Response Surface Methodology Application for Bacteriophage–Antibiotic Antibiofilm Activity Optimization
Abstract
:1. Introduction
- k—is the number of input variables;
- xi—is the input variable;
- y—is the response (output variable);
- β0—is the constant term;
- βi—is the coefficient of the linear variables;
- βii—is the coefficient of the quadratic parameter;
- βij—is the interaction coefficient between the input variables;
- ε—is the error associated with the experiments.
2. Materials and Methods
2.1. Bacterial Strain and Bacteriophage Used
2.2. Antibiotics Used in This Study
2.3. Antibiofilm Activity
2.4. Data Normalization
- cA_var
- —is the value of antibiotic concentration (this value was varied in the range of 0 to 1024), µg/mL;
- cA_max
- —is the maximum value of antibiotic concentration (this value is equal to 1024), µg/mL;
- cP_var
- —is the value of phage concentration (this value was varied in the range of 0 to 108), PFU/mL;
- cP_max
- —is the maximum value of phage concentration (this value is equal to 108), PFU/mL.
| Antibiotic Concentration | Normalized Antibiotic Concentration | Phage Concentration | Normalized Phage Concentration |
|---|---|---|---|
| [µg/mL] | cA [-] | [PFU/mL] | cP [-] |
| 1024 | 1 | 108 | 1 |
| 512 | 0.5 | 107 | 0.1 |
| 256 | 0.25 | 106 | 0.01 |
| 128 | 0.125 | 105 | 0.001 |
| 64 | 0.0625 | 104 | 0.0001 |
| 32 | 0.03125 | 103 | 0.00001 |
| 16 | 0.015625 | 0 | 0 |
| 8 | 0.007813 | ||
| 4 | 0.003906 | ||
| 2 | 0.001953 | ||
| 1 | 0.000977 | ||
| 0 | 0 |
2.5. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Mah, T.-F. Biofilm-specific antibiotic resistance. Futur. Microbiol. 2012, 7, 1061–1072. [Google Scholar] [CrossRef] [PubMed]
- Olsen, I. Biofilm-Specific Antibiotic Tolerance and Resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 877–886. [Google Scholar] [CrossRef]
- Grygorcewicz, B.; Wojciuk, B.; Roszak, M.; Łubowska, N.; Błażejczak, P.; Jursa-Kulesza, J.; Rakoczy, R.; Masiuk, H.; Dołęgowska, B. Environmental Phage-Based Cocktail and Antibiotic Combination Effects on Acinetobacter baumannii Biofilm in a Human Urine Model. Microb. Drug Resist. 2021, 27, 25–35. [Google Scholar] [CrossRef]
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Barr, J.J.; Reed, S.L.; Rohwer, F.; Benler, S.; et al. Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails To Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection. Antimicrob. Agents Chemother. 2017, 61, e00954-17. [Google Scholar] [CrossRef] [PubMed]
- Galtier, M.; De Sordi, L.; Sivignon, A.; De Vallée, A.; Maura, D.; Neut, C.; Rahmouni, O.; Wannerberger, K.; Darfeuille-Michaud, A.; Desreumaux, P.; et al. Bacteriophages targeting adherent invasive Escherichia coli strains as a promising new treatment for Crohn’s disease. J. Crohn’s Colitis 2017, 11, 840–847. [Google Scholar] [CrossRef]
- Augustyniak, A.; Grygorcewicz, B.; Nawrotek, P. Isolation of multidrug resistant coliforms and their bacteriophages from swine slurry. Turk. J. Veter- Anim. Sci. 2018, 42, 319–325. [Google Scholar] [CrossRef]
- Comeau, A.M.; Tétart, F.; Trojet, S.N.; Prère, M.-F.; Krisch, H.M. Phage-Antibiotic Synergy (PAS): β-Lactam and Quinolone Antibiotics Stimulate Virulent Phage Growth. PLoS ONE 2007, 2, e799. [Google Scholar] [CrossRef]
- Grygorcewicz, B.; Roszak, M.; Golec, P.; Śleboda-Taront, D.; Łubowska, N.; Górska, M.; Jursa-Kulesza, J.; Rakoczy, R.; Wojciuk, B.; Dołęgowska, B. Antibiotics Act with vB_AbaP_AGC01 Phage against Acinetobacter baumannii in Human Heat-Inactivated Plasma Blood and Galleria mellonella Models. Int. J. Mol. Sci. 2020, 21, 4390. [Google Scholar] [CrossRef]
- Kamal, F.; Dennis, J.J. Burkholderia cepacia Complex Phage-Antibiotic Synergy (PAS): Antibiotics Stimulate Lytic Phage Activity. Appl. Environ. Microbiol. 2015, 81, 1132–1138. [Google Scholar] [CrossRef]
- Nouraldin, A.A.M.; Baddour, M.M.; Harfoush, R.A.H.; Essa, S.A.M. Bacteriophage-antibiotic synergism to control planktonic and biofilm producing clinical isolates of Pseudomonas aeruginosa. Alex. J. Med. 2016, 52, 99–105. [Google Scholar] [CrossRef]
- Jo, A.; Ding, T.; Ahn, J. Synergistic antimicrobial activity of bacteriophages and antibiotics against Staphylococcus aureus. Food Sci. Biotechnol. 2016, 25, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.G.; Green, S.I.; Min, L.; Clark, J.R.; Salazar, K.C.; Terwilliger, A.L.; Kaplan, H.B.; Trautner, B.W.; Ramig, R.F.; Maresso, A.W. Phage-Antibiotic Synergy Is Driven by a Unique Combination of Antibacterial Mechanism of Action and Stoichiometry. mBio 2020, 11, e01462-20. [Google Scholar] [CrossRef]
- Pandey, N.; Thakur, C. Statistical Comparison of Response Surface Methodology–Based Central Composite Design and Hybrid Central Composite Design for Paper Mill Wastewater Treatment by Electrocoagulation. Process. Integr. Optim. Sustain. 2020, 4, 343–359. [Google Scholar] [CrossRef]
- Kumari, M.; Gupta, S.K. Response surface methodological (RSM) approach for optimizing the removal of trihalomethanes (THMs) and its precursor’s by surfactant modified magnetic nanoadsorbents (sMNP)—An endeavor to diminish probable cancer risk. Sci. Rep. 2019, 9, 18339. [Google Scholar] [CrossRef]
- Boyacı, H. A new approach for determination of enzyme kinetic constants using response surface methodology. Biochem. Eng. J. 2005, 25, 55–62. [Google Scholar] [CrossRef]
- Damena, T.; Alansi, T. Review: Low Cost, Environmentally Friendly Humic Acid Coated Magnetite Nanoparticles (HA-MNP) and Its Application for the Remediation of Phosphate from Aqueous Media. J. Encapsulation Adsorpt. Sci. 2018, 8, 256–279. [Google Scholar] [CrossRef]
- Sarrai, A.E.; Hanini, S.; Merzouk, N.K.; Tassalit, D.; Szabó, T.; Hernádi, K.; Nagy, L. Using Central Composite Experimental Design to Optimize the Degradation of Tylosin from Aqueous Solution by Photo-Fenton Reaction. Materials 2016, 9, 428. [Google Scholar] [CrossRef]
- Arslan-Alaton, I.; Tureli, G.; Olmez-Hanci, T. Treatment of azo dye production wastewaters using Photo-Fenton-like advanced oxidation processes: Optimization by response surface methodology. J. Photochem. Photobiol. A Chem. 2009, 202, 142–153. [Google Scholar] [CrossRef]
- Li, Z.; Zhu, W.; Wang, B.; Wang, Q.; Du, J.; Sun, B. Optimization of cooling water jacket structure of high-speed electric spindle based on response surface method. Case Stud. Therm. Eng. 2023, 48, 103158. [Google Scholar] [CrossRef]
- Lee, E.Y.; Wong, S.Y.; Phang, S.J.; Wong, V.-L.; Cheah, K.H. Additively manufactured photoreactor with immobilized thermoset acrylic-graphitic carbon nitride nanosheets for water remediation: Response surface methods and adsorption modelling studies. Chem. Eng. J. 2023, 455, 140633. [Google Scholar] [CrossRef]
- Markowska-Szczupak, A.; Wesołowska, A.; Borowski, T.; Sołoducha, D.; Paszkiewicz, O.; Kordas, M.; Rakoczy, R. Effect of pine essential oil and rotating magnetic field on antimicrobial performance. Sci. Rep. 2022, 12, 9712. [Google Scholar] [CrossRef] [PubMed]
- Tazik, M.; Dehghani, M.H.; Yaghmaeian, K.; Nazmara, S.; Salari, M.; Mahvi, A.H.; Nasseri, S.; Soleimani, H.; Karri, R.R. 4-Chlorophenol adsorption from water solutions by activated carbon functionalized with amine groups: Response surface method and artificial neural networks. Sci. Rep. 2023, 13, 7831. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.R.; Pathak, A.; Poluri, K.M. Comparative analysis of the physicochemical and anti-biofilm properties of iota and lambda carrageenan-capped silver nanocomposites synthesized using response surface methodology. N. J. Chem. 2023, 47, 16643–16658. [Google Scholar] [CrossRef]
- Gharbani, P.; Jam, N.; Doshmanfekan, H.; Mehrizad, A. Optimization of synergic antibacterial activity of Punica granatum L. and Areca nut (P.G.L.A.N) extracts through response surface methodology. Sci. Rep. 2023, 13, 6098. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Suo, J.; Cui, Y.; Huang, S.-Y.; Zhu, X.-Q.; Wang, Y.; Liu, H.-B.; Dai, L.; He, J.-K.; Li, B.-B.; et al. Optimization of Antimicrobial Activity of Surfactin and Polylysine Against Salmonella enteritidis in Milk Evaluated by a Response Surface Methodology. Foodborne Pathog. Dis. 2011, 8, 439–443. [Google Scholar] [CrossRef]
- Grygorcewicz, B.; Roszak, M.; Rakoczy, R.; Augustyniak, A.; Konopacki, M.; Jabłońska, J.; Serwin, N.; Cecerska-Heryć, E.; Kordas, M.; Galant, K.; et al. PhageScore-based analysis of Acinetobacter baumannii infecting phages antibiotic interaction in liquid medium. Arch. Microbiol. 2022, 204, 421. [Google Scholar] [CrossRef]
- Kaźmierczak, N.; Grygorcewicz, B.; Roszak, M.; Bochentyn, B.; Piechowicz, L. Comparative Assessment of Bacteriophage and Antibiotic Activity against Multidrug-Resistant Staphylococcus aureus Biofilms. Int. J. Mol. Sci. 2022, 23, 1274. [Google Scholar] [CrossRef]
- Kaźmierczak, N.; Grygorcewicz, B.; Piechowicz, L. Biofilm Formation and Prevalence of Biofilm-Related Genes Among Clinical Strains of Multidrug-Resistant Staphylococcus aureus. Microb. Drug Resist. 2021, 27, 956–964. [Google Scholar] [CrossRef]
- Keijok, W.J.; Pereira, R.H.A.; Alvarez, L.A.C.; Prado, A.R.; da Silva, A.R.; Ribeiro, J.; de Oliveira, J.P.; Guimarães, M.C.C. Controlled biosynthesis of gold nanoparticles with Coffea arabica using factorial design. Sci. Rep. 2019, 9, 16019. [Google Scholar] [CrossRef]
- Vukotic, G.; Obradovic, M.; Novovic, K.; Di Luca, M.; Jovcic, B.; Fira, D.; Neve, H.; Kojic, M.; McAuliffe, O. Characterization, Antibiofilm, and Depolymerizing Activity of Two Phages Active on Carbapenem-Resistant Acinetobacter baumannii. Front. Med. 2020, 7, 426. [Google Scholar] [CrossRef]
- Fong, S.A.; Drilling, A.; Morales, S.; Cornet, M.E.; Woodworth, B.A.; Fokkens, W.J.; Psaltis, A.J.; Vreugde, S.; Wormald, P.-J. Activity of Bacteriophages in Removing Biofilms of Pseudomonas aeruginosa Isolates from Chronic Rhinosinusitis Patients. Front. Cell. Infect. Microbiol. 2017, 7, 418. [Google Scholar] [CrossRef] [PubMed]
- Kwiatek, M.; Parasion, S.; Rutyna, P.; Mizak, L.; Gryko, R.; Niemcewicz, M.; Olender, A.; Łobocka, M. Isolation of bacteriophages and their application to control Pseudomonas aeruginosa in planktonic and biofilm models. Res. Microbiol. 2017, 168, 194–207. [Google Scholar] [CrossRef]
- Liao, K.; Lehman, S.; Tweardy, D.; Donlan, R.; Trautner, B. Bacteriophages are synergistic with bacterial interference for the prevention of Pseudomonas aeruginosa biofilm formation on urinary catheters. J. Appl. Microbiol. 2012, 113, 1530–1539. [Google Scholar] [CrossRef]
- Ashrit, P.; Sadanandan, B.; Natraj, L.K.; Shetty, K.; Vaniyamparambath, V.; Raghu, A.V. A microplate-based Response Surface Methodology model for growth optimization and biofilm formation on polystyrene polymeric material in a Candida albicans and Escherichia coli co-culture. Polym. Adv. Technol. 2022, 33, 2872–2885. [Google Scholar] [CrossRef]
- Sadanandan, B.; Vaniyamparambath, V.; Lokesh, K.N.; Shetty, K.; Joglekar, A.P.; Ashrit, P.; Hemanth, B. Candida albicans biofilm formation and growth optimization for functional studies using response surface methodology. J. Appl. Microbiol. 2021, 132, 3277–3292. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.; Xiao, Y.; Huang, W.-M.; Chen, C.-H.; Baltrusaitis, J. DBD plasma-assisted ethanol steam reforming for green H2 production: Process optimization through response surface methodology (RSM). Int. J. Hydrogen Energy 2023, 48, 553–565. [Google Scholar] [CrossRef]
- Huang, X.; Lv, Z.; Zhao, B.; Zhang, H.; Yao, X.; Shuai, Y. Optimization of operating parameters for methane steam reforming thermochemical process using Response Surface Methodology. Int. J. Hydrogen Energy 2022, 47, 28313–28321. [Google Scholar] [CrossRef]
- Raheem, A.T.; Aziz, A.R.A.; Zulkifli, S.A.; Baharom, M.B.; Rahem, A.T.; Ayandotun, W.B. Optimisation of operating parameters on the performance characteristics of a free piston engine linear generator fuelled by CNG–H2 blends using the response surface methodology (RSM). Int. J. Hydrogen Energy 2021, 47, 1996–2016. [Google Scholar] [CrossRef]
- Chaudhry, W.N.; Concepción-Acevedo, J.; Park, T.; Andleeb, S.; Bull, J.J.; Levin, B.R. Synergy and Order Effects of Antibiotics and Phages in Killing Pseudomonas aeruginosa Biofilms. PLoS ONE 2017, 12, e0168615. [Google Scholar] [CrossRef]
- Ryan, E.M.; Alkawareek, M.Y.; Donnelly, R.F.; Gilmore, B.F. Synergistic phage-antibiotic combinations for the control of Escherichia coli biofilms in vitro. FEMS Immunol. Med. Microbiol. 2012, 65, 395–398. [Google Scholar] [CrossRef]
- Wang, X.; Loh, B.; Altamirano, F.G.; Yu, Y.; Hua, X.; Leptihn, S. Colistin-phage combinations decrease antibiotic resistance in Acinetobacter baumannii via changes in envelope architecture. Emerg. Microbes Infect. 2021, 10, 2205–2219. [Google Scholar] [CrossRef] [PubMed]
- Kebriaei, R.; Lehman, S.M.; Shah, R.M.; Stamper, K.C.; Kunz Coyne, A.J.; Holger, D.; El Ghali, A.; Rybak, M.J. Opti-mization of Phage-Antibiotic Combinations against Staphylococcus Aureus Biofilms. Microbiol. Spectr. 2023, 11, e04918-22. [Google Scholar] [CrossRef] [PubMed]
- Popova, A.V.; Shneider, M.M.; Arbatsky, N.P.; Kasimova, A.A.; Senchenkova, S.N.; Shashkov, A.S.; Dmitrenok, A.S.; Chizhov, A.O.; Mikhailova, Y.V.; Shagin, D.A.; et al. Specific Interaction of Novel Friunavirus Phages Encoding Tailspike Depolymerases with Corresponding Acinetobacter baumannii Capsular Types. J. Virol. 2021, 95, 10–1128. [Google Scholar] [CrossRef]
- Popova, A.V.; Lavysh, D.G.; Klimuk, E.I.; Edelstein, M.V.; Bogun, A.G.; Shneider, M.M.; Goncharov, A.E.; Leonov, S.V.; Severinov, K.V. Novel Fri1-like Viruses Infecting Acinetobacter baumannii—vB_AbaP_AS11 and vB_AbaP_AS12—Characterization, Comparative Genomic Analysis, and Host-Recognition Strategy. Viruses 2017, 9, 188. [Google Scholar] [CrossRef] [PubMed]
- Knezevic, P.; Curcin, S.; Aleksic, V.; Petrusic, M.; Vlaski, L. Phage-antibiotic synergism: A possible approach to combatting Pseudomonas aeruginosa. Res. Microbiol. 2013, 164, 55–60. [Google Scholar] [CrossRef]
- Manohar, P.; Royam, M.M.; Loh, B.; Bozdogan, B.; Nachimuthu, R.; Leptihn, S. Synergistic Effects of Phage–Antibiotic Combinations against Citrobacter amalonaticus. ACS Infect. Dis. 2022, 8, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Kever, L.; Hardy, A.; Luthe, T.; Hünnefeld, M.; Gätgens, C.; Milke, L.; Wiechert, J.; Wittmann, J.; Moraru, C.; Marienhagen, J.; et al. Aminoglycoside Antibiotics Inhibit Phage Infection by Blocking an Early Step of the Infection Cycle. mBio 2022, 13, e0078322. [Google Scholar] [CrossRef] [PubMed]

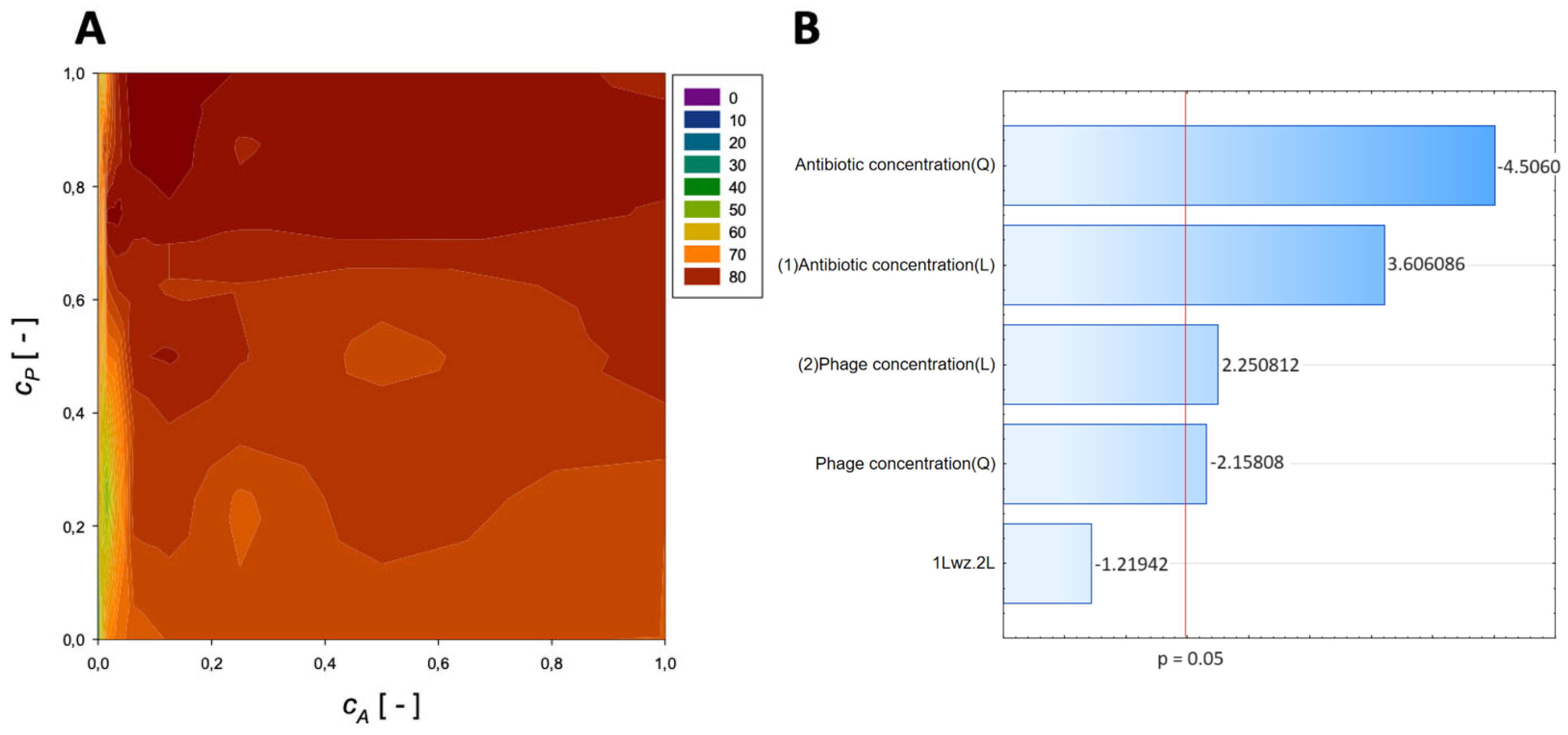

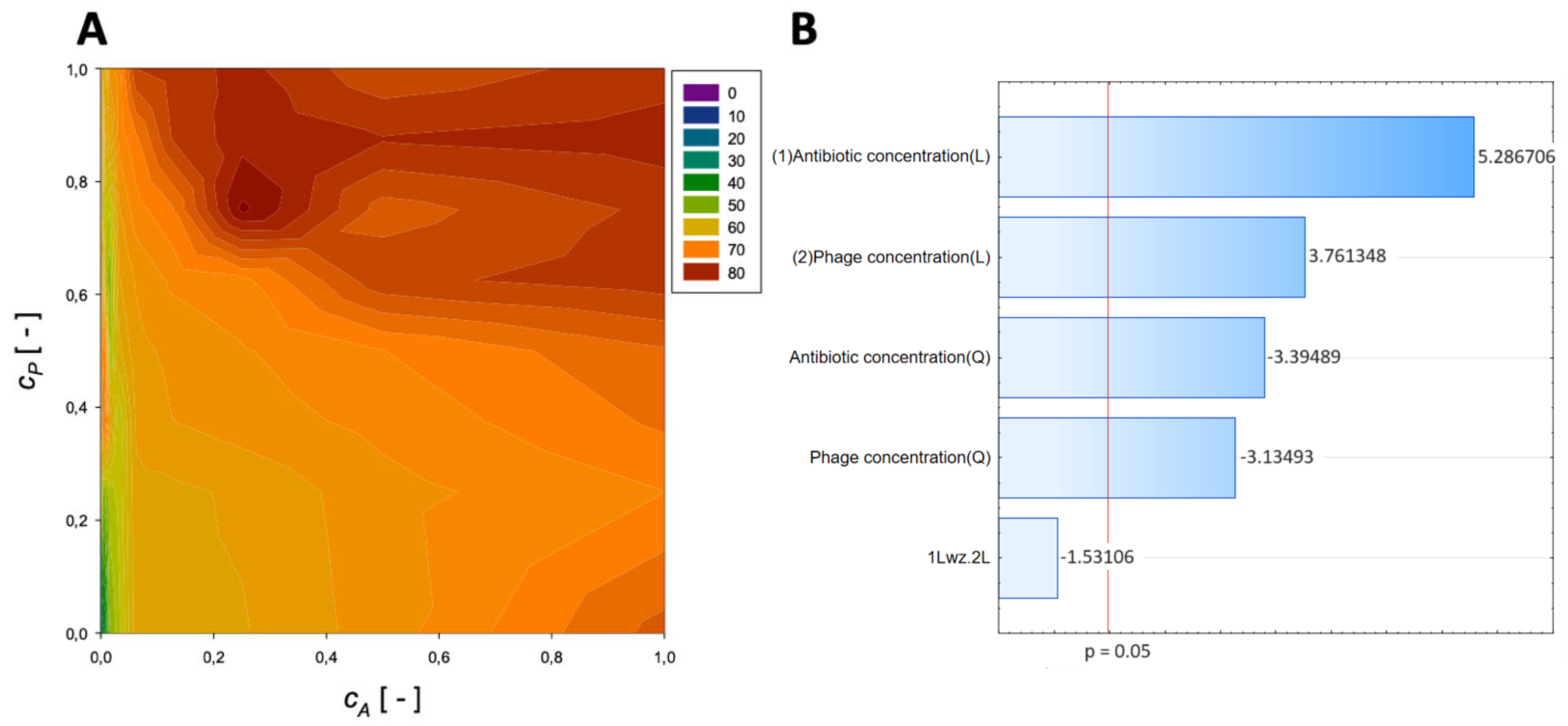
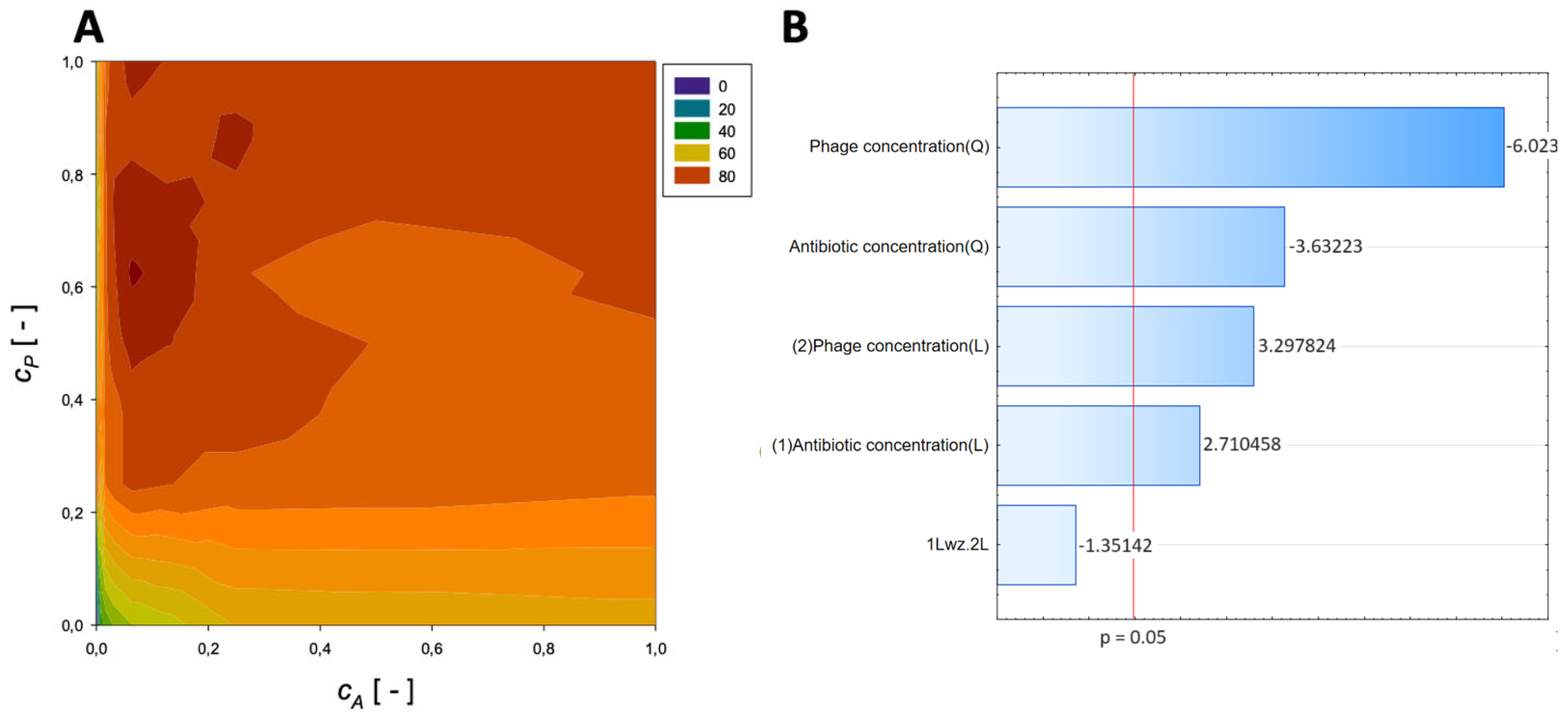

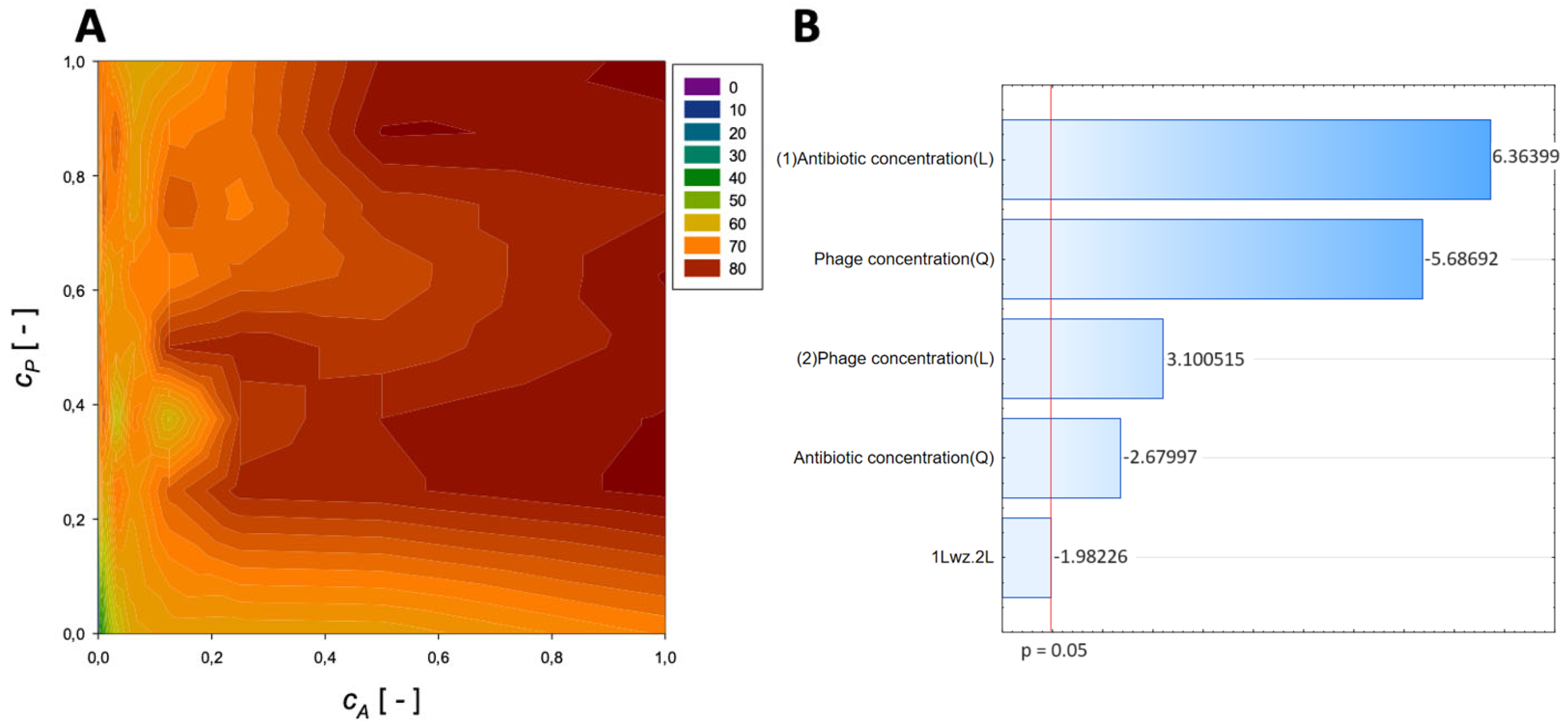
| Antibiotic | Normalized Antibiotic Concentration, cA [-] | Normalized Phage Concentration, cP [-] | Maximum Reduction [%] |
|---|---|---|---|
| Amikacin | 0.00195 | 0.38 | 84.18 |
| Meropenem | 0.0625 | 1 | 85.79 |
| Fosfomycin | 1 | 1 | 85.52 |
| Ceftazidime | 0.25 | 0.75 | 84.45 |
| Imipenem | 0.0625 | 0.63 | 88.74 |
| Gentamicin | 0 | 1 | 68.36 |
| Colistin | 1 | 0.25 | 84.72 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grygorcewicz, B.; Gliźniewicz, M.; Olszewska, P.; Miłek, D.; Czajkowski, A.; Serwin, N.; Cecerska-Heryć, E.; Rakoczy, R. Response Surface Methodology Application for Bacteriophage–Antibiotic Antibiofilm Activity Optimization. Microorganisms 2023, 11, 2352. https://doi.org/10.3390/microorganisms11092352
Grygorcewicz B, Gliźniewicz M, Olszewska P, Miłek D, Czajkowski A, Serwin N, Cecerska-Heryć E, Rakoczy R. Response Surface Methodology Application for Bacteriophage–Antibiotic Antibiofilm Activity Optimization. Microorganisms. 2023; 11(9):2352. https://doi.org/10.3390/microorganisms11092352
Chicago/Turabian StyleGrygorcewicz, Bartłomiej, Marta Gliźniewicz, Patrycja Olszewska, Dominika Miłek, Artur Czajkowski, Natalia Serwin, Elżbieta Cecerska-Heryć, and Rafał Rakoczy. 2023. "Response Surface Methodology Application for Bacteriophage–Antibiotic Antibiofilm Activity Optimization" Microorganisms 11, no. 9: 2352. https://doi.org/10.3390/microorganisms11092352





