The Evolving Microbiome of Dental Caries
Abstract
:1. Introduction
2. Background
2.1. Epidemiology of Dental Caries
2.2. Diet and Dental Caries
2.3. Plaque pH and Host Factors
3. Microbiome of Dental Caries
3.1. History of Caries Microbiology
3.2. Development of the Tooth-Associated Microbiome Leading to Caries
3.2.1. Pioneer Species
3.2.2. Early Colonizers
3.2.3. Early Colonizers and the Acidogenic Stage of Plaque Biofilm Development
3.2.4. Secondary Colonizers and the Aciduric Stage of Plaque Biofilm Development
3.2.5. The Mature Biofilm
3.3. Culture and Genetic Analysis of the Cariogenic Microbiome
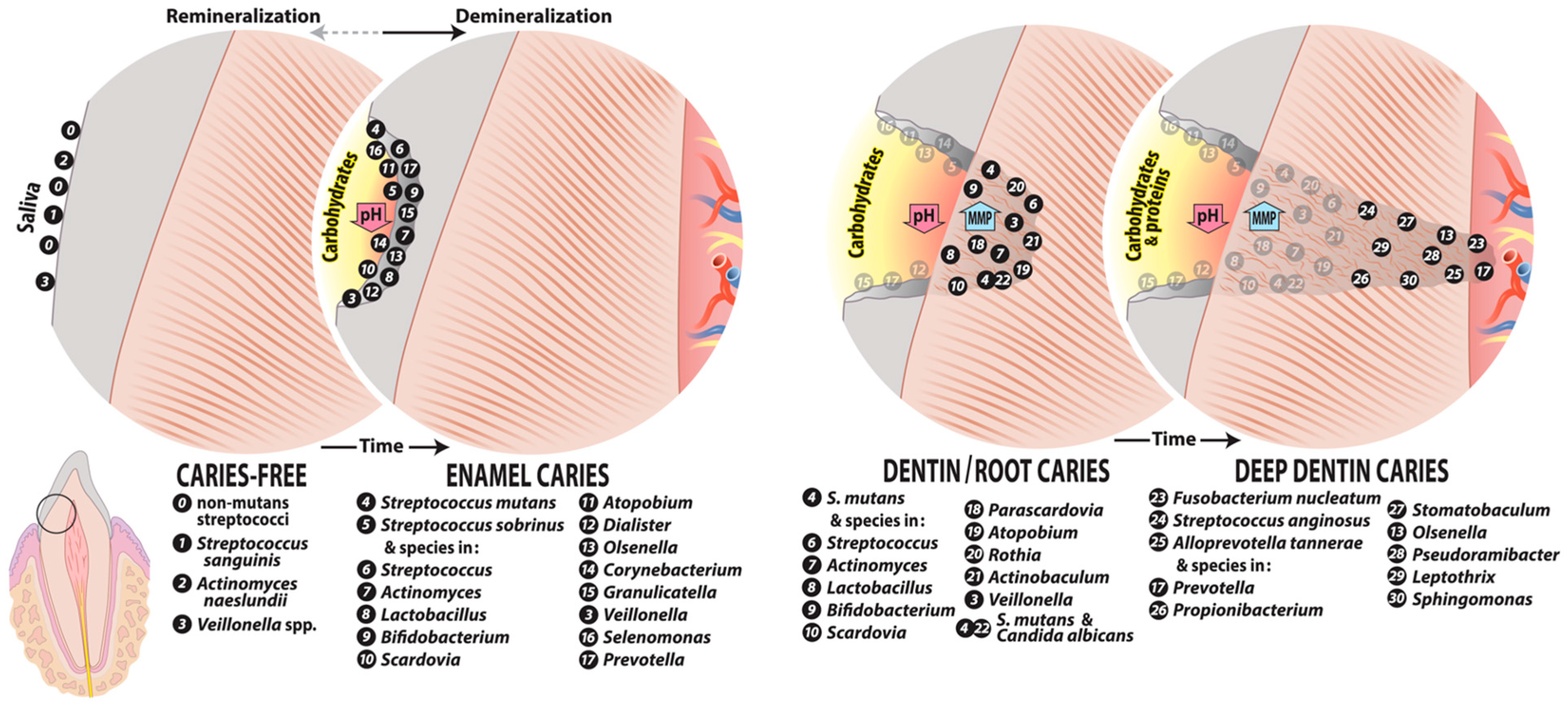
4. Caries Microbiome at Different Lesion Sites
4.1. Initial Caries
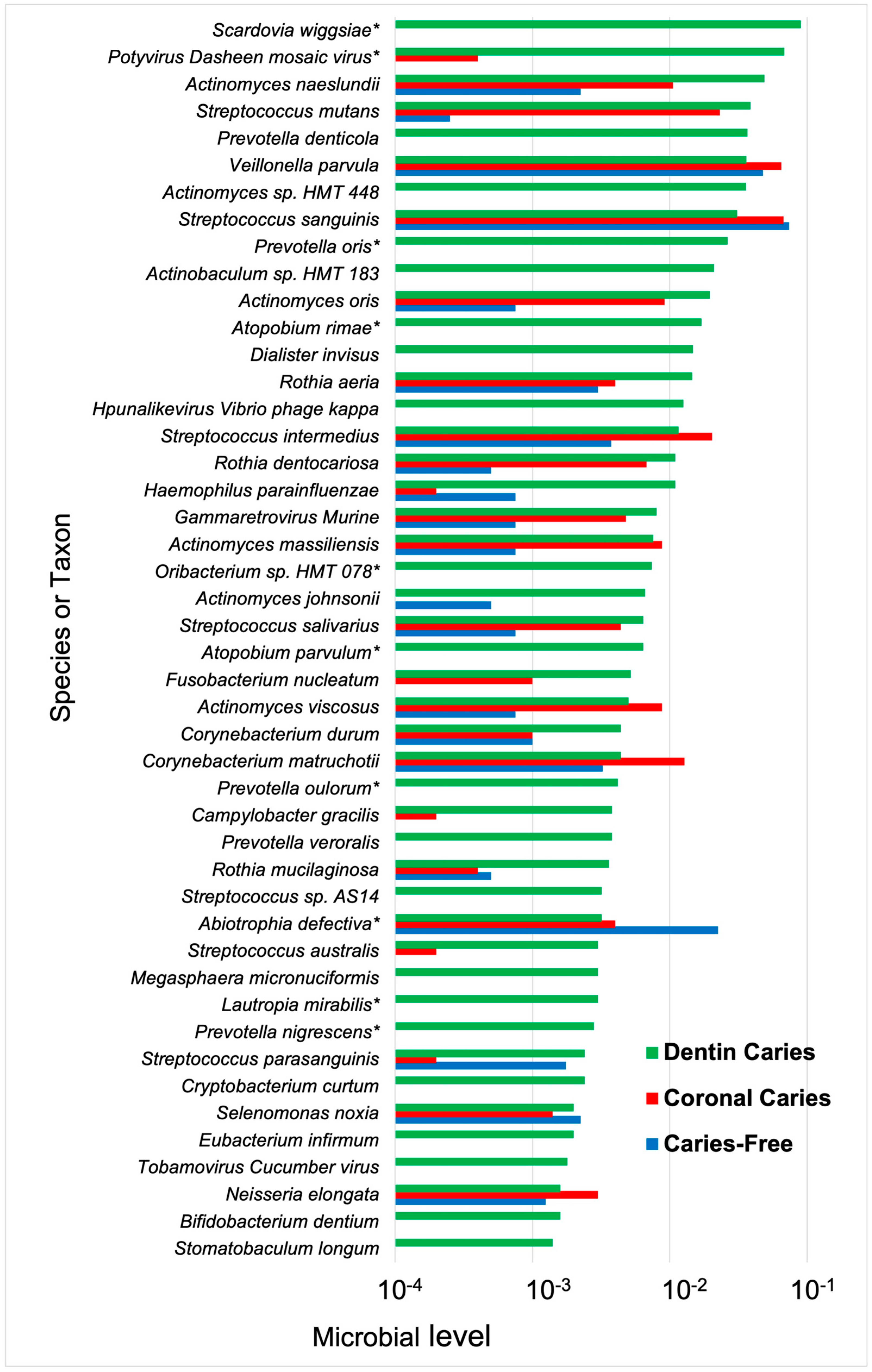
4.2. Dentin Caries
4.2.1. Root Caries
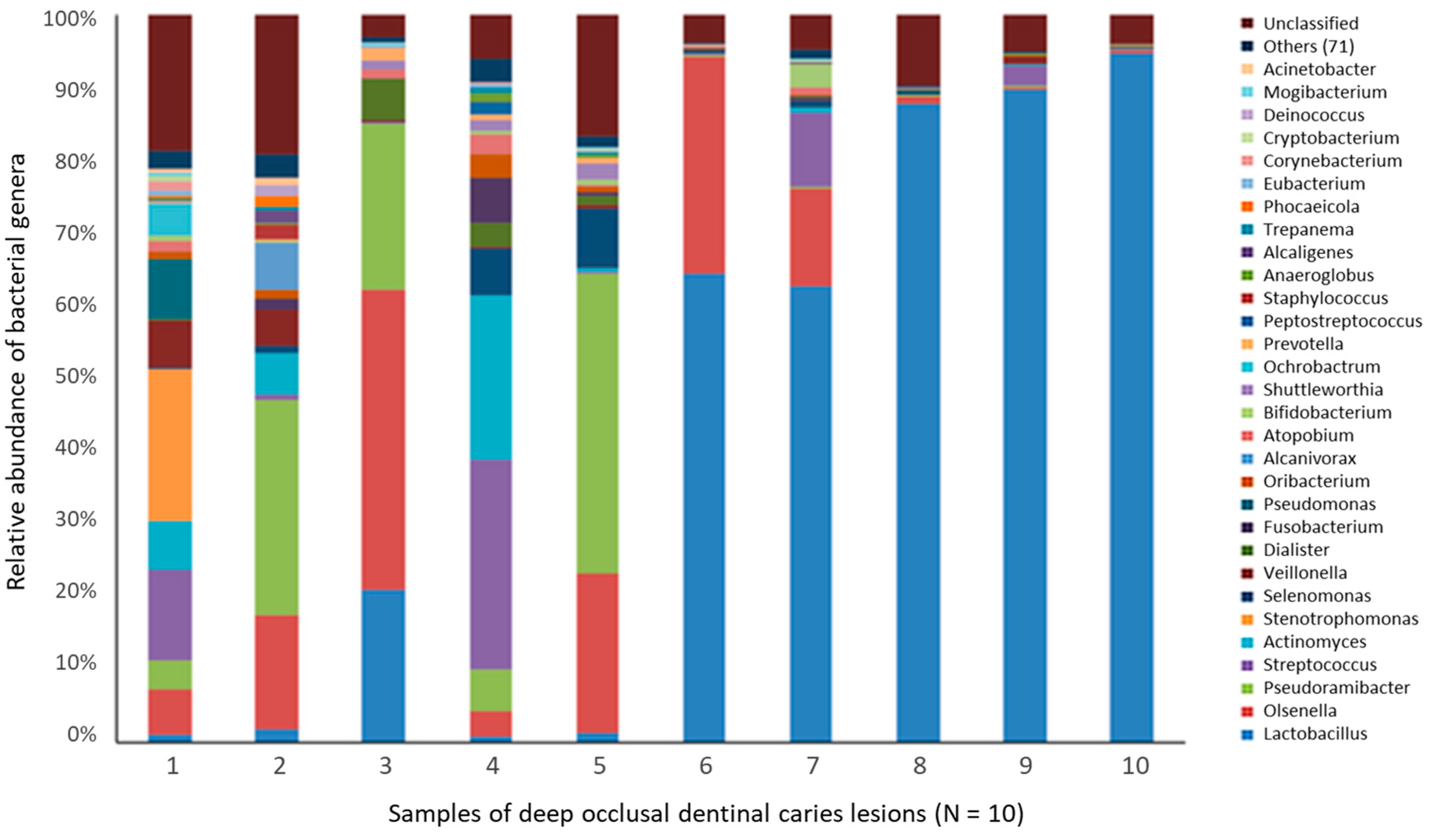
4.2.2. Deep Dentin Caries
4.3. Caries Microbiome of Aggressive Lesions
4.4. Yeasts and Dental Caries
5. Principal Caries-Associated Bacteria
5.1. Streptococcus mutans
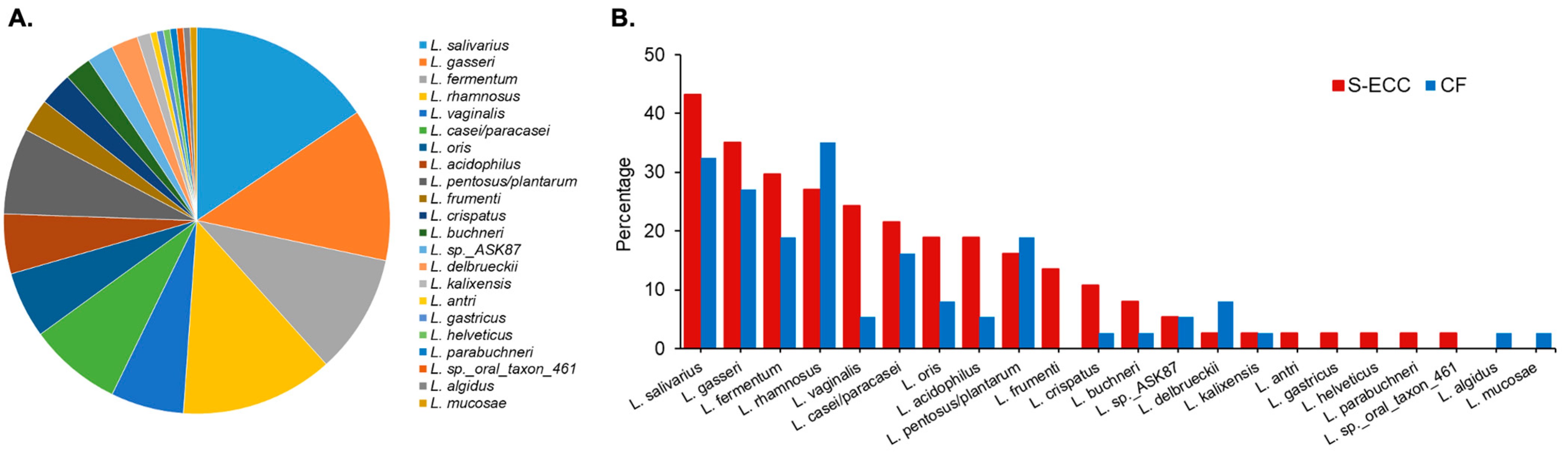
5.2. Lactobacillus Species
5.3. Actinomycetaceae
6. Microbiome: Beyond Microbial Composition and the Oral Microbiome: Functional Genomics
6.1. Metagenome
6.2. Metatranscriptome
6.3. Multi-Omics
7. Treatment of Caries as a Microbial Disease
7.1. Fluoride and Anticaries Activity
7.1.1. Fluoride as a Preventative Anticaries Agent
7.1.2. Antimicrobial Properties of Fluoride
7.1.3. Chemical Action of Fluoride to Strengthen Teeth
7.2. Silver Nanoparticles
7.3. Approaches for Caries Control Targeting the Microbiome
7.4. Challenges to Antimicrobial Approaches for Caries Management
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Milgrom, P. Tooth decay in our poorest children: What can we do? J. Indiana Dent. Assoc. 2000, 79, 24–26. [Google Scholar] [PubMed]
- Pitts, N.B.; Zero, D.T.; Marsh, P.D.; Ekstrand, K.; Weintraub, J.A.; Ramos-Gomez, F.; Tagami, J.; Twetman, S.; Tsakos, G.; Ismail, A. Dental caries. Nat. Rev. Dis. Primers 2017, 3, 17030. [Google Scholar] [CrossRef] [PubMed]
- Meier, T.; Deumelandt, P.; Christen, O.; Stangl, G.I.; Riedel, K.; Langer, M. Global burden of sugar-related dental diseases in 168 countries and corresponding health care costs. J. Dent. Res. 2017, 96, 845–854. [Google Scholar] [CrossRef] [PubMed]
- GBD 2017 Oral Disorders Collaborators; Bernabe, E.; Marcenes, W.; Hernandez, C.R.; Bailey, J.; Abreu, L.G.; Alipour, V.; Amini, S.; Arabloo, J.; Arefi, Z.; et al. Global, regional, and national levels and trends in burden of oral conditions from 1990 to 2017: A systematic analysis for the global burden of disease 2017 study. J. Dent. Res. 2020, 99, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Agency for Healthcare Research and Quality. MEPS HC-188B: 2016 Dental Visits; Agency for Healthcare Research and Quality: Rockville, MD, USA, 2018. [Google Scholar]
- Fantom, N.; Serajuddin, U. The World Bank’s Classification of Countries by Income; Policy Research Working Paper 7528; The World Bank: Washington, DC, USA, 2016. [Google Scholar]
- Hescot, P. The new definition of oral health and relationship between oral health and quality of life. Chin. J. Dent. Res. 2017, 20, 189–192. [Google Scholar] [CrossRef]
- Kassebaum, N.J.; Smith, A.G.C.; Bernabe, E.; Fleming, T.D.; Reynolds, A.E.; Vos, T.; Murray, C.J.L.; Marcenes, W.; GBD Oral Health Collaborators. Global, regional, and national prevalence, incidence, and disability-adjusted life years for oral conditions for 195 countries, 1990–2015: A systematic analysis for the global burden of diseases, injuries, and risk factors. J. Dent. Res. 2017, 96, 380–387. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Oral Health Surveillance Report: Trends in Dental Caries and Sealants, Tooth Retention, and Edentulism, United States, 1999–2004 to 2011–2016; Centers for Disease Control and Prevention, US Dept of Health and Human Services: Atlanta, GA, USA, 2019. [Google Scholar]
- Frencken, J.E.; Sharma, P.; Stenhouse, L.; Green, D.; Laverty, D.; Dietrich, T. Global epidemiology of dental caries and severe periodontitis—A comprehensive review. J. Clin. Periodontol. 2017, 44 (Suppl. S18), S94–S105. [Google Scholar] [CrossRef]
- FDI World Dental Federation. The Challenge of Oral Disease—A Call for Global Action. The Oral Health Atlas, 2nd ed.; Benzian, H., Williams, D., Eds.; FDI World Dental Federation: Geneva, Switzerland, 2015; pp. 16–21. [Google Scholar]
- U.S. Department of Health & Human Services. Ten great public health achievements–United States, 1900–1999. MMWR Morb. Mortal. Wkly. Rep. 1999, 48, 241–243. [Google Scholar]
- Kim, H.N.; Kong, W.S.; Lee, J.H.; Kim, J.B. Reduction of dental caries among children and adolescents from a 15-year community water fluoridation program in a township area, Korea. Int. J. Environ. Res. Public Health 2019, 16, 1306. [Google Scholar] [CrossRef]
- Schluter, P.J.; Hobbs, M.; Atkins, H.; Mattingley, B.; Lee, M. Association between community water fluoridation and severe dental caries experience in 4-year-old New Zealand children. JAMA Pediatr. 2020, 174, 969–976. [Google Scholar] [CrossRef]
- Foley, M.A.; Sexton, C.; Spencer, A.J.; Lalloo, R.; Do, L.G. Water fluoridation, dental caries and parental ratings of child oral health. Community Dent. Oral Epidemiol. 2021, 50, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Davies, G.M.; Neville, J.; Jones, K.; White, S. Why are caries levels reducing in five-year-olds in England? Br. Dent. J. 2017, 223, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Arheiam, A.A.; Harris, R.V.; Baker, S.R. Changes in dental caries and sugar intake before and during the conflict in Libya: A natural experiment. Community Dent. Oral Epidemiol. 2020, 48, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Thornton-Evans, G.; Griffin, S.O.; Wei, L.; Junger, M.; Espinoza, L. Increased dental use may affect changes in treated and untreated dental caries in young children. JDR Clin. Trans. Res. 2019, 4, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Dye, B.A.; Mitnik, G.L.; Iafolla, T.J.; Vargas, C.M. Trends in dental caries in children and adolescents according to poverty status in the United States from 1999 through 2004 and from 2011 through 2014. J. Am. Dent. Assoc. 2017, 148, 550–565. [Google Scholar] [CrossRef] [PubMed]
- Dye, B.A.; Tan, S.; Smith, V.; Lewis, B.G.; Barker, L.K.; Thornton-Evans, G.; Eke, P.I.; Beltran-Aguilar, E.D.; Horowitz, A.M.; Li, C.H. Trends in Oral Health Status: United States, 1988–1994 and 1999–2004; Vital and Health Statistics; National Center for Health Statistics: Atlanta, GA, USA, 2007; Volume 11. [Google Scholar]
- Dye, B.A.; Thornton-Evans, G.; Li, X.; Iafolla, T. Dental Caries and Tooth Loss in Adults in the United States, 2011–2012; NCHS Data Brief; US Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Health Statistics: Hyattsville, MA, USA, 2015; pp. 1–8. [Google Scholar]
- Lagerweij, M.D.; van Loveren, C. Declining caries trends: Are we satisfied? Curr. Oral Health Rep. 2015, 2, 212–217. [Google Scholar] [CrossRef]
- World Health Organization. Global Oral Health Status Report: Towards Universal Health Coverage for Oral Health by 2030; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Bernabe, E.; Sheiham, A. Age, period and cohort trends in caries of permanent teeth in four developed countries. Am. J. Public Health 2014, 104, 115–121. [Google Scholar] [CrossRef]
- Petersen, P.E.; Ogawa, H. Promoting oral health and quality of life of older people—The need for public health action. Oral Health Prev. Dent. 2018, 16, 113–124. [Google Scholar] [CrossRef]
- Fleming, E.; Afful, J. Prevalence of Total and Untreated Dental Caries among Youth: United States, 2015–2016; NCHS Data Brief; US Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Health Statistics: Hyattsville, MA, USA, 2018; pp. 1–8. [Google Scholar]
- Gupta, N.; Vujicic, M.; Yarbrough, C.; Harrison, B. Disparities in untreated caries among children and adults in the U.S., 2011–2014. BMC Oral Health 2018, 18, 30. [Google Scholar] [CrossRef]
- Peres, M.A.; Ju, X.; Mittinty, M.; Spencer, A.J.; Do, L.G. Modifiable factors explain socioeconomic inequalities in children’s dental caries. J. Dent. Res. 2019, 98, 1211–1218. [Google Scholar] [CrossRef]
- Jenson, L.; Budenz, A.W.; Featherstone, J.D.; Ramos-Gomez, F.J.; Spolsky, V.W.; Young, D.A. Clinical protocols for caries management by risk assessment. J. Calif. Dent. Assoc. 2007, 35, 714–723. [Google Scholar] [CrossRef] [PubMed]
- Bratthall, D.; Hansel Petersson, G. Cariogram—A multifactorial risk assessment model for a multifactorial disease. Community Dent. Oral Epidemiol. 2005, 33, 256–264. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Oral Health Surveys: Basic Methods, 5th ed.; World Health Organization: Geneva, Switzerland, 2013; pp. 42–47. [Google Scholar]
- Frazao, P. Epidemiology of dental caries: When structure and context matter. Braz. Oral Res. 2012, 26 (Suppl. S1), 108–114. [Google Scholar] [CrossRef] [PubMed]
- Kutsch, V.K. Dental caries: An updated medical model of risk assessment. J. Prosthet. Dent. 2014, 111, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Pitts, N. “ICDAS”—An international system for caries detection and assessment being developed to facilitate caries epidemiology, research and appropriate clinical management. Community Dent. Health 2004, 21, 193–198. [Google Scholar]
- Ismail, A.I.; Sohn, W.; Tellez, M.; Amaya, A.; Sen, A.; Hasson, H.; Pitts, N.B. The International Caries Detection and Assessment System (ICDAS): An integrated system for measuring dental caries. Community Dent. Oral Epidemiol. 2007, 35, 170–178. [Google Scholar] [CrossRef]
- Pitts, N.B.; Ekstrand, K.R.; Foundation, I. International Caries Detection and Assessment System (ICDAS) and its International Caries Classification and Management System (ICCMS)—Methods for staging of the caries process and enabling dentists to manage caries. Community Dent. Oral Epidemiol. 2013, 41, e41–e52. [Google Scholar] [CrossRef]
- Adler, C.J.; Dobney, K.; Weyrich, L.S.; Kaidonis, J.; Walker, A.W.; Haak, W.; Bradshaw, C.J.; Townsend, G.; Soltysiak, A.; Alt, K.W.; et al. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nat. Genet. 2013, 45, 450–455. [Google Scholar] [CrossRef]
- Touger-Decker, R.; van Loveren, C. Sugars and dental caries. Am. J. Clin. Nutr. 2003, 78, 881S–892S. [Google Scholar] [CrossRef]
- Toverud, G. Dental caries in Norwegian children during and after the last World War; a preliminary report. Proc. R. Soc. Med. 1949, 42, 249–258. [Google Scholar] [CrossRef]
- Sognnaes, R.F. Analysis of wartime reduction of dental caries in European children; with special regard to observations in Norway. Am. J. Dis. Child. 1948, 75, 792–821. [Google Scholar] [CrossRef] [PubMed]
- Krasse, B. The Vipeholm Dental Caries Study: Recollections and reflections 50 years later. J. Dent. Res. 2001, 80, 1785–1788. [Google Scholar] [CrossRef] [PubMed]
- American Academy of Pediatrics; Committee on Native American Child Health, Canadian Paediatric Society; First Nations, Inuit and Métis Committee. Early childhood caries in indigenous communities. Pediatrics 2011, 127, 1190–1198. [Google Scholar] [CrossRef] [PubMed]
- Punitha, V.C.; Amudhan, A.; Sivaprakasam, P.; Rathanaprabu, V. Role of dietary habits and diet in caries occurrence and severity among urban adolescent school children. J. Pharm. Bioallied Sci. 2015, 7, S296–S300. [Google Scholar] [CrossRef] [PubMed]
- Stephan, R.M. Intra-oral hydrogen-ion concentrations associated with dental caries activity. J. Dent. Res. 1944, 23, 257–266. [Google Scholar] [CrossRef]
- Flemming, H.C.; Neu, T.R.; Wozniak, D.J. The EPS matrix: The “house of biofilm cells”. J. Bacteriol. 2007, 189, 7945–7947. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.; Nicoll, A.D.; Adair, P.M.; Pine, C.M. Risk factors for dental caries in young children: A systematic review of the literature. Community Dent. Health 2004, 21, 71–85. [Google Scholar] [PubMed]
- Vieira, A.R.; Modesto, A.; Marazita, M.L. Caries: Review of human genetics research. Caries Res. 2014, 48, 491–506. [Google Scholar] [CrossRef]
- Wang, X.; Shaffer, J.R.; Weyant, R.J.; Cuenco, K.T.; DeSensi, R.S.; Crout, R.; McNeil, D.W.; Marazita, M.L. Genes and their effects on dental caries may differ between primary and permanent dentitions. Caries Res. 2010, 44, 277–284. [Google Scholar] [CrossRef]
- Colombo, N.H.; Ribas, L.F.; Pereira, J.A.; Kreling, P.F.; Kressirer, C.A.; Tanner, A.C.; Duque, C. Antimicrobial peptides in saliva of children with severe early childhood caries. Arch. Oral Biol. 2016, 69, 40–46. [Google Scholar] [CrossRef]
- Fourie, N.H.; Wang, D.; Abey, S.K.; Sherwin, L.B.; Joseph, P.V.; Rahim-Williams, B.; Ferguson, E.G.; Henderson, W.A. The microbiome of the oral mucosa in irritable bowel syndrome. Gut Microbes 2016, 7, 286–301. [Google Scholar] [CrossRef]
- Kilian, M.; Chapple, I.L.; Hannig, M.; Marsh, P.D.; Meuric, V.; Pedersen, A.M.; Tonetti, M.S.; Wade, W.G.; Zaura, E. The oral microbiome—An update for oral healthcare professionals. Br. Dent. J. 2016, 221, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Jin, Y.; Chen, G.; Ma, X.; Zhang, L. Gut microbiota dysbiosis drives the development of colorectal cancer. Digestion 2021, 102, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, I.B.; Quigley, E.M.; Ohman, L.; Simren, M.; O’Toole, P.W. The microbiota link to irritable bowel syndrome: An emerging story. Gut Microbes 2012, 3, 572–576. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.R.; Pop, M.; Deboy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic analysis of the human distal gut microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef] [PubMed]
- Kianoush, N.; Adler, C.J.; Nguyen, K.A.; Browne, G.V.; Simonian, M.; Hunter, N. Bacterial profile of dentine caries and the impact of pH on bacterial population diversity. PLoS ONE 2014, 9, e92940. [Google Scholar] [CrossRef]
- Black, G.V. The Formation of Poisons by Micro-Organisms. A Biological Study of the Germ Theory of Disease; P Blakiston, Son & Co.: Philadelphia, PA, USA, 1884. [Google Scholar]
- Miller, W.D. The Micro-Organisms of the Human Mouth. The Local and General Diseas Which Are Caused by Them; The S. S. White Dental MFG. Co.: Philadelphia, PA, USA, 1890. [Google Scholar]
- Howe, P.R.; Hatch, R.E. A study of the microorganisms of dental caries. J. Med. Res. 1917, 36, 481–492.5. [Google Scholar] [CrossRef] [PubMed]
- Rosebury, T. Acid production and tolerance of lactobacilli from dental caries and other sources as measured by the glass electrode. J. Bacteriol. 1932, 24, 321–334. [Google Scholar] [CrossRef]
- Clarke, J.K. On the bacterial factor in the etiology of dental caries. Br. J. Exp. Pathol. 1924, 5, 141–147. [Google Scholar]
- Orland, F.J.; Blayney, J.R.; Harrison, R.W.; Reyniers, J.A.; Trexler, P.C.; Wagner, M.; Gordon, H.A.; Luckey, T.D. Use of the germfree animal technic in the study of experimental dental caries. I. Basic observations on rats reared free of all microorganisms. J. Dent. Res. 1954, 33, 147–174. [Google Scholar] [CrossRef]
- Keyes, P.H. The infectious and transmissible nature of experimental dental caries. Findings and implications. Arch. Oral Biol. 1960, 1, 304–320. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, R.J.; Keyes, P.H. Demonstration of the etiologic role of streptococci in experimental caries in the hamster. J. Am. Dent. Assoc. 1960, 61, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Edwardsson, S. Characteristics of caries-inducing human streptococci resembling Streptococcus mutans. Arch. Oral Biol. 1968, 13, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Loesche, W.J.; Rowan, J.; Straffon, L.H.; Loos, P.J. Association of Streptococcus mutans with human dental decay. Infect. Immun. 1975, 11, 1252–1260. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, T.; Sandham, H.J.; Bradley, E.L., Jr. Changes in Streptococcus mutans and lactobacilli in plaque in relation to the initiation of dental caries in Negro children. Arch. Oral Biol. 1973, 18, 555–566. [Google Scholar] [CrossRef] [PubMed]
- van Houte, J.; Green, D.B. Relationship between the concentration of bacteria in saliva and the colonization of teeth in humans. Infect. Immun. 1974, 9, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Schamschula, R.G.; Charlton, G. A study of caries aetiology in New South Wales schoolchildren. I. The streptococcal flora of plaque and caries prevalence. Aust. Dent. J. 1971, 16, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Marsh, P.D. Dental plaque as a biofilm and a microbial community—Implications for health and disease. BMC Oral Health 2006, 6 (Suppl. S1), S14. [Google Scholar] [CrossRef]
- Bradshaw, D.J.; McKee, A.S.; Marsh, P.D. Effects of carbohydrate pulses and pH on population shifts within oral microbial communities in vitro. J. Dent. Res. 1989, 68, 1298–1302. [Google Scholar] [CrossRef]
- Costalonga, M.; Herzberg, M.C. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol. Lett. 2014, 162, 22–38. [Google Scholar] [CrossRef]
- Takahashi, N.; Nyvad, B. The role of bacteria in the caries process: Ecological perspectives. J. Dent. Res. 2011, 90, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Nyvad, B. Ecological hypothesis of dentin and root caries. Caries Res. 2016, 50, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Kolenbrander, P.E.; Palmer, R.J., Jr.; Periasamy, S.; Jakubovics, N.S. Oral multispecies biofilm development and the key role of cell–cell distance. Nat. Rev. Microbiol. 2010, 8, 471. [Google Scholar] [CrossRef] [PubMed]
- Schweigel, H.; Wicht, M.; Schwendicke, F. Salivary and pellicle proteome: A datamining analysis. Sci. Rep. 2016, 6, 38882. [Google Scholar] [CrossRef] [PubMed]
- Chawhuaveang, D.D.; Yu, O.Y.; Yin, I.X.; Lam, W.Y.; Mei, M.L.; Chu, C.H. Acquired salivary pellicle and oral diseases: A literature review. J. Dent. Sci. 2021, 16, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Marsh, P.D.; Do, T.; Beighton, D.; Devine, D.A. Influence of saliva on the oral microbiota. Periodontology 2000 2016, 70, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, R.J.; Hay, D.I.; Childs, W.C., 3rd; Davis, G. Role of cryptic receptors (cryptitopes) in bacterial adhesion to oral surfaces. Arch. Oral Biol. 1990, 35, 107S–114S. [Google Scholar] [CrossRef] [PubMed]
- Jack, A.A.; Daniels, D.E.; Jepson, M.A.; Vickerman, M.M.; Lamont, R.J.; Jenkinson, H.F.; Nobbs, A.H. Streptococcus gordonii comCDE (competence) operon modulates biofilm formation with Candida albicans. Microbiology 2015, 161, 411–421. [Google Scholar] [CrossRef]
- Oguchi, R.; Takahashi, Y.; Shimazu, K.; Urano-Tashiro, Y.; Kawarai, T.; Konishi, K.; Karibe, H. Contribution of Streptococcus gordonii Hsa Adhesin to Biofilm Formation. Jpn. J. Infect. Dis. 2017, 70, 399–404. [Google Scholar] [CrossRef]
- Gibbins, H.L.; Yakubov, G.E.; Proctor, G.B.; Wilson, S.; Carpenter, G.H. What interactions drive the salivary mucosal pellicle formation? Colloids Surf. B Biointerfaces 2014, 120, 184–192. [Google Scholar] [CrossRef]
- Nadell, C.D.; Xavier, J.B.; Levin, S.A.; Foster, K.R. The evolution of quorum sensing in bacterial biofilms. PLoS Biol. 2008, 6, e14. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.C.; Rostami, N.; Casement, J.; Vollmer, W.; Rickard, A.H.; Jakubovics, N.S. ArcR modulates biofilm formation in the dental plaque colonizer Streptococcus gordonii. Mol. Oral Microbiol. 2018, 33, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Bowen, W.H.; Burne, R.A.; Wu, H.; Koo, H. Oral biofilms: Pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 2018, 26, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Sethi, A.; Mohanty, B.; Ramasubbu, N.; Gooley, P.R. Structure of amylase-binding protein A of Streptococcus gordonii: A potential receptor for human salivary alpha-amylase enzyme. Protein Sci. 2015, 24, 1013–1018. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Li, M.; Gregory, R.L. Bacterial interactions in dental biofilm. Virulence 2011, 2, 435–444. [Google Scholar] [CrossRef]
- Haase, E.M.; Kou, Y.; Sabharwal, A.; Liao, Y.C.; Lan, T.; Lindqvist, C.; Scannapieco, F.A. Comparative genomics and evolution of the amylase-binding proteins of oral streptococci. BMC Microbiol. 2017, 17, 94. [Google Scholar] [CrossRef]
- Burne, R.A.; Marquis, R.E. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol. Lett. 2000, 193, 1–6. [Google Scholar] [CrossRef]
- Nascimento, M.M.; Browngardt, C.; Xiaohui, X.; Klepac-Ceraj, V.; Paster, B.J.; Burne, R.A. The effect of arginine on oral biofilm communities. Mol. Oral Microbiol. 2014, 29, 45–54. [Google Scholar] [CrossRef]
- Huang, X.; Palmer, S.; Ahn, S.J.; Richards, V.P.; Williams, M.L.; Nascimento, M.M.; Burne, R.A. Characterization of a highly arginolytic Streptococcus species that potently antagonizes Streptococcus mutans. Appl. Environ. Microbiol. 2016, 82, 2187–2201. [Google Scholar] [CrossRef]
- Mashima, I.; Nakazawa, F. Identification of Veillonella tobetsuensis in tongue biofilm by using a species-specific primer pair. Anaerobe 2013, 22, 77–81. [Google Scholar] [CrossRef]
- Mashima, I.; Nakazawa, F. The interaction between Streptococcus spp. and Veillonella tobetsuensis in the early stages of oral biofilm formation. J. Bacteriol. 2015, 197, 2104–2111. [Google Scholar] [CrossRef] [PubMed]
- Listgarten, M.A. Structure of the microbial flora associated with periodontal health and disease in man. A light and electron microscopic study. J. Periodontol. 1976, 47, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Kolenbrander, P.E.; Andersen, R.N.; Blehert, D.S.; Egland, P.G.; Foster, J.S.; Palmer, R.J., Jr. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 2002, 66, 486–505. [Google Scholar] [CrossRef] [PubMed]
- Forssten, S.D.; Bjorklund, M.; Ouwehand, A.C. Streptococcus mutans, caries and simulation models. Nutrients 2010, 2, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Moye, Z.D.; Zeng, L.; Burne, R.A. Fueling the caries process: Carbohydrate metabolism and gene regulation by Streptococcus mutans. J. Oral Microbiol. 2014, 6, 24878. [Google Scholar] [CrossRef] [PubMed]
- Jakubovics, N.S. Intermicrobial interactions as a driver for community composition and stratification of oral biofilms. J. Mol. Biol. 2015, 427, 3662–3675. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yoshida, Y.; Cisar, J.O. Genetic basis of coaggregation receptor polysaccharide biosynthesis in Streptococcus sanguinis and related species. Mol. Oral Microbiol. 2014, 29, 24–31. [Google Scholar] [CrossRef]
- Simon-Soro, A.; Mira, A. Solving the etiology of dental caries. Trends Microbiol. 2015, 23, 76–82. [Google Scholar] [CrossRef]
- Loesche, W.J.; Syed, S.A. The predominant cultivable flora of carious plaque and carious dentine. Caries Res. 1973, 7, 201–216. [Google Scholar] [CrossRef]
- Svensater, G.; Borgstrom, M.; Bowden, G.H.; Edwardsson, S. The acid-tolerant microbiota associated with plaque from initial caries and healthy tooth surfaces. Caries Res. 2003, 37, 395–403. [Google Scholar] [CrossRef]
- Marchant, S.; Brailsford, S.R.; Twomey, A.C.; Roberts, G.J.; Beighton, D. The predominant microflora of nursing caries lesions. Caries Res. 2001, 35, 397–406. [Google Scholar] [CrossRef]
- Manganiello, A.D.; Socransky, S.S.; Smith, C.; Propas, D.; Oram, V.; Dogon, I.L. Attempts to increase viable count recovery of human supragingival dental plaque. J. Periodontal Res. 1977, 12, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.; Yu, W.H.; Lakshmanan, A.; Wade, W.G. The human oral microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef] [PubMed]
- Becker, M.R.; Paster, B.J.; Leys, E.J.; Moeschberger, M.L.; Kenyon, S.G.; Galvin, J.L.; Boches, S.K.; Dewhirst, F.E.; Griffen, A.L. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 2002, 40, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Corby, P.M.; Lyons-Weiler, J.; Bretz, W.A.; Hart, T.C.; Aas, J.A.; Boumenna, T.; Goss, J.; Corby, A.L.; Junior, H.M.; Weyant, R.J.; et al. Microbial risk indicators of early childhood caries. J. Clin. Microbiol. 2005, 43, 5753–5759. [Google Scholar] [CrossRef] [PubMed]
- Aas, J.A.; Paster, B.J.; Stokes, L.N.; Olsen, I.; Dewhirst, F.E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 2005, 43, 5721–5732. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ge, Y.; Saxena, D.; Caufield, P.W. Genetic profiling of the oral microbiota associated with severe early-childhood caries. J. Clin. Microbiol. 2007, 45, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Gross, E.L.; Beall, C.J.; Kutsch, S.R.; Firestone, N.D.; Leys, E.J.; Griffen, A.L. Beyond Streptococcus mutans: Dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS ONE 2012, 7, e47722. [Google Scholar] [CrossRef]
- Munson, M.A.; Banerjee, A.; Watson, T.F.; Wade, W.G. Molecular analysis of the microflora associated with dental caries. J. Clin. Microbiol. 2004, 42, 3023–3029. [Google Scholar] [CrossRef]
- Kanasi, E.; Dewhirst, F.E.; Chalmers, N.I.; Kent, R., Jr.; Moore, A.; Hughes, C.V.; Pradhan, N.; Loo, C.Y.; Tanner, A.C. Clonal analysis of the microbiota of severe early childhood caries. Caries Res. 2010, 44, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Tanner, A.C.; Mathney, J.M.; Kent, R.L.; Chalmers, N.I.; Hughes, C.V.; Loo, C.Y.; Pradhan, N.; Kanasi, E.; Hwang, J.; Dahlan, M.A.; et al. Cultivable anaerobic microbiota of severe early childhood caries. J. Clin. Microbiol. 2011, 49, 1464–1474. [Google Scholar] [CrossRef] [PubMed]
- Tanner, A.C.R.; Kressirer, C.A.; Rothmiller, S.; Johansson, I.; Chalmers, N.I. The caries microbiome: Implications for reversing dysbiosis. Adv. Dent. Res. 2018, 29, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Schulze-Schweifing, K.; Banerjee, A.; Wade, W.G. Comparison of bacterial culture and 16S rRNA community profiling by clonal analysis and pyrosequencing for the characterization of the dentine caries-associated microbiome. Front. Cell. Infect. Microbiol. 2014, 4, 164. [Google Scholar] [CrossRef] [PubMed]
- Buermans, H.P.; den Dunnen, J.T. Next generation sequencing technology: Advances and applications. Biochim. Biophys. Acta 2014, 1842, 1932–1941. [Google Scholar] [CrossRef] [PubMed]
- Ji, B.; Nielsen, J. From next-generation sequencing to systematic modeling of the gut microbiome. Front. Genet. 2015, 6, 219. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.T.; Derome, N.; Boyle, B.; Culley, A.I.; Charette, S.J. Next-generation sequencing (NGS) in the microbiological world: How to make the most of your money. J. Microbiol. Methods 2017, 138, 60–71. [Google Scholar] [CrossRef]
- Hardie, J.M.; Thomson, P.L.; South, R.J.; Marsh, P.D.; Bowden, G.H.; McKee, A.S.; Fillery, E.D.; Slack, G.L. A longitudinal epidemiological study on dental plaque and the development of dental caries–interim results after two years. J. Dent. Res. 1977, 56, C90–C98. [Google Scholar] [CrossRef]
- van Houte, J.; Sansone, C.; Joshipura, K.; Kent, R. In vitro acidogenic potential and mutans streptococci of human smooth-surface plaque associated with initial caries lesions and sound enamel. J. Dent. Res. 1991, 70, 1497–1502. [Google Scholar] [CrossRef]
- Boyar, R.M.; Thylstrup, A.; Holmen, L.; Bowden, G.H. The microflora associated with the development of initial enamel decalcification below orthodontic bands in vivo in children living in a fluoridated-water area. J. Dent. Res. 1989, 68, 1734–1738. [Google Scholar] [CrossRef]
- Tanner, A.C.; Sonis, A.L.; Lif Holgerson, P.; Starr, J.R.; Nunez, Y.; Kressirer, C.A.; Paster, B.J.; Johansson, I. White-spot lesions and gingivitis microbiotas in orthodontic patients. J. Dent. Res. 2012, 91, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Torlakovic, L.; Klepac-Ceraj, V.; Ogaard, B.; Cotton, S.L.; Paster, B.J.; Olsen, I. Microbial community succession on developing lesions on human enamel. J. Oral Microbiol. 2012, 4, e16125. [Google Scholar] [CrossRef] [PubMed]
- Aas, J.A.; Griffen, A.L.; Dardis, S.R.; Lee, A.M.; Olsen, I.; Dewhirst, F.E.; Leys, E.J.; Paster, B.J. Bacteria of dental caries in primary and permanent teeth in children and young adults. J. Clin. Microbiol. 2008, 46, 1407–1417. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Barraza, J.P.; Arthur, R.A.; Hara, A.; Lewis, K.; Liu, Y.; Scisci, E.L.; Hajishengallis, E.; Whiteley, M.; Koo, H. Spatial mapping of polymicrobial communities reveals a precise biogeography associated with human dental caries. Proc. Natl. Acad. Sci. USA 2020, 117, 12375–12386. [Google Scholar] [CrossRef]
- Syed, S.A.; Loesche, W.J.; Pape, H.L., Jr.; Grenier, E. Predominant cultivable flora isolated from human root surface caries plaque. Infect. Immun. 1975, 11, 727–731. [Google Scholar] [CrossRef]
- van Houte, J.; Lopman, J.; Kent, R. The final pH of bacteria comprising the predominant flora on sound and carious human root and enamel surfaces. J. Dent. Res. 1996, 75, 1008–1014. [Google Scholar] [CrossRef]
- Brailsford, S.R.; Shah, B.; Simons, D.; Gilbert, S.; Clark, D.; Ines, I.; Adams, S.E.; Allison, C.; Beighton, D. The predominant aciduric microflora of root-caries lesions. J. Dent. Res. 2001, 80, 1828–1833. [Google Scholar] [CrossRef]
- Ikebe, K.; Imazato, S.; Izutani, N.; Matsuda, K.; Ebisu, S.; Nokubi, T.; Walls, A.W. Association of salivary Streptococcus mutans levels determined by rapid detection system using monoclonal antibodies with prevalence of root surface caries. Am. J. Dent. 2008, 21, 283–287. [Google Scholar]
- Mantzourani, M.; Fenlon, M.; Beighton, D. Association between Bifidobacteriaceae and the clinical severity of root caries lesions. Oral Microbiol. Immunol. 2009, 24, 32–37. [Google Scholar] [CrossRef]
- Hashimoto, K.; Sato, T.; Shimauchi, H.; Takahashi, N. Profiling of dental plaque microflora on root caries lesions and the protein-denaturing activity of these bacteria. Am. J. Dent. 2011, 24, 295–299. [Google Scholar]
- Shen, S.; Samaranayake, L.P.; Yip, H.K.; Dyson, J.E. Bacterial and yeast flora of root surface caries in elderly, ethnic Chinese. Oral Dis. 2002, 8, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Preza, D.; Olsen, I.; Willumsen, T.; Boches, S.K.; Cotton, S.L.; Grinde, B.; Paster, B.J. Microarray analysis of the microflora of root caries in elderly. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2009, 28, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Qin, B.; Du, M.; Zhong, H.; Xu, Q.; Li, Y.; Zhang, P.; Fan, M. Extensive description and comparison of human supra-gingival microbiome in root caries and health. PLoS ONE 2015, 10, e0117064. [Google Scholar] [CrossRef]
- Dame-Teixeira, N.; Parolo, C.C.; Maltz, M.; Tugnait, A.; Devine, D.; Do, T. Actinomyces spp. gene expression in root caries lesions. J. Oral Microbiol. 2016, 8, 32383. [Google Scholar] [CrossRef]
- Lima, K.C.; Coelho, L.T.; Pinheiro, I.V.; Rocas, I.N.; Siqueira, J.F., Jr. Microbiota of dentinal caries as assessed by reverse-capture checkerboard analysis. Caries Res. 2011, 45, 21–30. [Google Scholar] [CrossRef]
- Liu, G.; Wu, C.; Abrams, W.R.; Li, Y. Structural and functional characteristics of the microbiome in deep-dentin caries. J. Dent. Res. 2020, 99, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Chhour, K.L.; Nadkarni, M.A.; Byun, R.; Martin, F.E.; Jacques, N.A.; Hunter, N. Molecular analysis of microbial diversity in advanced caries. J. Clin. Microbiol. 2005, 43, 843–849. [Google Scholar] [CrossRef] [PubMed]
- Kuribayashi, M.; Kitasako, Y.; Matin, K.; Sadr, A.; Shida, K.; Tagami, J. Intraoral pH measurement of carious lesions with qPCR of cariogenic bacteria to differentiate caries activity. J. Dent. 2012, 40, 222–228. [Google Scholar] [CrossRef]
- Kianoush, N.; Nguyen, K.A.; Browne, G.V.; Simonian, M.; Hunter, N. pH gradient and distribution of streptococci, lactobacilli, prevotellae, and fusobacteria in carious dentine. Clin. Oral Investig. 2014, 18, 659–669. [Google Scholar] [CrossRef]
- Do, T.; Dame-Teixeira, N.; Naginyte, M.; Marsh, P.D. Root surface biofilms and caries. Monogr. Oral Sci. 2017, 26, 26–34. [Google Scholar] [CrossRef]
- Kressirer, C.A.; Chen, T.; Lake Harriman, K.; Frias-Lopez, J.; Dewhirst, F.E.; Tavares, M.A.; Tanner, A.C. Functional profiles of coronal and dentin caries in children. J. Oral Microbiol. 2018, 10, 1495976. [Google Scholar] [CrossRef] [PubMed]
- Milnes, A.R.; Bowden, G.H. The microflora associated with developing lesions of nursing caries. Caries Res. 1985, 19, 289–297. [Google Scholar] [CrossRef]
- Berkowitz, R.J.; Jordan, H.V. Similarity of bacteriocins of Streptococcus mutans from mother and infant. Arch. Oral Biol. 1975, 20, 725–730. [Google Scholar] [CrossRef]
- Li, Y.; Caufield, P.W. The fidelity of initial acquisition of mutans streptococci by infants from their mothers. J. Dent. Res. 1995, 74, 681–685. [Google Scholar] [CrossRef] [PubMed]
- Kohler, B.; Lundberg, A.B.; Birkhed, D.; Papapanou, P.N. Longitudinal study of intrafamilial mutans streptococci ribotypes. Eur. J. Oral Sci. 2003, 111, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Tanner, A.C.; Kent, R.L., Jr.; Holgerson, P.L.; Hughes, C.V.; Loo, C.Y.; Kanasi, E.; Chalmers, N.I.; Johansson, I. Microbiota of severe early childhood caries before and after therapy. J. Dent. Res. 2011, 90, 1298–1305. [Google Scholar] [CrossRef] [PubMed]
- Hughes, C.V.; Dahlan, M.; Papadopolou, E.; Loo, C.Y.; Pradhan, N.S.; Lu, S.C.; Mathney, J.M.; Bravoco, A.; Kent, R.L., Jr.; Tanner, A.C. Aciduric microbiota and mutans streptococci in severe and recurrent severe early childhood caries. Pediatr. Dent. 2012, 34, e16–e23. [Google Scholar]
- Warren, J.J.; Blanchette, D.; Dawson, D.V.; Marshall, T.A.; Phipps, K.R.; Starr, D.; Drake, D.R. Factors associated with dental caries in a group of American Indian children at age 36 months. Community Dent. Oral Epidemiol. 2016, 44, 154–161. [Google Scholar] [CrossRef]
- Batliner, T.; Wilson, A.R.; Tiwari, T.; Glueck, D.; Henderson, W.; Thomas, J.; Braun, P.; Cudeii, D.; Quissell, D.; Albino, J. Oral health status in Navajo Nation Head Start children. J. Public Health Dent. 2014, 74, 317–325. [Google Scholar] [CrossRef]
- Agnello, M.; Marques, J.; Cen, L.; Mittermuller, B.; Huang, A.; Chaichanasakul Tran, N.; Shi, W.; He, X.; Schroth, R.J. Microbiome associated with severe caries in Canadian First Nations children. J. Dent. Res. 2017, 96, 1378–1385. [Google Scholar] [CrossRef]
- Villhauer, A.L.; Lynch, D.J.; Warren, J.J.; Dawson, D.V.; Blanchette, D.R.; Drake, D.R. Genotypic characterization and comparison of Streptococcus mutans in American Indian and Southeast Iowa children. Clin. Exp. Dent. Res. 2017, 3, 235–243. [Google Scholar] [CrossRef]
- Johansson, I.; Witkowska, E.; Kaveh, B.; Lif Holgerson, P.; Tanner, A.C. The microbiome in populations with a low and high prevalence of caries. J. Dent. Res. 2016, 95, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, L.; Lif Holgerson, P.; Esberg, A.; Johansson, I. Microbial complexes and caries in 17-year-olds with and without Streptococcus mutans. J. Dent. Res. 2018, 97, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.Z.; Zijnge, V.; Cicek, A.; de Soet, J.J.; Harmsen, H.J.; Huysmans, M.C. Shifts in the microbial population in relation to in situ caries progression. Caries Res. 2012, 46, 427–431. [Google Scholar] [CrossRef]
- Falsetta, M.L.; Klein, M.I.; Colonne, P.M.; Scott-Anne, K.; Gregoire, S.; Pai, C.H.; Gonzalez-Begne, M.; Watson, G.; Krysan, D.J.; Bowen, W.H.; et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect. Immun. 2014, 82, 1968–1981. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Huang, X.; Alkhers, N.; Alzamil, H.; Alzoubi, S.; Wu, T.T.; Castillo, D.A.; Campbell, F.; Davis, J.; Herzog, K.; et al. Candida albicans and early childhood caries: A systematic review and meta-analysis. Caries Res. 2018, 52, 102–112. [Google Scholar] [CrossRef]
- Janus, M.M.; Crielaard, W.; Volgenant, C.M.; van der Veen, M.H.; Brandt, B.W.; Krom, B.P. Candida albicans alters the bacterial microbiome of early in vitro oral biofilms. J. Oral Microbiol. 2017, 9, 1270613. [Google Scholar] [CrossRef]
- Diaz, P.I.; Xie, Z.; Sobue, T.; Thompson, A.; Biyikoglu, B.; Ricker, A.; Ikonomou, L.; Dongari-Bagtzoglou, A. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect. Immun. 2012, 80, 620–632. [Google Scholar] [CrossRef]
- Metwalli, K.H.; Khan, S.A.; Krom, B.P.; Jabra-Rizk, M.A. Streptococcus mutans, Candida albicans, and the human mouth: A sticky situation. PLoS Pathog. 2013, 9, e1003616. [Google Scholar] [CrossRef]
- He, J.; Kim, D.; Zhou, X.; Ahn, S.J.; Burne, R.A.; Richards, V.P.; Koo, H. RNA-Seq reveals enhanced sugar metabolism in Streptococcus mutans co-cultured with Candida albicans within mixed-species biofilms. Front. Microbiol. 2017, 8, 1036. [Google Scholar] [CrossRef]
- Kneist, S.; Borutta, A.; Sigusch, B.W.; Nietzsche, S.; Kupper, H.; Kostrzewa, M.; Callaway, A. First-time isolation of Candida dubliniensis from plaque and carious dentine of primary teeth. Eur. Arch. Paediatr. Dent. 2015, 16, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Al-Ahmad, A.; Auschill, T.M.; Dakhel, R.; Wittmer, A.; Pelz, K.; Heumann, C.; Hellwig, E.; Arweiler, N.B. Prevalence of Candida albicans and Candida dubliniensis in caries-free and caries-active children in relation to the oral microbiota-a clinical study. Clin. Oral Investig. 2016, 20, 1963–1971. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Grier, A.; Faustoferri, R.C.; Alzoubi, S.; Gill, A.L.; Feng, C.; Liu, Y.; Quivey, R.G.; Kopycka-Kedzierawski, D.T.; Koo, H.; et al. Association between oral candida and bacteriome in children with severe ECC. J. Dent. Res. 2018, 97, 1468–1476. [Google Scholar] [CrossRef] [PubMed]
- van Houte, J. Role of micro-organisms in caries etiology. J. Dent. Res. 1994, 73, 672–681. [Google Scholar] [CrossRef] [PubMed]
- Colby, S.M.; Russell, R.R.B. Sugar metabolism by mutans streptococci. J. Appl. Microbiol. 1997, 83, 80S–88S. [Google Scholar] [CrossRef] [PubMed]
- Argimon, S.; Konganti, K.; Chen, H.; Alekseyenko, A.V.; Brown, S.; Caufield, P.W. Comparative genomics of oral isolates of Streptococcus mutans by in silico genome subtraction does not reveal accessory DNA associated with severe early childhood caries. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2014, 21C, 269–278. [Google Scholar] [CrossRef]
- Argimon, S.; Caufield, P.W. Distribution of putative virulence genes in Streptococcus mutans strains does not correlate with caries experience. J. Clin. Microbiol. 2011, 49, 984–992. [Google Scholar] [CrossRef]
- Palmer, S.R.; Miller, J.H.; Abranches, J.; Zeng, L.; Lefebure, T.; Richards, V.P.; Lemos, J.A.; Stanhope, M.J.; Burne, R.A. Phenotypic heterogeneity of genomically-diverse isolates of Streptococcus mutans. PLoS ONE 2013, 8, e61358. [Google Scholar] [CrossRef]
- Esberg, A.; Sheng, N.; Marell, L.; Claesson, R.; Persson, K.; Boren, T.; Stromberg, N. Streptococcus mutans adhesin biotypes that match and predict individual caries development. EBioMedicine 2017, 24, 205–215. [Google Scholar] [CrossRef]
- Aviles-Reyes, A.; Miller, J.H.; Simpson-Haidaris, P.J.; Lemos, J.A.; Abranches, J. Cnm is a major virulence factor of invasive Streptococcus mutans and part of a conserved three-gene locus. Mol. Oral Microbiol. 2014, 29, 11–23. [Google Scholar] [CrossRef]
- Rainey, K.; Michalek, S.M.; Wen, Z.T.; Wu, H. Glycosyltransferase mediated biofilm matrix dynamics and virulence of Streptococcus mutans. Appl. Environ. Microbiol. 2019, 85, e02247-18. [Google Scholar] [CrossRef] [PubMed]
- Kleinberg, I. A mixed-bacteria ecological approach to understanding the role of the oral bacteria in dental caries causation: An alternative to Streptococcus mutans and the specific-plaque hypothesis. Crit. Rev. Oral Biol. Med. 2002, 13, 108–125. [Google Scholar] [CrossRef] [PubMed]
- Bowden, G.H. Does assessment of microbial composition of plaque/saliva allow for diagnosis of disease activity of individuals? Community Dent. Oral Epidemiol. 1997, 25, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Coeuret, V.; Dubernet, S.; Bernardeau, M.; Gueguen, M.; Vernoux, J.P. Isolation, characterisation and identification of lactobacilli focusing mainly on cheeses and other dairy products. Lait 2003, 83, 269–306. [Google Scholar] [CrossRef]
- Samot, J.; Lebreton, J.; Badet, C. Adherence capacities of oral lactobacilli for potential probiotic purposes. Anaerobe 2011, 17, 69–72. [Google Scholar] [CrossRef]
- van Pijkeren, J.P.; O’Toole, P.W. Comparative and functional genomics of the genus Lactobacillus. In Lactobacillus Molecular Biology from Genomics to Probiotics; Ljungh, A., Wadstrom, T., Eds.; Caister Academic Press: Norfold, UK, 2009; pp. 59–82. [Google Scholar]
- Badet, C.; Thebaud, N.B. Ecology of lactobacilli in the oral cavity: A review of literature. Open Microbiol. J. 2008, 2, 38–48. [Google Scholar] [CrossRef]
- Plonka, K.A.; Pukallus, M.L.; Barnett, A.G.; Walsh, L.J.; Holcombe, T.H.; Seow, W.K. Mutans streptococci and lactobacilli colonization in predentate children from the neonatal period to seven months of age. Caries Res. 2012, 46, 213–220. [Google Scholar] [CrossRef]
- Nelun Barfod, M.; Magnusson, K.; Lexner, M.O.; Blomqvist, S.; Dahlen, G.; Twetman, S. Oral microflora in infants delivered vaginally and by caesarean section. Int. J. Paediatr. Dent./Br. Paedodontic Soc. Int. Assoc. Dent. Child. 2011, 21, 401–406. [Google Scholar] [CrossRef]
- Holgerson, P.L.; Vestman, N.R.; Claesson, R.; Ohman, C.; Domellof, M.; Tanner, A.C.; Hernell, O.; Johansson, I. Oral microbial profile discriminates breast-fed from formula-fed infants. J. Pediatr. Gastroenterol. Nutr. 2013, 56, 127–136. [Google Scholar] [CrossRef]
- Teanpaisan, R.; Chaethong, W.; Piwat, S.; Thitasomakul, S. Vertical transmission of mutans streptococci and lactobacilli in Thai families. Pediatr. Dent. 2012, 34, e24–e29. [Google Scholar]
- Teanpaisan, R.; Thitasomakul, S.; Piwat, S.; Thearmontree, A.; Pithpornchaiyakul, W.; Chankanka, O. Longitudinal study of the presence of mutans streptococci and lactobacilli in relation to dental caries development in 3–24 month old Thai children. Int. Dent. J. 2007, 57, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Loesche, W.J.; Eklund, S.; Earnest, R.; Burt, B. Longitudinal investigation of bacteriology of human fissure decay: Epidemiological studies in molars shortly after eruption. Infect. Immun. 1984, 46, 765–772. [Google Scholar] [CrossRef] [PubMed]
- Roeters, F.J.; van der Hoeven, J.S.; Burgersdijk, R.C.; Schaeken, M.J. Lactobacilli, mutants streptococci and dental caries: A longitudinal study in 2-year-old children up to the age of 5 years. Caries Res. 1995, 29, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Kanasi, E.; Johansson, I.; Lu, S.C.; Kressin, N.R.; Nunn, M.E.; Kent, R., Jr.; Tanner, A.C. Microbial risk markers for childhood caries in pediatricians’ offices. J. Dent. Res. 2010, 89, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Caufield, P.W.; Li, Y.; Dasanayake, A.; Saxena, D. Diversity of lactobacilli in the oral cavities of young women with dental caries. Caries Res. 2007, 41, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Argimon, S.; Li, Y.; Gu, H.; Zhou, X.; Caufield, P.W. Determining the genetic diversity of lactobacilli from the oral cavity. J. Microbiol. Methods 2010, 82, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Argimon, S.; Casey, S.; Saraithong, P.; Schon, C.; Li, Y.; Caufield, P. Comparison of bacterial culture and culture-independent 16S rRNA gene sequencing for the study of Lactobacillus diversity in children with severe early childhood caries. In Proceedings of the 5th ASM Conference on Beneficial Microbes, Washington, DC, USA, 27–30 September 2014. [Google Scholar]
- Kneist, S.; Schmidt, F.; Callaway, A.; Willershausen, B.; Rupf, S.; Wicht, M.; Thiede, B. Diversity of Lactobacillus species in deep carious lesions of primary molars. Eur. Arch. Paediatr. Dent. 2010, 11, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Rocas, I.N.; Alves, F.R.; Rachid, C.T.; Lima, K.C.; Assuncao, I.V.; Gomes, P.N.; Siqueira, J.F., Jr. Microbiome of deep dentinal caries lesions in teeth with symptomatic irreversible pulpitis. PLoS ONE 2016, 11, e0154653. [Google Scholar] [CrossRef]
- Caufield, P.W.; Schon, C.N.; Saraithong, P.; Li, Y.; Argimon, S. Oral lactobacilli and dental caries: A model for niche adaptation in humans. J. Dent. Res. 2015, 94, 110S–118S. [Google Scholar] [CrossRef]
- Twetman, S.; Fritzon, B.; Jensen, B.; Hallberg, U.; Stahl, B. Pre- and post-treatment levels of salivary mutans streptococci and lactobacilli in pre-school children. Int. J. Paediatr. Dent./Br. Paedodontic Soc. Int. Assoc. Dent. Child. 1999, 9, 93–98. [Google Scholar] [CrossRef]
- Klinke, T.; Urban, M.; Luck, C.; Hannig, C.; Kuhn, M.; Kramer, N. Changes in Candida spp., mutans streptococci and lactobacilli following treatment of early childhood caries: A 1-year follow-up. Caries Res. 2014, 48, 24–31. [Google Scholar] [CrossRef]
- Callaway, A.; Kostrzewa, M.; Willershausen, B.; Schmidt, F.; Thiede, B.; Kupper, H.; Kneist, S. Identification of lactobacilli from deep carious lesions by means of species-specific PCR and MALDI-TOF mass spectrometry. Clin. Lab. 2013, 59, 1373–1379. [Google Scholar] [CrossRef]
- Dasanayake, A.P.; Chhun, N.; Tanner, A.C.; Craig, R.G.; Lee, M.J.; Moore, A.F.; Norman, R.G. Periodontal pathogens and gestational diabetes mellitus. J. Dent. Res. 2008, 87, 328–333. [Google Scholar] [CrossRef]
- Harper, D.S.; Loesche, W.J. Growth and acid tolerance of human dental plaque bacteria. Arch. Oral Biol. 1984, 29, 843–848. [Google Scholar] [CrossRef]
- Behbehani, M.J.; Jordan, H.V.; Heeley, J.D. Oral colonization and pathogenicity of Actinomyces israelii in gnotobiotic rats. J. Dent. Res. 1983, 62, 69–74. [Google Scholar] [CrossRef]
- van Houte, J.; van Palenstein-Helderman, W.H. Cariogenic potential of Bifidobacterium in gnotobiotic rats. J. Dent. Res. 1980, 59, 1176. [Google Scholar] [CrossRef]
- Kressirer, C.A.; Smith, D.J.; King, W.F.; Dobeck, J.M.; Starr, J.K.; Tanner, A.C. Scardovia wiggsiae and its potential role as a caries pathogen. J. Oral Biosci. 2017, 59, 135–141. [Google Scholar] [CrossRef]
- Mantzourani, M.; Gilbert, S.C.; Sulong, H.N.; Sheehy, E.C.; Tank, S.; Fenlon, M.; Beighton, D. The isolation of bifidobacteria from occlusal carious lesions in children and adults. Caries Res. 2009, 43, 308–313. [Google Scholar] [CrossRef]
- Kaur, R.; Gilbert, S.C.; Sheehy, E.C.; Beighton, D. Salivary levels of bifidobacteria in caries-free and caries-active children. Int. J. Paediatr. Dent./Br. Paedodontic Soc. Int. Assoc. Dent. Child. 2013, 23, 32–38. [Google Scholar] [CrossRef]
- de Matos, B.M.; Brighenti, F.L.; Do, T.; Beighton, D.; Koga-Ito, C.Y. Acidogenicity of dual-species biofilms of bifidobacteria and Streptococcus mutans. Clin. Oral Investig. 2017, 21, 1769–1776. [Google Scholar] [CrossRef]
- Alcaraz, L.D.; Belda-Ferre, P.; Cabrera-Rubio, R.; Romero, H.; Simon-Soro, A.; Pignatelli, M.; Mira, A. Identifying a healthy oral microbiome through metagenomics. Clin. Microbiol. Infect. 2012, 18 (Suppl. S4), 54–57. [Google Scholar] [CrossRef]
- Peterson, S.N.; Snesrud, E.; Schork, N.J.; Bretz, W.A. Dental caries pathogenicity: A genomic and metagenomic perspective. Int. Dent. J. 2011, 61 (Suppl. S1), 11–22. [Google Scholar] [CrossRef]
- Vaishampayan, P.A.; Kuehl, J.V.; Froula, J.L.; Morgan, J.L.; Ochman, H.; Francino, M.P. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol. Evol. 2010, 2, 53–66. [Google Scholar] [CrossRef]
- Thomas, T.; Gilbert, J.; Meyer, F. Metagenomics—A guide from sampling to data analysis. Microb. Inform. Exp. 2012, 2, 3. [Google Scholar] [CrossRef]
- Wang, W.L.; Xu, S.Y.; Ren, Z.G.; Tao, L.; Jiang, J.W.; Zheng, S.S. Application of metagenomics in the human gut microbiome. World J. Gastroenterol. 2015, 21, 803–814. [Google Scholar] [CrossRef]
- Jovel, J.; Patterson, J.; Wang, W.; Hotte, N.; O’Keefe, S.; Mitchel, T.; Perry, T.; Kao, D.; Mason, A.L.; Madsen, K.L.; et al. Characterization of the Gut Microbiome Using 16S or Shotgun Metagenomics. Front. Microbiol. 2016, 7, 459. [Google Scholar] [CrossRef]
- Xu, P.; Gunsolley, J. Application of metagenomics in understanding oral health and disease. Virulence 2014, 5, 424–432. [Google Scholar] [CrossRef]
- Wang, J.; Qi, J.; Zhao, H.; He, S.; Zhang, Y.; Wei, S.; Zhao, F. Metagenomic sequencing reveals microbiota and its functional potential associated with periodontal disease. Sci. Rep. 2013, 3, 1843. [Google Scholar] [CrossRef]
- Xie, G.; Chain, P.S.; Lo, C.C.; Liu, K.L.; Gans, J.; Merritt, J.; Qi, F. Community and gene composition of a human dental plaque microbiota obtained by metagenomic sequencing. Mol. Oral Microbiol. 2010, 25, 391–405. [Google Scholar] [CrossRef]
- Belda-Ferre, P.; Alcaraz, L.D.; Cabrera-Rubio, R.; Romero, H.; Simon-Soro, A.; Pignatelli, M.; Mira, A. The oral metagenome in health and disease. ISME J. 2012, 6, 46–56. [Google Scholar] [CrossRef]
- Takahashi, N.; Yamada, T. Effects of pH on the glucose and lactate metabolisms by the washed cells of Actinomyces naeslundii under anaerobic and aerobic conditions. Oral Microbiol. Immunol. 1999, 14, 60–65. [Google Scholar] [CrossRef]
- Lemos, J.A.; Abranches, J.; Burne, R.A. Responses of cariogenic streptococci to environmental stresses. Curr. Issues Mol. Biol. 2005, 7, 95–107. [Google Scholar]
- Jorth, P.; Turner, K.H.; Gumus, P.; Nizam, N.; Buduneli, N.; Whiteley, M. Metatranscriptomics of the human oral microbiome during health and disease. mBio 2014, 5, e01012–e01014. [Google Scholar] [CrossRef]
- Takahashi, N. Oral microbiome metabolism: From “Who Are They?” to “What Are They Doing?”. J. Dent. Res. 2015, 94, 1628–1637. [Google Scholar] [CrossRef]
- Simon-Soro, A.; Guillen-Navarro, M.; Mira, A. Metatranscriptomics reveals overall active bacterial composition in caries lesions. J. Oral Microbiol. 2014, 6, 25443. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Darveau, R.P.; Curtis, M.A. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 2012, 10, 717–725. [Google Scholar] [CrossRef]
- Peterson, S.N.; Meissner, T.; Su, A.I.; Snesrud, E.; Ong, A.C.; Schork, N.J.; Bretz, W.A. Functional expression of dental plaque microbiota. Front. Cell. Infect. Microbiol. 2014, 4, 108. [Google Scholar] [CrossRef]
- May, A.; Brandt, B.W.; El-Kebir, M.; Klau, G.W.; Zaura, E.; Crielaard, W.; Heringa, J.; Abeln, S. metaModules identifies key functional subnetworks in microbiome-related disease. Bioinformatics 2016, 32, 1678–1685. [Google Scholar] [CrossRef]
- Belstrom, D.; Constancias, F.; Liu, Y.; Yang, L.; Drautz-Moses, D.I.; Schuster, S.C.; Kohli, G.S.; Jakobsen, T.H.; Holmstrup, P.; Givskov, M. Metagenomic and metatranscriptomic analysis of saliva reveals disease-associated microbiota in patients with periodontitis and dental caries. NPJ Biofilms Microbiomes 2017, 3, 23. [Google Scholar] [CrossRef]
- Duran-Pinedo, A.E.; Frias-Lopez, J. Beyond microbial community composition: Functional activities of the oral microbiome in health and disease. Microbes Infect. 2015, 17, 505–516. [Google Scholar] [CrossRef]
- Jagtap, P.; McGowan, T.; Bandhakavi, S.; Tu, Z.J.; Seymour, S.; Griffin, T.J.; Rudney, J.D. Deep metaproteomic analysis of human salivary supernatant. Proteomics 2012, 12, 992–1001. [Google Scholar] [CrossRef]
- Takahashi, N.; Washio, J.; Mayanagi, G. Metabolomics of supragingival plaque and oral bacteria. J. Dent. Res. 2010, 89, 1383–1388. [Google Scholar] [CrossRef]
- Edlund, A.; Yang, Y.; Yooseph, S.; Hall, A.P.; Nguyen, D.D.; Dorrestein, P.C.; Nelson, K.E.; He, X.; Lux, R.; Shi, W.; et al. Meta-omics uncover temporal regulation of pathways across oral microbiome genera during in vitro sugar metabolism. ISME J. 2015, 9, 2605–2619. [Google Scholar] [CrossRef] [PubMed]
- Edlund, A.; Garg, N.; Mohimani, H.; Gurevich, A.; He, X.; Shi, W.; Dorrestein, P.C.; McLean, J.S. Metabolic fingerprints from the human oral microbiome reveal a vast knowledge gap of secreted small peptidic molecules. mSystems 2017, 2, e00058-17. [Google Scholar] [CrossRef] [PubMed]
- Axelsson, P.; Lindhe, J. The effect of a plaque control program on gingivitis and dental caries in schoolchildren. J. Dent. Res. 1977, 56, C142–C148. [Google Scholar] [CrossRef]
- Marsh, P.D. In sickness and in health-what does the oral microbiome mean to us? An ecological perspective. Adv. Dent. Res. 2018, 29, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Loesche, W.J.; Bradbury, D.R.; Woolfolk, M.P. Reduction of dental decay in rampant caries individuals following short-term kanamycin treatment. J. Dent. Res. 1977, 56, 254–265. [Google Scholar] [CrossRef]
- de Paola, P.F.; Jordan, H.V.; Berg, J. Temporary suppression of Streptococcus mutans in humans through topical application of vancomycin. J. Dent. Res. 1974, 53, 108–114. [Google Scholar] [CrossRef]
- Li, Y.; Tanner, A. Effect of antimicrobial interventions on the oral microbiota associated with early childhood caries. Pediatr. Dent. 2015, 37, 226–244. [Google Scholar]
- Huang, X.; Schulte, R.M.; Burne, R.A.; Nascimento, M.M. Characterization of the arginolytic microflora provides insights into pH homeostasis in human oral biofilms. Caries Res. 2015, 49, 165–176. [Google Scholar] [CrossRef]
- Koopman, J.E.; Roling, W.F.; Buijs, M.J.; Sissons, C.H.; ten Cate, J.M.; Keijser, B.J.; Crielaard, W.; Zaura, E. Stability and resilience of oral microcosms toward acidification and Candida outgrowth by arginine supplementation. Microb. Ecol. 2015, 69, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.; Murata, R.M.; Duarte, S. Antimicrobial traits of tea- and cranberry-derived polyphenols against Streptococcus mutans. Caries Res. 2011, 45, 327–335. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, D.L.; Araujo Lima, R.; Zanin, I.C.; Klein, M.I.; Janal, M.N.; Duarte, S. Effect of twice-daily blue light treatment on matrix-rich biofilm development. PLoS ONE 2015, 10, e0131941. [Google Scholar] [CrossRef] [PubMed]
- Twetman, S.; Dhar, V. Evidence of Effectiveness of Current Therapies to Prevent and Treat Early Childhood Caries. Pediatr. Dent. 2015, 37, 246–253. [Google Scholar]
- O’Mullane, D.M.; Baez, R.J.; Jones, S.; Lennon, M.A.; Petersen, P.E.; Rugg-Gunn, A.J.; Whelton, H.; Whitford, G.M. Fluoride and oral health. Community Dent. Health 2016, 33, 69–99. [Google Scholar] [PubMed]
- Slade, G.D.; Grider, W.B.; Maas, W.R.; Sanders, A.E. Water fluoridation and dental caries in U.S. children and adolescents. J. Dent. Res. 2018, 97, 1122–1128. [Google Scholar] [CrossRef] [PubMed]
- Allukian, M., Jr.; Carter-Pokras, O.D.; Gooch, B.F.; Horowitz, A.M.; Iida, H.; Jacob, M.; Kleinman, D.V.; Kumar, J.; Maas, W.R.; Pollick, H.; et al. Science, politics, and communication: The case of community water fluoridation in the US. Ann. Epidemiol. 2018, 28, 401–410. [Google Scholar] [CrossRef]
- Kilian, M.; Larsen, M.J.; Fejerskov, O.; Thylstrup, A. Effects of fluoride on the initial colonization of teeth in vivo. Caries Res. 1979, 13, 319–329. [Google Scholar] [CrossRef]
- van Loveren, C.; Gerardu, V.A.; Sissons, C.H.; van Bekkum, M.; ten Cate, J.M. Effect of various rinsing protocols after use of amine fluoride/stannous fluoride toothpaste on the bacterial composition of dental plaque. Caries Res. 2009, 43, 462–467. [Google Scholar] [CrossRef]
- Hamilton, I.R. Effects of fluoride on enzymatic regulation of bacterial carbohydrate metabolism. Caries Res. 1977, 11 (Suppl. S1), 262–291. [Google Scholar] [CrossRef]
- Marquis, R.E.; Clock, S.A.; Mota-Meira, M. Fluoride and organic weak acids as modulators of microbial physiology. FEMS Microbiol. Rev. 2003, 26, 493–510. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, I.R. Biochemical effects of fluoride on oral bacteria. J. Dent. Res. 1990, 69, 660–667, discussion 682–663. [Google Scholar] [CrossRef]
- Bowden, G.H. Effects of fluoride on the microbial ecology of dental plaque. J. Dent. Res. 1990, 69, 653–659, discussion 682–653. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, D.J.; Marsh, P.D.; Hodgson, R.J.; Visser, J.M. Effects of glucose and fluoride on competition and metabolism within in vitro dental bacterial communities and biofilms. Caries Res. 2002, 36, 81–86. [Google Scholar] [CrossRef]
- Ahumada Ostengo, M.d.C.; Wiese, B.; Nader-Macias, M.E. Inhibitory effect of sodium fluoride and chlorhexidine on the growth of oral lactobacilli. Can. J. Microbiol. 2005, 51, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Twetman, S.; Mattiasson, A.; Varela; Bratthall, D. Mutans streptococci in saliva and dental caries in children living in a high and a low fluoride area. Oral Microbiol. Immunol. 1990, 5, 169–171. [Google Scholar] [CrossRef] [PubMed]
- Marinho, V.C.; Chong, L.Y.; Worthington, H.V.; Walsh, T. Fluoride mouthrinses for preventing dental caries in children and adolescents. Cochrane Database Syst. Rev. 2016, 7, CD002284. [Google Scholar] [CrossRef] [PubMed]
- Thurnheer, T.; Belibasakis, G.N. Effect of sodium fluoride on oral biofilm microbiota and enamel demineralization. Arch. Oral Biol. 2018, 89, 77–83. [Google Scholar] [CrossRef]
- Koopman, J.E.; van der Kaaij, N.C.; Buijs, M.J.; Elyassi, Y.; van der Veen, M.H.; Crielaard, W.; Ten Cate, J.M.; Zaura, E. The effect of fixed orthodontic appliances and fluoride mouthwash on the oral microbiome of adolescents—A randomized controlled clinical trial. PLoS ONE 2015, 10, e0137318. [Google Scholar] [CrossRef]
- Reilly, C.; Rasmussen, K.; Selberg, T.; Stevens, J.; Jones, R.S. Biofilm community diversity after exposure to 0.4% stannous fluoride gels. J. Appl. Microbiol. 2014, 117, 1798–1809. [Google Scholar] [CrossRef]
- Reilly, C.; Goettl, M.; Steinmetz, M.; Nikrad, J.; Jones, R.S. Short-term effects of povidone iodine and sodium fluoride therapy on plaque levels and microbiome diversity. Oral Dis. 2016, 22, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Marinho, V.C.; Worthington, H.V.; Walsh, T.; Clarkson, J.E. Fluoride varnishes for preventing dental caries in children and adolescents. Cochrane Database Syst. Rev. 2013, 7, CD002279. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, H.P.; Binguis, D.; Douglas, J.; McKeown, L.; Switzer, B.; Figueiredo, R.; Laporte, A. A 2-year community-randomized controlled trial of fluoride varnish to prevent early childhood caries in Aboriginal children. Community Dent. Oral Epidemiol. 2008, 36, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Holve, S. An observational study of the association of fluoride varnish applied during well child visits and the prevention of early childhood caries in American Indian children. Matern. Child Health J. 2008, 12 (Suppl. S1), 64–67. [Google Scholar] [CrossRef]
- Weintraub, J.A.; Ramos-Gomez, F.; Jue, B.; Shain, S.; Hoover, C.I.; Featherstone, J.D.; Gansky, S.A. Fluoride varnish efficacy in preventing early childhood caries. J. Dent. Res. 2006, 85, 172–176. [Google Scholar] [CrossRef] [PubMed]
- American Academy of Pediatric Dentistry. Guideline on caries-risk assessment and management for infants, children, and adolescents. Pediatr. Dent. 2016, 38, 142–149. [Google Scholar]
- Paul, S.; Baranya Shrikrishna, S.; Suman, E.; Shenoy, R.; Rao, A. Effect of fluoride varnish and chlorhexidine-thymol varnish on mutans streptococci levels in human dental plaque: A double-blinded randomized controlled trial. Int. J. Paediatr. Dent./Br. Paedodontic Soc. Int. Assoc. Dent. Child. 2014, 24, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Paek, A.E.; Li, Y.; Wang, Z.; So, P.; Janal, M.N.; Herman, N.G.; Hopkins, A.; Chinn, C. Caries outcome following an intensive fluoride varnish treatment regimen for children at high risk for early childhood caries. Int. J. Paediatr. Dent./Br. Paedodontic Soc. Int. Assoc. Dent. Child. 2018, 28, 291–299. [Google Scholar] [CrossRef]
- Ramos-Gomez, F.J.; Gansky, S.A.; Featherstone, J.D.; Jue, B.; Gonzalez-Beristain, R.; Santo, W.; Martinez, E.; Weintraub, J.A. Mother and youth access (MAYA) maternal chlorhexidine, counselling and paediatric fluoride varnish randomized clinical trial to prevent early childhood caries. Int. J. Paediatr. Dent./Br. Paedodontic Soc. Int. Assoc. Dent. Child. 2012, 22, 169–179. [Google Scholar] [CrossRef]
- Lobo, P.L.; de Carvalho, C.B.; Fonseca, S.G.; de Castro, R.S.; Monteiro, A.J.; Fonteles, M.C.; Fonteles, C.S. Sodium fluoride and chlorhexidine effect in the inhibition of mutans streptococci in children with dental caries: A randomized, double-blind clinical trial. Oral Microbiol. Immunol. 2008, 23, 486–491. [Google Scholar] [CrossRef]
- Weinstein, P.; Spiekerman, C.; Milgrom, P. Randomized equivalence trial of intensive and semiannual applications of fluoride varnish in the primary dentition. Caries Res. 2009, 43, 484–490. [Google Scholar] [CrossRef] [PubMed]
- Berger, T.J.; Spadaro, J.A.; Chapin, S.E.; Becker, R.O. Electrically generated silver ions: Quantitative effects on bacterial and mammalian cells. Antimicrob. Agents Chemother. 1976, 9, 357–358. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.S.; Zhang, S.; Mei, M.L.; Lo, E.C.; Chu, C.H. Caries remineralisation and arresting effect in children by professionally applied fluoride treatment—A systematic review. BMC Oral Health 2016, 16, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Crystal, Y.O.; Niederman, R. Evidence-based dentistry update on silver diamine fluoride. Dent. Clin. N. Am. 2019, 63, 45–68. [Google Scholar] [CrossRef] [PubMed]
- Llodra, J.C.; Rodriguez, A.; Ferrer, B.; Menardia, V.; Ramos, T.; Morato, M. Efficacy of silver diamine fluoride for caries reduction in primary teeth and first permanent molars of schoolchildren: 36-month clinical trial. J. Dent. Res. 2005, 84, 721–724. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.Y.; Lo, E.C.; Chu, C.H.; Lin, H.C. Randomized trial on fluorides and sealants for fissure caries prevention. J. Dent. Res. 2012, 91, 753–758. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.P.; Lo, E.C.; Dyson, J.E.; Luo, Y.; Corbet, E.F. A randomized trial on root caries prevention in elders. J. Dent. Res. 2010, 89, 1086–1090. [Google Scholar] [CrossRef]
- Rosenblatt, A.; Stamford, T.C.; Niederman, R. Silver diamine fluoride: A caries “silver-fluoride bullet”. J. Dent. Res. 2009, 88, 116–125. [Google Scholar] [CrossRef]
- Suzuki, T.; Sobue, S.; Suginaka, H. Mechanism of antiplaque action of diamine silver fluoride. J. Osaka Univ. Dent. Sch. 1976, 16, 87–95. [Google Scholar]
- Hiraishi, N.; Yiu, C.K.; King, N.M.; Tagami, J.; Tay, F.R. Antimicrobial efficacy of 3.8% silver diamine fluoride and its effect on root dentin. J. Endod. 2010, 36, 1026–1029. [Google Scholar] [CrossRef]
- Knight, G.M.; McIntyre, J.M.; Craig, G.G.; Mulyani; Zilm, P.S.; Gully, N.J. Inability to form a biofilm of Streptococcus mutans on silver fluoride- and potassium iodide-treated demineralized dentin. Quintessence Int. 2009, 40, 155–161. [Google Scholar] [PubMed]
- Knight, G.M.; McIntyre, J.M.; Craig, G.G.; Mulyani; Zilm, P.S.; Gully, N.J. Differences between normal and demineralized dentine pretreated with silver fluoride and potassium iodide after an in vitro challenge by Streptococcus mutans. Aust. Dent. J. 2007, 52, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.H.; Mei, L.; Seneviratne, C.J.; Lo, E.C. Effects of silver diamine fluoride on dentine carious lesions induced by Streptococcus mutans and Actinomyces naeslundii biofilms. Int. J. Paediatr. Dent./Br. Paedodontic Soc. Int. Assoc. Dent. Child. 2012, 22, 2–10. [Google Scholar] [CrossRef]
- Mei, M.L.; Li, Q.L.; Chu, C.H.; Lo, E.C.; Samaranayake, L.P. Antibacterial effects of silver diamine fluoride on multi-species cariogenic biofilm on caries. Ann. Clin. Microbiol. Antimicrob. 2013, 12, 4. [Google Scholar] [CrossRef] [PubMed]
- Hamama, H.H.; Yiu, C.K.; Burrow, M.F. Effect of silver diamine fluoride and potassium iodide on residual bacteria in dentinal tubules. Aust. Dent. J. 2015, 60, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Yu, O.Y.; Zhao, I.S.; Mei, M.L.; Lo, E.C.M.; Chu, C.H. Caries-arresting effects of silver diamine fluoride and sodium fluoride on dentine caries lesions. J. Dent. 2018, 78, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Marinho, V.C.; Worthington, H.V.; Walsh, T.; Chong, L.Y. Fluoride gels for preventing dental caries in children and adolescents. Cochrane Database Syst. Rev. 2015, 6, CD002280. [Google Scholar] [CrossRef]
- Walsh, T.; Worthington, H.; Glenny, A.; Appelbe, P.; Marinho, V.; Shi, X. Fluoride toothpastes of different concentrations for preventing dental caries in children and adolescents. Cochrane Database Syst. Rev. 2010, 1, CD007868. [Google Scholar] [CrossRef]
- Rugg-Gunn, A. Dental caries: Strategies to control this preventable disease. Acta Med. Acad. 2013, 42, 117–130. [Google Scholar] [CrossRef]
- Yamaga, R.; Nishino, M.; Yoshida, S.; Yokomizo, I. Diammine silver fluoride and its clinical application. J. Osaka Univ. Dent. Sch. 1972, 12, 1–20. [Google Scholar]
- Sinha, N.; Gupta, A.; Logani, A.; Shah, N. Remineralizing efficacy of silver diamine fluoride and glass ionomer type VII for their proposed use as indirect pulp capping materials—Part II (A clinical study). J. Conserv. Dent. 2011, 14, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Mei, M.L.; Lo, E.C.M.; Chu, C.H. Arresting dentine caries with silver diamine fluoride: What’s behind it? J. Dent. Res. 2018, 97, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Y.; Psoterc, W.J.; Nguyena, O.M.; Bromage, T.G.; Walters, M.A.; Hu, B.; Rabieh, S.; Kumararaja, F.C. ORIG Assessment of the silver penetration and distribution in carious lesions of deciduous teeth treated with silver diamine fluoride. Caries Res. 2019, 53, 431–440. [Google Scholar] [CrossRef]
- Crystal, Y.O.; Marghalani, A.A.; Ureles, S.D.; Wright, J.T.; Sulyanto, R.; Divaris, K.; Fontana, M.; Graham, L. Use of silver diamine fluoride for dental caries management in children and adolescents, including those with special health care needs. Pediatr. Dent. 2017, 39, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Clemens, J.; Gold, J.; Chaffin, J. Effect and acceptance of silver diamine fluoride treatment on dental caries in primary teeth. J. Public Health Dent. 2018, 78, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Holtz, R.D.; Souza Filho, A.G.; Brocchi, M.; Martins, D.; Duran, N.; Alves, O.L. Development of nanostructured silver vanadates decorated with silver nanoparticles as a novel antibacterial agent. Nanotechnology 2010, 21, 185102. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Sierra, J.F.; Ruiz, F.; Pena, D.C.; Martinez-Gutierrez, F.; Martinez, A.E.; Guillen Ade, J.; Tapia-Perez, H.; Castanon, G.M. The antimicrobial sensitivity of Streptococcus mutans to nanoparticles of silver, zinc oxide, and gold. Nanomed. Nanotechnol. Biol. Med. 2008, 4, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, V.E., Jr.; Filho, A.V.; Ribeiro Targino, A.G.; Pelagio Flores, M.A.; Galembeck, A.; Caldas, A.F., Jr.; Rosenblatt, A. A New “Silver-Bullet” to treat caries in children—Nano Silver Fluoride: A randomised clinical trial. J. Dent. 2014, 42, 945–951. [Google Scholar] [CrossRef]
- de Castro, D.T.; Valente, M.L.; da Silva, C.H.; Watanabe, E.; Siqueira, R.L.; Schiavon, M.A.; Alves, O.L.; Dos Reis, A.C. Evaluation of antibiofilm and mechanical properties of new nanocomposites based on acrylic resins and silver vanadate nanoparticles. Arch. Oral Biol. 2016, 67, 46–53. [Google Scholar] [CrossRef]
- de Castro, D.T.a.; do Nascimento, C.; Alves, O.L.; de Souza Santos, E.; Agnelli, J.A.M.; Dos Reis, A.C. Analysis of the oral microbiome on the surface of modified dental polymers. Arch. Oral Biol. 2018, 93, 107–114. [Google Scholar] [CrossRef]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, H.H.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar]
- Keeney, K.M.; Yurist-Doutsch, S.; Arrieta, M.C.; Finlay, B.B. Effects of antibiotics on human microbiota and subsequent disease. Annu. Rev. Microbiol. 2014, 68, 217–235. [Google Scholar] [CrossRef]
- Taubman, M.A.; Nash, D.A. The scientific and public-health imperative for a vaccine against dental caries. Nat. Rev. Immunol. 2006, 6, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Batista, M.T.; Ferreira, E.L.; Pereira, G.S.; Stafford, P.; Maeda, D.; Rodrigues, J.F.; Brady, L.J.; Johnston, S.A.; Ferreira, L.C.S.; Ferreira, R.C.C. LT adjuvant modulates epitope specificity and improves the efficacy of murine antibodies elicited by sublingual vaccination with the N-terminal domain of Streptococcus mutans P1. Vaccine 2017, 35, 7273–7282. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Hu, Y.; Yang, M.; Liu, H.; Jiang, G. Enhanced immune response to a dual-promoter anti-caries DNA vaccine orally delivered by attenuated Salmonella typhimurium. Immunobiology 2017, 222, 730–737. [Google Scholar] [CrossRef]
- St Michael, F.; Yang, Q.; Cairns, C.; Vinogradov, E.; Fleming, P.; Hayes, A.C.; Aubry, A.; Cox, A.D. Investigating the candidacy of the serotype specific rhamnan polysaccharide based glycoconjugates to prevent disease caused by the dental pathogen Streptococcus mutans. Glycoconj. J. 2018, 35, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, J.A.; Hilton, J.F.; White, J.M.; Hoover, C.I.; Wycoff, K.L.; Yu, L.; Larrick, J.W.; Featherstone, J.D. Clinical trial of a plant-derived antibody on recolonization of mutans streptococci. Caries Res. 2005, 39, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Szafranski, S.P.; Winkel, A.; Stiesch, M. The use of bacteriophages to biocontrol oral biofilms. J. Biotechnol. 2017, 250, 29–44. [Google Scholar] [CrossRef]
- Hoare, A.; Marsh, P.D.; Diaz, P.I. Ecological therapeutic opportunities for oral diseases. Microbiol. Spectr. 2017, 5, 10-1128. [Google Scholar] [CrossRef]
- Philip, N.; Suneja, B.; Walsh, L.J. Ecological approaches to dental caries prevention: Paradigm shift or shibboleth? Caries Res. 2018, 52, 153–165. [Google Scholar] [CrossRef]
- Lopez-Lopez, A.; Camelo-Castillo, A.; Ferrer, M.D.; Simon-Soro, A.; Mira, A. Health-associated niche inhabitants as oral probiotics: The case of Streptococcus dentisani. Front. Microbiol. 2017, 8, 379. [Google Scholar] [CrossRef] [PubMed]
- Burne, R.A. Getting to know “the known unknowns”: Heterogeneity in the oral microbiome. Adv. Dent. Res. 2018, 29, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Gruner, D.; Paris, S.; Schwendicke, F. Probiotics for managing caries and periodontitis: Systematic review and meta-analysis. J. Dent. 2016, 48, 16–25. [Google Scholar] [CrossRef]
- Garcia, S.S.; Blackledge, M.S.; Michalek, S.; Su, L.; Ptacek, T.; Eipers, P.; Morrow, C.; Lefkowitz, E.J.; Melander, C.; Wu, H. Targeting of Streptococcus mutans biofilms by a novel small molecule prevents dental caries and preserves the oral microbiome. J. Dent. Res. 2017, 96, 807–814. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Cui, T.; Zeng, J.; Chen, L.; Zhang, W.; Xu, X.; Cheng, L.; Li, M.; Li, J.; Zhou, X.; et al. Molecule targeting glucosyltransferase inhibits Streptococcus mutans biofilm formation and virulence. Antimicrob. Agents Chemother. 2016, 60, 126–135. [Google Scholar] [CrossRef]
- Chen, L.; Jia, L.; Zhang, Q.; Zhou, X.; Liu, Z.; Li, B.; Zhu, Z.; Wang, F.; Yu, C.; Zhang, Q.; et al. A novel antimicrobial peptide against dental-caries-associated bacteria. Anaerobe 2017, 47, 165–172. [Google Scholar] [CrossRef]
- Ding, Y.; Wang, W.; Fan, M.; Tong, Z.; Kuang, R.; Jiang, W.; Ni, L. Antimicrobial and anti-biofilm effect of Bac8c on major bacteria associated with dental caries and Streptococcus mutans biofilms. Peptides 2014, 52, 61–67. [Google Scholar] [CrossRef]
- Saputo, S.; Faustoferri, R.C.; Quivey, R.G., Jr. Vitamin D compounds are bactericidal against Streptococcus mutans and target the bacitracin-associated efflux system. Antimicrob. Agents Chemother. 2018, 62, e01675-17. [Google Scholar] [CrossRef]
- Eckert, R.; Qi, F.; Yarbrough, D.K.; He, J.; Anderson, M.H.; Shi, W. Adding selectivity to antimicrobial peptides: Rational design of a multidomain peptide against Pseudomonas spp. Antimicrob. Agents Chemother. 2006, 50, 1480–1488. [Google Scholar] [CrossRef]
- Eckert, R.; He, J.; Yarbrough, D.K.; Qi, F.; Anderson, M.H.; Shi, W. Targeted killing of Streptococcus mutans by a pheromone-guided “smart” antimicrobial peptide. Antimicrob. Agents Chemother. 2006, 50, 3651–3657. [Google Scholar] [CrossRef]
- Sullivan, R.; Santarpia, P.; Lavender, S.; Gittins, E.; Liu, Z.; Anderson, M.H.; He, J.; Shi, W.; Eckert, R. Clinical efficacy of a specifically targeted antimicrobial peptide mouth rinse: Targeted elimination of Streptococcus mutans and prevention of demineralization. Caries Res. 2011, 45, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; McLean, J.S.; Yang, Y.; Eckert, R.; Kaplan, C.W.; Kyme, P.; Sheikh, O.; Varnum, B.; Lux, R.; Shi, W.; et al. Precision-guided antimicrobial peptide as a targeted modulator of human microbial ecology. Proc. Natl. Acad. Sci. USA 2015, 112, 7569–7574. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, C.W.; Sim, J.H.; Shah, K.R.; Kolesnikova-Kaplan, A.; Shi, W.; Eckert, R. Selective membrane disruption: Mode of action of C16G2, a specifically targeted antimicrobial peptide. Antimicrob. Agents Chemother. 2011, 55, 3446–3452. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Reipa, V.; Liu, G.; Meng, Y.; Wang, X.; Mineart, K.P.; Prabhu, V.M.; Shi, W.; Lin, N.J.; He, X.; et al. pH-sensitive compounds for selective inhibition of acid-producing bacteria. ACS Appl. Mater. Interfaces 2018, 10, 8566–8573. [Google Scholar] [CrossRef] [PubMed]
- Horev, B.; Klein, M.I.; Hwang, G.; Li, Y.; Kim, D.; Koo, H.; Benoit, D.S. pH-activated nanoparticles for controlled topical delivery of farnesol to disrupt oral biofilm virulence. ACS Nano 2015, 9, 2390–2404. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Liu, Y.; Kim, D.; Li, Y.; Hwang, G.; Naha, P.C.; Cormode, D.P.; Koo, H. Nanocatalysts promote Streptococcus mutans biofilm matrix degradation and enhance bacterial killing to suppress dental caries in vivo. Biomaterials 2016, 101, 272–284. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, Y.; Kim, D.; Ren, Z.; Oh, M.J.; Cormode, D.P.; Hara, A.T.; Zero, D.T.; Koo, H. Ferumoxytol nanoparticles target biofilms causing tooth decay in the human mouth. Nano Lett. 2021, 21, 9442–9449. [Google Scholar] [CrossRef]
- Selwitz, R.H.; Ismail, A.I.; Pitts, N.B. Dental caries. Lancet 2007, 369, 51–59. [Google Scholar] [CrossRef]
- Divaris, K. Predicting dental caries outcomes in children: A “risky” concept. J. Dent. Res. 2016, 95, 248–254. [Google Scholar] [CrossRef]
- Tanner, A.C.; Kressirer, C.A.; Faller, L.L. Understanding caries from the oral microbiome perspective. J. Calif. Dent. Assoc. 2016, 44, 437–446. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.; Allan, R.N.; Howlin, R.P.; Stoodley, P.; Hall-Stoodley, L. Targeting microbial biofilms: Current and prospective therapeutic strategies. Nat. Rev. Microbiol. 2017, 15, 740–755. [Google Scholar] [CrossRef] [PubMed]
- Belda-Ferre, P.; Williamson, J.; Simon-Soro, A.; Artacho, A.; Jensen, O.N.; Mira, A. The human oral metaproteome reveals potential biomarkers for caries disease. Proteomics 2015, 15, 3497–3507. [Google Scholar] [CrossRef] [PubMed]
- Teng, F.; Yang, F.; Huang, S.; Bo, C.; Xu, Z.Z.; Amir, A.; Knight, R.; Ling, J.; Xu, J. Prediction of early childhood caries via spatial-temporal variations of oral microbiota. Cell Host Microbe 2015, 18, 296–306. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spatafora, G.; Li, Y.; He, X.; Cowan, A.; Tanner, A.C.R. The Evolving Microbiome of Dental Caries. Microorganisms 2024, 12, 121. https://doi.org/10.3390/microorganisms12010121
Spatafora G, Li Y, He X, Cowan A, Tanner ACR. The Evolving Microbiome of Dental Caries. Microorganisms. 2024; 12(1):121. https://doi.org/10.3390/microorganisms12010121
Chicago/Turabian StyleSpatafora, Grace, Yihong Li, Xuesong He, Annie Cowan, and Anne C. R. Tanner. 2024. "The Evolving Microbiome of Dental Caries" Microorganisms 12, no. 1: 121. https://doi.org/10.3390/microorganisms12010121





