Freshwater Sponges as a Neglected Reservoir of Bacterial Biodiversity
Abstract
:1. Introduction
2. Prokaryotic Communities Associated with Freshwater Sponges: Main Findings by Culture-Independent Approaches
2.1. Ephydatia spp.
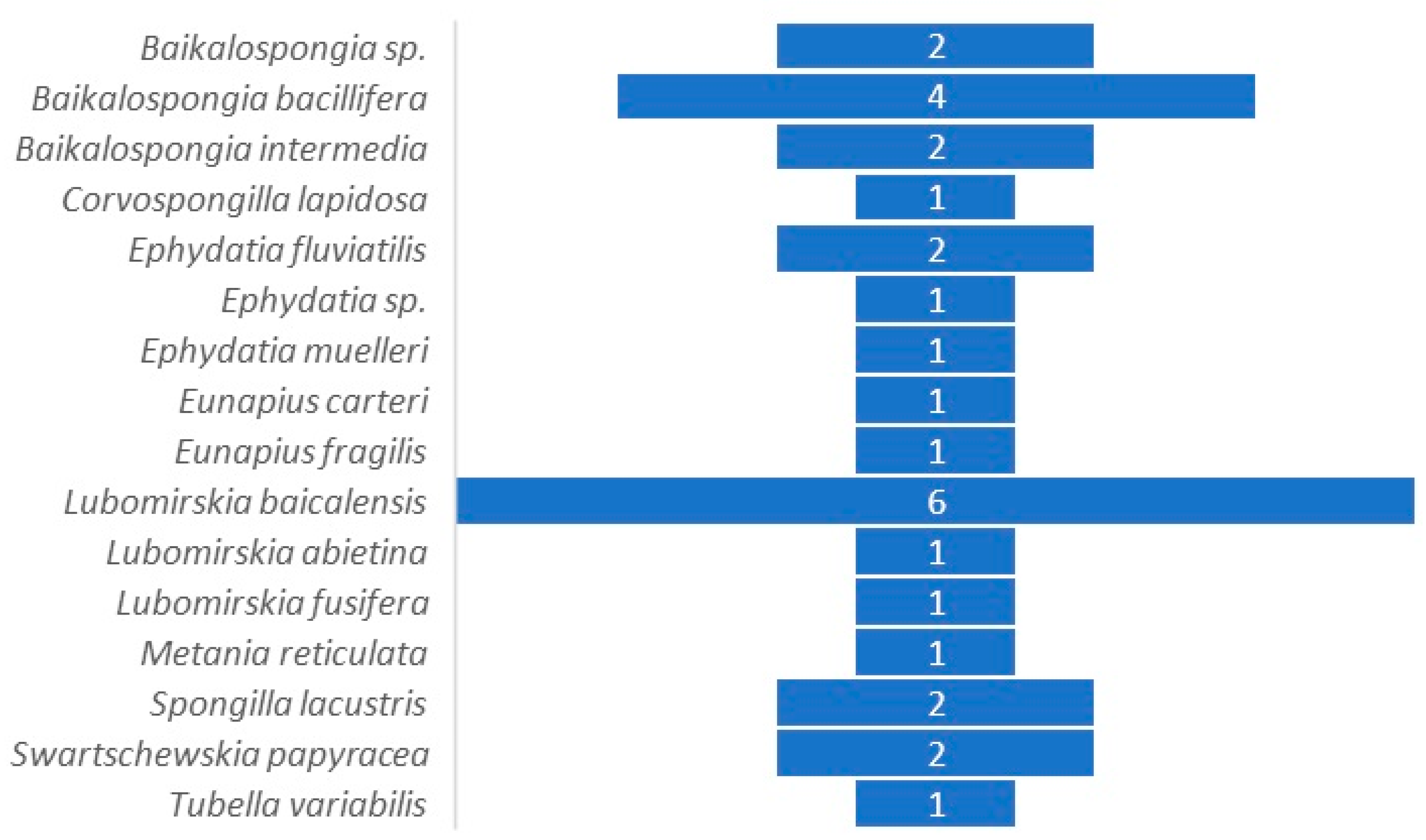
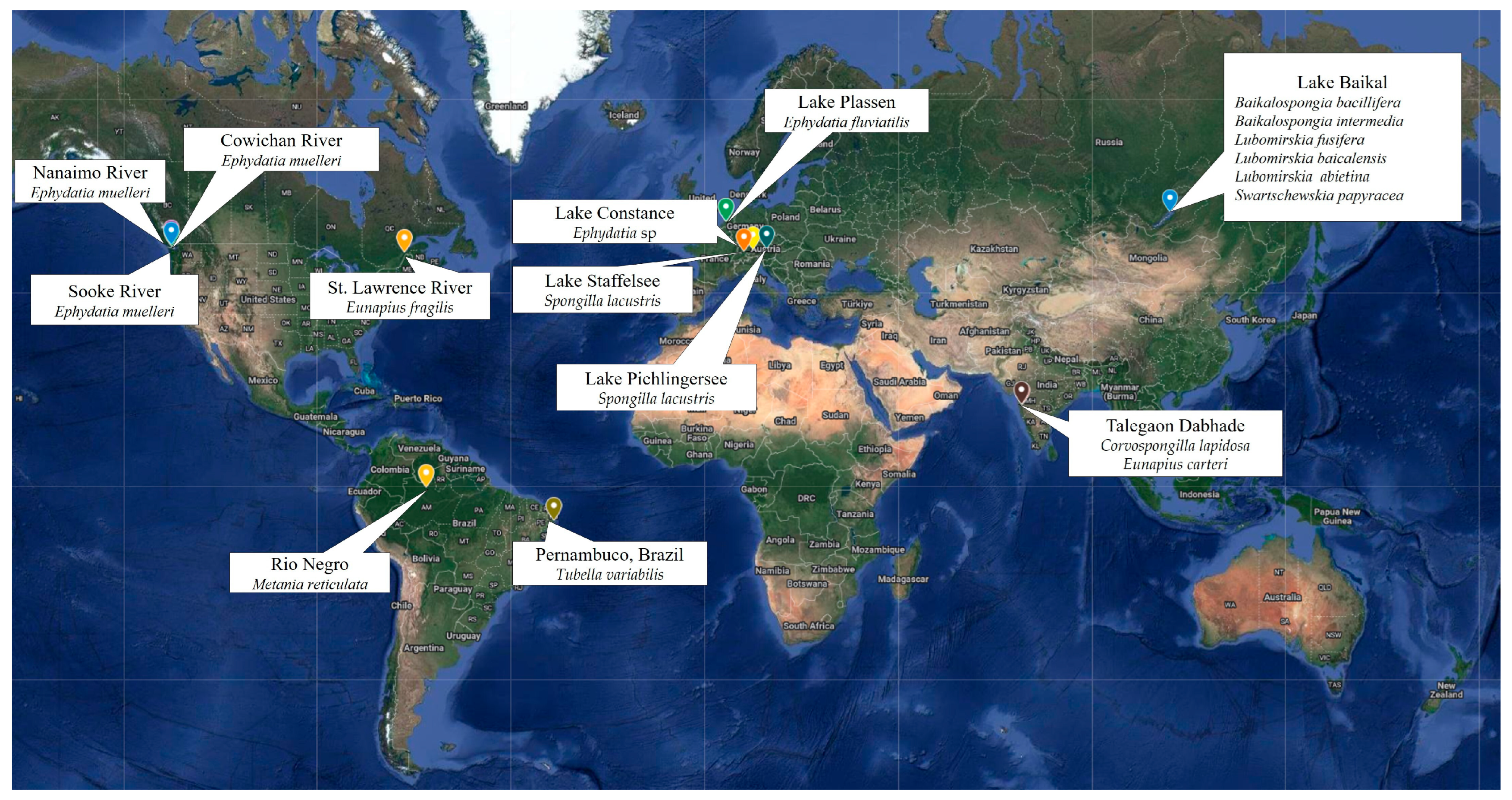
| Sponge Species | Freshwater Sampling Site | Reference(s) |
|---|---|---|
| Baikalospongia sp. | Lake Baikal (Russia) | [30,31] |
| Baikalospongia bacillifera (Dybowsky, 1880) | Lake Baikal (Russia) | [22] |
| Baikalospongia intermedia (Dybowski, 1880) | Lake Baikal (Russia) | [32] |
| Corvospongilla lapidosa (Annandale, 1908) | Talegaon Dabhade and Pashan (India) | [20] |
| Ephydatia fluviatilis (Linnaeus, 1759) | Vinkeveense Plassen Lake (The Netherlands) | [25,28] |
| Ephydatia muelleri (Lieberkühn, 1856) | Six locations in the Northern Hemisphere | [22] |
| Sooke, Cowichan, and Nanaimo Rivers (Canada) | [2] | |
| Eunapius carteri (Bowerbank, 1863) | Talegaon Dabhade and Pashan (India) | [20] |
| Lubomirskia baicalensis (Pallas, 1776) | Lake Baikal (Russia) | [15,30,31,32,33] |
| L. abietina (Swartschewsky, 1901) | Lake Baikal (Russia) | [33] |
| Spongilla lacustris (Linnaeus, 1759) | Lake Staffelsee (Germany) | [1] |
| Pichlinger See Lake (Upper Austria) | [23] | |
| Swartschewskia papyracea (Dybowsky, 1880) | Lake Baikal (Russia) | [32] |
| Tubella variabilis (Bonetto and Ezcurra de Drago, 1973) | Artificial channel (Brazil) | [21] |
2.2. Eunapius carteri and Corvospongilla lapidosa
2.3. Lubomirskiidae Family from the Lake Baikal
2.4. Spongilla lacustris
2.5. Tubella variabilis
3. Bacterial Isolates from Freshwater Sponges: Description and Biotechnological Potential
3.1. Characterization of the Cultivable Fraction of the Freshwater Sponges-Associated Bacterial Communities
3.1.1. Baikalospongia bacillifera, B. intermedia, Lubomirskia fusifera, L. baicalensis and Swartschewskia papyracea from Lake Baikal
3.1.2. Ephydatia sp.
3.1.3. Eunapius fragilis
3.1.4. Spongilla lacustris
3.1.5. Tubella variabilis
3.2. Biotechnological Relevant Bacteria from Freshwater Sponges
3.2.1. Potential for the Production of Bioactive Compounds by Bacterial Isolates
3.2.2. Biosynthesis of Enzymes by Bacterial Isolates
3.2.3. Polyketide Synthase Encoding Genes within the Associated Bacterial Communities
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gernert, C.; Glöckner, F.O.; Krohne, G.; Hentschel, U. Microbial diversity of the freshwater sponge Spongilla lacustris. Microb. Ecol. 2005, 50, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Sugden, S.; Holert, J.; Cardenas, E.; Mohn, W.W.; Stein, L.Y. Microbiome of the freshwater sponge Ephydatia muelleri shares compositional and functional similarities with those of marine sponges. ISME J. 2022, 16, 2503–2512. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.J. The functional roles of marine sponges. Estuar. Coast. Shelf Sci. 2008, 79, 341–353. [Google Scholar] [CrossRef]
- de Goeij, J.M.; van Oevelen, D.; Vermeij, M.J.; Osinga, R.; Middelburg, J.J.; de Goeij, A.F.; Admiraal, W. Surviving in a marine desert: The sponge loop retains resources within coral reefs. Science 2013, 342, 108–110. [Google Scholar] [CrossRef] [PubMed]
- Folkers, M.; Rombouts, T. Sponges revealed: A synthesis of their overlooked ecological functions within aquatic ecosystems. In YOUMARES 9—The Oceans: Our Research, Our Future; Jungblut, S., Liebich, V., Bode-Dalby, M., Eds.; Springer: Cham, Switzerland, 2020. [Google Scholar] [CrossRef]
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Moitinho-Silva, L.; Lurgi, M.; Björk, J.R.; Easson, C.; Astudillo-García, C.; Olson, J.B.; Erwin, P.M.; López-Legentil, S.; Luter, H.; et al. Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 2016, 7, 11870. [Google Scholar] [CrossRef] [PubMed]
- Hentschel, U.; Fieseler, L.; Wehrl, M.; Gernert, C.; Steinert, M.; Hacker, J.; Horn, M. Microbial diversity of marine sponges. In Marine Molecular Biotechnology; Mueller, W., Ed.; Springer: Berlin/Heidelberg, Germany, 2003; pp. 59–88. [Google Scholar]
- Engelberts, J.P.; Robbins, S.J.; de Goeij, J.M.; Aranda, M.; Bell, S.C.; Webster, N.S. Characterization of a sponge microbiome using an integrative genome-centric approach. ISME J. 2020, 14, 1100–1110. [Google Scholar] [CrossRef]
- Webster, N.S.; Thomas, T. The sponge hologenome. MBio 2016, 7, e00135-16. [Google Scholar] [CrossRef]
- Manconi, R.; Pronzato, R. Global diversity of sponges (Porifera: Spongillina) in freshwater. Hydrobiologia 2008, 595, 27–33. [Google Scholar] [CrossRef]
- Erpenbeck, D.; Galitz, A.; Wörheide, G.; Albrecht, C.; Pronzato, R.; Manconi, R. Having the balls to colonize—The Ephydatia fluviatilis group and the origin of (ancient) Lake “endemic” sponge lineages. J. Gt. Lakes Res. 2020, 46, 1140–1145. [Google Scholar] [CrossRef]
- Wilkinson, C.R. Nutrient translocation from green algal symbionts to the freshwater sponge Ephydatia fluviatilis. Hydrobiologia 1980, 75, 241–250. [Google Scholar] [CrossRef]
- Parfenova, V.; Terkina, I.; Kostornova, T.Y.; Nikulina, I.G.; Chernykh, V.I.; Maksimova, E.A. Microbial community of freshwater sponges in Lake Baikal. Biol. Bull. 2008, 35, 374–379. [Google Scholar] [CrossRef]
- Kaluzhnaya, O.; Krivich, A.; Itskovich, V. Diversity of 16S rRNA genes in metagenomic community of the freshwater sponge Lubomirskia baicalensis. Russ. J. Genet. 2012, 48, 855–858. [Google Scholar] [CrossRef]
- Belikov, S.; Belkova, N.; Butina, T.; Chernogor, L.; Kley, A.M.; Nalian, A.; Rorex, C.; Khanaev, I.; Maikova, O.; Feranchuk, S. Diversity and shifts of the bacterial community associated with Baikal sponge mass mortalities. PLoS ONE 2019, 14, e0213926. [Google Scholar] [CrossRef] [PubMed]
- Chernogor, L.; Klimenko, E.; Khanaev, I.; Belikov, S. Microbiome analysis of healthy and diseased sponges Lubomirskia baicalensis by using cell cultures of primmorphs. Peer J. 2020, 8, e9080. [Google Scholar] [CrossRef] [PubMed]
- Petrushin, I.; Belikov, S.; Chernogor, L. Cooperative interaction of Janthinobacterium sp. Slb01 and Flavobacterium sp. slb02 in the diseased sponge Lubomirskia baicalensis. Int. J. Mol. Sci. 2020, 21, 8128. [Google Scholar] [CrossRef] [PubMed]
- Kulakova, N.V.; Sakirko, M.V.; Adelshin, R.V.; Khanaev, I.V.; Nebesnykh, I.A.; Pérez, T. Brown rot syndrome and changes in the bacterial сommunity of the Baikal sponge Lubomirskia baicalensis. Microb. Ecol. 2018, 75, 1024–1034. [Google Scholar] [CrossRef]
- Gaikwad, S.; Shouche, Y.S.; Gade, W.N. Microbial community structure of two freshwater sponges using Illumina MiSeq sequencing revealed high microbial diversity. AMB Express 2016, 6, 40. [Google Scholar] [CrossRef]
- Laport, M.S.; Pinheiro, U.; da Costa Rachid, C.T.C. Freshwater sponge Tubella variabilis presents richer microbiota than marine sponge species. Front. Microbiol. 2019, 10, 2799. [Google Scholar] [CrossRef]
- Kenny, N.J.; Francis, W.R.; Rivera-Vicéns, R.E.; Juravel, K.; de Mendoza, A.; Díez-Vives, C.; Lister, R.; Bezares-Calderón, L.A.; Grombacher, L.; Roller, M.; et al. Tracing animal genomic evolution with the chromosomal-level assembly of the freshwater sponge Ephydatia muelleri. Nat. Commun. 2020, 11, 3676. [Google Scholar] [CrossRef]
- Graffius, S.; Garzón, J.F.G.; Zehl, M.; Pjevac, P.; Kirkegaard, R.; Flieder, M.; Loy, A.; Rattei, T.; Ostrovsky, A.; Zotchev, S.B. Secondary metabolite production potential in a microbiome of the freshwater sponge Spongilla lacustris. Microbiol. Spectr. 2023, 11, e0435322. [Google Scholar] [CrossRef] [PubMed]
- Wiens, M.; Korzhev, M.; Perovic-Ottstadt, S.; Luthringer, B.; Brandt, D.; Klein, S.; Muller, W.E.G. Toll-like receptors are part of the innate immune defense system of sponges (Demospongiae: Porifera). Mol. Biol. Evol. 2007, 24, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Keller-Costa, T.; Jousset, A.; Van Overbeek, L.; Van Elsas, J.D.; Costa, R. The freshwater sponge Ephydatia fluviatilis harbours diverse Pseudomonas species (Gammaproteobacteria, Pseudomonadales) with broad-spectrum antimicrobial activity. PLoS ONE 2014, 9, e88429. [Google Scholar] [CrossRef] [PubMed]
- Dembitsky, V.M.; Rezanka, T.; Srebnik, M. Lipid compounds of freshwater sponges: Family Spongillidae class Demospongiae. Chem. Phys. Lip. 2003, 123, 117–155. [Google Scholar] [CrossRef] [PubMed]
- Rozas, E.E.; Mendes, M.A.; Nascimento, C.A.O.; Rodrigues, J.C.V.; Albano, R.M.; Custódio, M.R. Reduction of RBL–2H3 cells degranulation by nitroaromatic compounds from a Bacillus strain associated to the Amazonian sponge Metania reticulata. J. Mar. Biol. Assoc. U. K. 2016, 96, 567–572. [Google Scholar] [CrossRef]
- Costa, R.; Keller-Costa, T.; Gomes, N.C.M.; da Rocha, U.N.; van Overbeek, L.; van Elsas, J.D. Evidence for selective bacterial community structuring in the freshwater sponge Ephydatia fluviatilis. Microb. Ecol. 2013, 65, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Kaluzhnaya, O.V.; Itskovich, V.B.; McCormack, G.P. Phylogenetic diversity of bacteria associated with the endemic freshwater sponge Lubomirskia baicalensis. World J. Microbiol. Biotechnol. 2011, 27, 1955–1959. [Google Scholar] [CrossRef]
- Gladkikh, A.S.; Kalyuzhnaya, O.V.; Belykh, O.I.; Ahn, T.S.; Parfenova, V.V. Analysis of bacterial communities of two Lake Baikal endemic sponge species. Mikrobiologiia 2014, 83, 787–797. [Google Scholar] [CrossRef]
- Jung, D.; Seo, E.-Y.; Epstein, S.S.; Joung, Y.; Han, J.; Parfenova, V.V.; Belykh, O.I.; Gladkikh, A.S.; Ahn, T.S. Application of a new cultivation technology, I-tip, for studying microbial diversity in freshwater sponges of Lake Baikal, Russia. FEMS Microbiol. Ecol. 2014, 90, 417–423. [Google Scholar] [CrossRef]
- Seo, E.-Y.; Jung, D.; Belykh, O.I.; Bukshuk, N.A.; Parfenova, V.V.; Joung, Y.; Kim, I.C.; Yim, J.H.; Ahn, T.-S. Comparison of bacterial diversity and species composition in three endemic Baikalian sponges. Ann. Limnol.—Int. J. Lim. 2016, 52, 27–32. [Google Scholar] [CrossRef]
- Kenny, N.J.; Please, B.; Riesgo, A.; Itskovich, V.B. Symbiosis, selection, and novelty: Freshwater adaptation in the unique sponges of Lake Baikal. Mol. Biol. Evol. 2019, 36, 2462–2480. [Google Scholar] [CrossRef] [PubMed]
- Rivera, A.S.; Hammel, J.U.; Haen, K.M.; Danka, E.S.; Cieniewicz, B.; Winters, I.P.; Posfai, D.; Wörheide, G.; Lavrov, D.V.; Knight, S.W.; et al. RNA interference in marine and freshwater sponges: Actin knockdown in Tethya wilhelma and Ephydatia muelleri by ingested dsRNA expressing bacteria. BMC Biotechnol. 2011, 11, 67. [Google Scholar] [CrossRef] [PubMed]
- Hustus, K.; Díez-Vives, C.; Mitsi, K.; Nutakki, J.; Kering, V.; Nguyen, I.T.; Spencer, M.G.; Leys, S.P.; Hill, M.S.; Riesgo, A. Algal symbionts of the freshwater sponge Ephydatia muelleri. Symbiosis 2023, 90, 259–273. [Google Scholar] [CrossRef]
- Hall, C.; Camilli, S.; Dwaah, H.; Kornegay, B.; Lacy, C.; Hill, M.S.; Hill, A.L. Freshwater sponge hosts and their green algae symbionts: A tractable model to understand intracellular symbiosis. Peer J. 2021, 9, e10654. [Google Scholar] [CrossRef] [PubMed]
- Müller, W.E.; Belikov, S.I.; Kaluzhnaya, O.V.; Perović-Ottstadt, S.; Fattorusso, E.; Ushijima, H.; Krasko, A.; Schröder, H.C. Cold stress defense in the freshwater sponge Lubomirskia baicalensis. Role of okadaic acid produced by symbiotic dinoflagellates. FEBS J. 2007, 274, 23–36. [Google Scholar] [CrossRef]
- Frost, T.M.; Williamson, C.E. In situ determination of the effect of symbiotic algae on the growth of the freshwater sponge Spongilla lacustris. Ecology 1980, 61, 1361–1370. [Google Scholar] [CrossRef]
- Sand-Jensen, K.; Pedersen, M.F. Photosynthesis by symbiotic algae in the fresh-water sponge, Spongilla lacustris. Limnol. Oceanogr. 1994, 39, 551–561. [Google Scholar] [CrossRef]
- Nicacio, G.; Pinheiro, U. Biodiversity of freshwater sponges (porifera: Spongillina) from northeast Brazil: New species and notes on systematics. Zootaxa 2015, 3981, 220–240. [Google Scholar] [CrossRef]
- Kohn, T.; Wiegand, S.; Boedeker, C.; Rast, P.; Heuer, A.; Jetten, M.S.M.; Schüler, M.; Becker, S.; Rohde, C.; Müller, R.W.; et al. Planctopirus ephydatiae, a novel Planctomycete isolated from a freshwater sponge. Syst. Appl. Microbiol. 2020, 43, 126022. [Google Scholar] [CrossRef]
- Clark, C.M.; Hernandez, A.; Mullowney, M.W.; Fitz-Henley, J.; Li, E.; Romanowski, S.B.; Pronzato, R.; Manconi, R.; Sanchez, L.M.; Murphy, B.T. Relationship between bacterial phylotype and specialized metabolite production in the culturable microbiome of two freshwater sponges. ISME Commun. 2022, 2, 22. [Google Scholar] [CrossRef]
- Lipko, I.A.; Kalyuzhnaya, O.V.; Kravchenko, O.S.; Parfenova, V.V. Identification of polyketide synthase genes in genome of Pseudomonas fluorescens strain 28Bb-06 from freshwater sponge Baikalospongia bacillifera. Mol. Biol. 2012, 46, 609–611. [Google Scholar] [CrossRef]
- Axenov-Gribanov, D.; Rebets, Y.; Tokovenko, B.; Voytsekhovskaya, I.; Timofeyev, M.; Luzhetskyy, A. The isolation and characterization of actinobacteria from dominant benthic macroinvertebrates endemic to Lake Baikal. Folia Microbiol. 2016, 61, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Corsaro, D.; Venditti, D.; Padula, M.; Valassina, M. Intracellular life. Crit. Rev. Microbiol. 1999, 25, 39–79. [Google Scholar] [CrossRef] [PubMed]
- Kaluzhnaya, O.V.; Kulakova, N.V.; Itskovich, V.B. Diversity of polyketide synthase (PKS) genes in metagenomic community of freshwater sponge Lubomirskia baicalensis. Mol. Biol. 2012, 46, 790–795. [Google Scholar] [CrossRef]
- Kaluzhnaya, O.V.; Itskovich, V.B. Distinctive features of the microbial diversity and the polyketide synthase genes spectrum in the community of the endemic Baikal sponge Swartschewskia papyracea. Russ. J. Genet. 2016, 52, 38–48. [Google Scholar] [CrossRef]
- Kaluzhnaya, O.V.; Itskovich, V.B. Diversity of potential producers of bioactive metabolites having polyketide nature in the Baikal sponge community of Rezinkovia echinate. Limnol. Freshw. Biol. 2020, 3, 423–428. [Google Scholar] [CrossRef]
- Kaluzhnaya, O.V.; Itskovich, V.B. Features of diversity of polyketide synthase genes in the community of freshwater sponge Baikalospongia fungiformis. Russ. J. Genet. 2022, 58, 336–346. [Google Scholar] [CrossRef]

| Sponge Species | Freshwater Sampling Site | Reference |
| Baikalospongia bacillifera (Dybowski, 1880) | Lake Baikal (Russia) | [14] |
| Baikalospongia intermedia (Dybowsky, 1880) | Lake Baikal (Russia) | [14] |
| Ephydatia sp. | Lake Constance (Germany) | [41] |
| Eunapius fragilis (Leidy, 1851) | St. Lawrence River (North America) | [42] |
| Lubomirskia fusifera (Soukatschoff, 1895) | Lake Baikal (Russia) | [14] |
| Lubomirskia baicalensis (Pallas, 1776) | Lake Baikal (Russia) | [14] |
| Spongilla lacustris (Linnaeus, 1759) | Pichlinger See (Austria) | [23] |
| Swartschewskia papyracea (Dybowsky, 1880) | Lake Baikal (Russia) | [14] |
| Tubella variabilis (Bonetto and Ezcurra de Drago, 1973) | Artificial channel (Brazil) | [21] |
| Sponge Species | Freshwater Sampling Site | Bacterial Isolate(s) ID | Activity | Reference |
|---|---|---|---|---|
| B. bacillifera | Lake Baikal | Pseudomonas strain 28Bb08 | Detection of PKS genes | [43] |
| Lake Baikal | Unidentified | Enzyme activity | [14] | |
| Lake Baikal | Streptomyces sp. IB2014/01-2; Pseudonocardia sp. IB2014/02-2 | Antibiotic activities | [44] | |
| E. fluviatilis | Vinkeveense Plassen Lake (The Netherlands) | Pseudomonas spp. | Antibiotic activities | [25] |
| L. baicalensis | Lake Baikal | Unidentified | Enzyme activity | [14] |
| M. reticulata | Negro River (Brazil) | Bacillus strain MERETb.762 | Antibiotic activities | [27] |
| S. lacustris | Pichlinger See, Upper Austria | Bacillus sp. SL112 | Antibiotic activities | [23] |
| Pichlinger See, Upper Austria | Gordonia sp. SL306 | Antibiotic activities | [23] | |
| Pichlinger See, Upper Austria | Streptomyces spp. SL203 and SL294 | Antibiotic activities | [23] | |
| T. variabilis | Artificial channel (Brazil) | Several genera | Antibiotic activities | [21] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lo Giudice, A.; Rizzo, C. Freshwater Sponges as a Neglected Reservoir of Bacterial Biodiversity. Microorganisms 2024, 12, 25. https://doi.org/10.3390/microorganisms12010025
Lo Giudice A, Rizzo C. Freshwater Sponges as a Neglected Reservoir of Bacterial Biodiversity. Microorganisms. 2024; 12(1):25. https://doi.org/10.3390/microorganisms12010025
Chicago/Turabian StyleLo Giudice, Angelina, and Carmen Rizzo. 2024. "Freshwater Sponges as a Neglected Reservoir of Bacterial Biodiversity" Microorganisms 12, no. 1: 25. https://doi.org/10.3390/microorganisms12010025







