0 |
| enhI/II, HBx, Pol. | G1613A, E80E, R841K | 0 | 1 | 5 | 0 | 48 | 0 | |
| enhI/II, HBx | C1653T, H94Y | 0 | 0 | 0 | 0 | 36 | 0 | 0 |
| enhI/II, HBx | T1753C, I127Y | 0 | 0 | 1 | 0 | 48 | 0 | 1 (G) |
| Basal Core Promoter, HBx | A1762T, K130M | 2 | 0 | 1 | 1 | 56 | 0 | 1 |
| Basal Core Promoter, HBx | G1764A, V131I | 2 | 0 | 1 | 1 | 56 | 1 | 1 |
| Basal Core Promoter, HBx | A1762T/G1764A | 2 | 0 | 1 | 1 | 54 | 0 | 1 |
| Basal Core Promoter, HBx | C1766T, F132Y | 0 | 0 | 0 | 0 | 1 | 3 | 0 |
| HBx | T1674C, S101P | 0 | 0 | 0 | 0 | 2 | 1 | 0 |
| HBx | A1727T, A118N | 0 | 0 | 0 | 0 | 1 (C) | 0 | 0 |
| HBx | 1767-1769, F132Y/I/R | 0 | 0 | 0 | 0 | 1(Y)/1 (I) | 2 (Y) | 0 |
| HBx | C1773T, L134L | 0 | 0 | 0 | 0 | 1/1 (A) | 7 | 0 |
| HBe | A1846T, S11S | 0 | 1 | 0 | 0 | 35 | 1 | 0 |
| HBe | G1896A, W28Stop | 0 | 0 | 0 | 3 | 32 | 3 | 1 |
| HBe | G1899A, G29D | 0 | 0 | 0 | 1 | 12 | 1 | 0 |
| HBc | T1961G, S21A | 0 | 0 | 0 | 1 | 2(T) | 0 | 0 |
| HBc | A2131T, E77D | 0 | 0 | 0 | 1 (D) | 3 (D) | 1 (Q) | 0 |
| HBc | 2139-40, A80T/I/G | 0 | 0 | 0 | 1(S) | 1 (T) | 0 | 0 |
| HBc | 2159-61 S87R | 0 | 0 | 0 | N (1)/G (1) | R (41) | N (1) | 1 (G) |
| HBc | A2189T, I97F | 0 | 0 | 0 | 1 | 1 | 1 | 0 |
| HBc | A2439C, E180A | 0 | 0 | 0 | 1 (G) | 1 (K) | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marchio, A.; Sitbounlang, P.; Deharo, E.; Paboriboune, P.; Pineau, P. Concealed for a Long Time on the Marches of Empires: Hepatitis B Virus Genotype I. Microorganisms 2023, 11, 2204. https://doi.org/10.3390/microorganisms11092204
Marchio A, Sitbounlang P, Deharo E, Paboriboune P, Pineau P. Concealed for a Long Time on the Marches of Empires: Hepatitis B Virus Genotype I. Microorganisms. 2023; 11(9):2204. https://doi.org/10.3390/microorganisms11092204
Chicago/Turabian StyleMarchio, Agnès, Philavanh Sitbounlang, Eric Deharo, Phimpha Paboriboune, and Pascal Pineau. 2023. "Concealed for a Long Time on the Marches of Empires: Hepatitis B Virus Genotype I" Microorganisms 11, no. 9: 2204. https://doi.org/10.3390/microorganisms11092204