Novel Methanotrophs of the Family Methylococcaceae from Different Geographical Regions and Habitats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling and Cultivation Conditions
2.2. Isolation of Three Mesophilic Methanotrophs
2.3. Growth Conditions, Including Carbon and Nitrogen Sources
2.4. Naphthalene Assay, Acetylene Inhibition and Heat Resistance Tests
2.5. Morphology and Electron Microscopy
2.6. Cellular Fatty Acid Analysis
2.7. DNA Isolation, 16S rRNA Gene and Functional Genes Analyses
2.8. Accession Numbers
3. Results and Discussion
3.1. Isolation of Three Novel Methanotrophs of the Family Methylococcaceae
3.2. Morphological and Physiological Properties
3.3. Phospholipids Fatty Acids

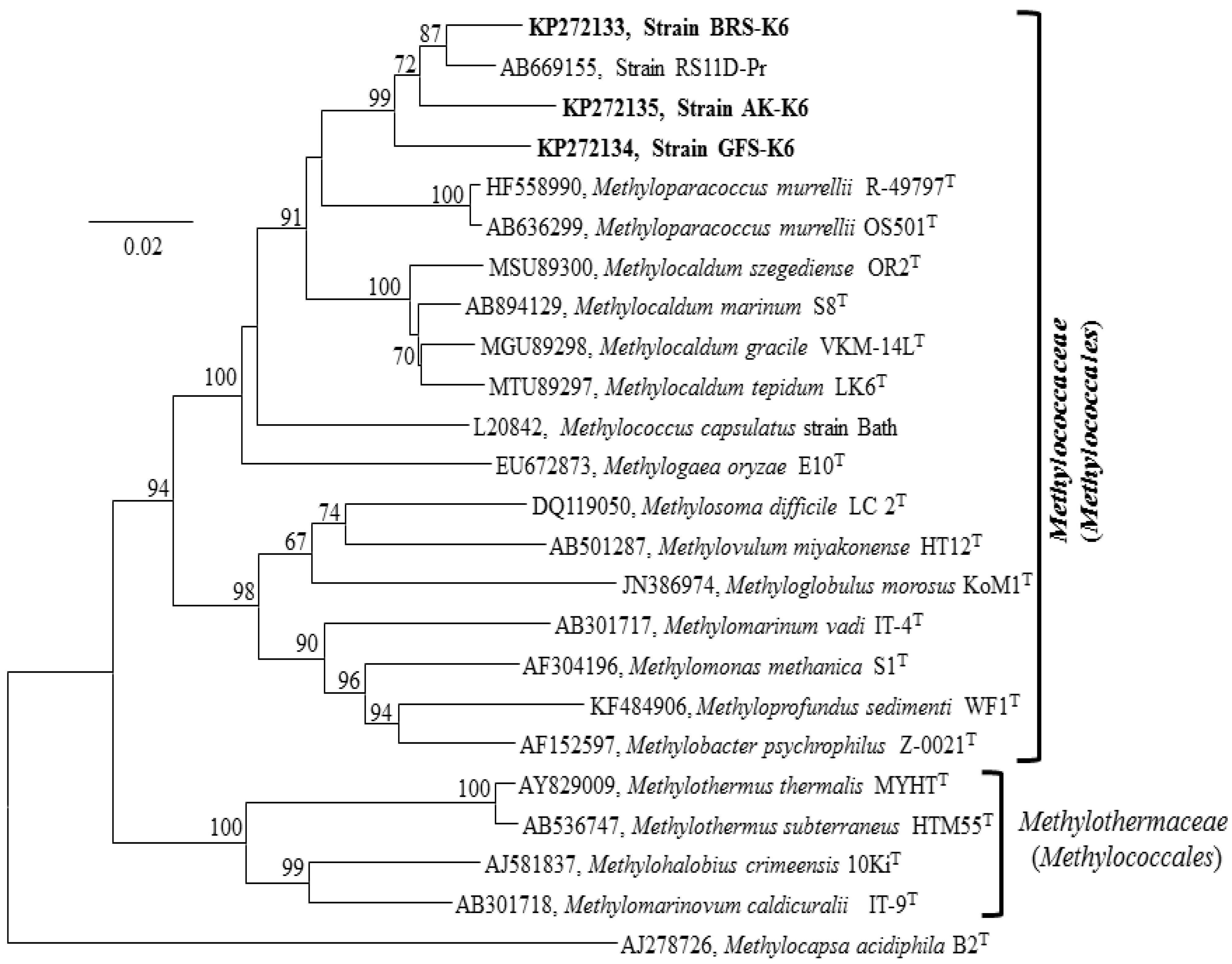
3.4. 16S rRNA, Functional Genes and Phylogenetic Analyses
| Strains | BRS-K6 | GFS-K6 | AK-K6 |
|---|---|---|---|
| BRS-K6 | 100 | - | - |
| GFS-K6 | 95.1 | 100 | - |
| AK-K6 | 95.5 | 94.8 | 100 |
| RS11D-Pr a | 97.8 | 95.4 | 96.0 |
| Methyloparacoccus murrellii R-49797T | 93.8 | 93.0 | 92.9 |
| Methyloparacoccus murrellii OS501T | 93.8 | 92.7 | 92.9 |
| Methylocaldum szegediense OR2T | 93.1 | 92.8 | 93.2 |
| Methylocaldum marinum S8T | 92.8 | 93.9 | 91.1 |
| Methylocaldum tepidum LK6T | 92.6 | 92.8 | 92.2 |
| Methylocaldum gracile VKM 14LT | 92.5 | 92.8 | 92.9 |
| Methylococcus capsulatus strain Bath | 92.2 | 91.2 | 91.0 |
| Methylogaea oryzae E10T | 91.4 | 91.4 | 90.8 |
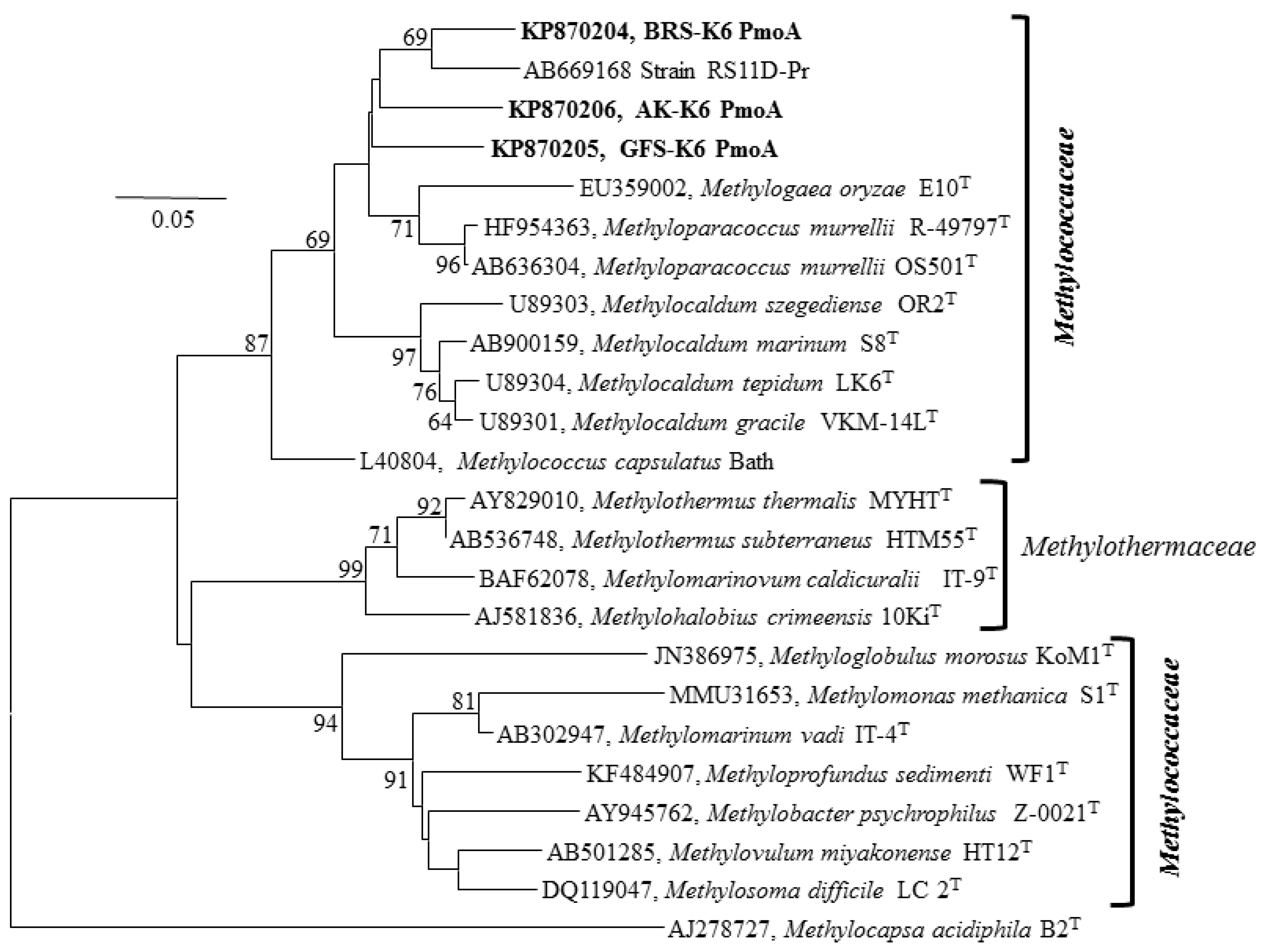
3.5. Environmental Perspective
3.6. Taxonomy
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgements
Author Contributions
Conflicts of Interest
References
- Conrad, R. The global methane cycle: Recent advances in understanding the microbial processes involved. Environ. Microbiol. Rep. 2009, 1, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Hanson, R.S.; Hanson, T.E. Methanotrophic bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar] [PubMed]
- McDonald, I.R.; Bodrossy, L.; Chen, Y.; Murrell, J.C. Molecular ecology techniques for the study of aerobic methanotrophs. Appl. Eenviron. Microbiol. 2008, 74, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Saidi-Mehrabad, A.; He, Z.; Tamas, I.; Sharp, C.E.; Brady, A.L.; Rochman, F.F.; Bodrossy, L.; Abell, G.C.; Penner, T.; Dong, X.; et al. Methanotrophic bacteria in oilsands tailings ponds of northern Alberta. ISME J. 2013, 7, 908–921. [Google Scholar] [CrossRef] [PubMed]
- Semrau, J.D.; Dispirito, A.A.; Murrell, J.C. Life in the extreme: Thermoacidophilic methanotrophy. Trends Microbiol. 2008, 16, 190–193. [Google Scholar] [CrossRef] [PubMed]
- Dedysh, S.N.; Belova, S.E.; Bodelier, P.L.; Smirnova, K.V.; Khmelenina, V.N.; Chidthaisong, A.; Trotsenko, Y.A.; Liesack, W.; Dunfield, P.F. Methylocystis heyeri sp. nov., a novel type II methanotrophic bacterium possessing “signature” fatty acids of type I methanotrophs. Int. J. Syst. Evol. Microbiol. 2007, 57, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Bodrossy, L.; Holmes, E.M.; Holmes, A.J.; Kovacs, K.L.; Murrell, J.C. Analysis of 16S rRNA and methane monooxygenase gene sequences reveals a novel group of thermotolerant and thermophilic methanotrophs, Methylocaldum gen. nov. Arch. Microbiol. 1997, 168, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Tsubota, J.; Eshinimaev, B.; Khmelenina, V.N.; Trotsenko, Y.A. Methylothermus thermalis gen. nov., sp. nov., a novel moderately thermophilic obligate methanotroph from a hot spring in Japan. Int. J. Syst. Evol. Microbiol. 2005, 55, 1877–1884. [Google Scholar] [CrossRef] [PubMed]
- Geymonat, E.; Ferrando, L.; Tarlera, S.E. Methylogaea oryzae gen. nov., sp. nov., a mesophilic methanotroph isolated from a rice paddy field. Int. J. Syst. Evol. Microbiol. 2011, 61, 2568–2572. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, H.; Suzuki, Y.; Abe, M.; Miyazaki, M.; Makita, H.; Inagaki, F.; Uematsu, K.; Takai, K. Methylothermus subterraneus sp. nov., a moderately thermophilic methanotroph isolated from a terrestrial subsurface hot aquifer. Int. J. Syst. Evol. Microbiol. 2011, 61, 2646–2653. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, H.; Abe, M.; Miyazaki, M.; Nunoura, T.; Furushima, Y.; Yamamoto, H.; Takai, K. Methylomarinovum caldicuralii gen. nov., sp. nov., a moderately thermophilic methanotroph isolated from a shallow submarine hydrothermal system, and proposal of the family Methylothermaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Hoefman, S.; van der Ha, D.; Iguchi, H.; Yurimoto, H.; Sakai, Y.; Boon, N.; Vandamme, P.; Heylen, K.; de Vos, P. Methyloparacoccus murrellii gen. nov., sp. nov., a methanotroph isolated from pond water. Int. J. Syst. Evol. Microbiol. 2014, 64, 2100–2107. [Google Scholar] [CrossRef] [PubMed]
- Dunfield, P.F.; Yuryev, A.; Senin, P.; Smirnova, A.V.; Stott, M.B.; Hou, S.; Ly, B.; Saw, J.H.; Zhou, Z.; Ren, Y.; et al. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 2007, 450, 879–882. [Google Scholar] [CrossRef] [PubMed]
- Pol, A.; Heijmans, K.; Harhangi, H.R.; Tedesco, D.; Jetten, M.S.; Op den Camp, H.J. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 2007, 450, 874–878. [Google Scholar] [CrossRef] [PubMed]
- Islam, T.; Jensen, S.; Reigstad, L.J.; Larsen, O.; Birkeland, N.K. Methane oxidation at 55 degrees C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc. Natl. Acad. Sci. USA 2008, 105, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Op den Camp, H.J.; Islam, T.; Stott, M.B.; Harhangi, H.R.; Hynes, A.; Schouten, S.; Jetten, M.S.; Birkeland, N.K.; Pol, A.; Dunfield, P.F. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 2009, 1, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Sharp, C.E.; Smirnova, A.V.; Graham, J.M.; Stott, M.B.; Khadka, R.; Moore, T.R.; Grasby, S.E.; Strack, M.; Dunfield, P.F. Distribution and diversity of Verrucomicrobia methanotrophs in geothermal and acidic environments. Environ. Microbiol. 2014, 16, 1867–1878. [Google Scholar] [CrossRef] [PubMed]
- Van Teeseling, M.C.; Pol, A.; Harhangi, H.R.; van der Zwart, S.; Jetten, M.S.; Op den Camp, H.J.; van Niftrik, L. Expanding the verrucomicrobial methanotrophic world: Description of three novel species of Methylacidimicrobium gen. nov. Appl. Environ. Microbiol. 2014, 80, 6782–6791. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.P.; Sly, L.I.; Nichols, P.D.; Hayward, A.C. Revised taxonomy of the methanotrophs: description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group I methanotrophs. Int. J. Syst. Bacteriol. 1993, 43, 735–753. [Google Scholar] [CrossRef]
- Deutzmann, J.S.; Hoppert, M.; Schink, B. Characterization and phylogeny of a novel methanotroph, Methyloglobulus morosus gen. nov., spec. nov. Syst. Appl. Microbiol. 2014, 37, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Tavormina, P.L.; Hatzenpichler, R.; McGlynn, S.; Chadwick, G.; Dawson, K.S.; Connon, S.A.; Orphan, V.J. Methyloprofundus sedimenti gen. nov., sp. nov., an obligate methanotroph from ocean sediment belonging to the “deep sea-1” clade of marine methanotrophs. Int. J. Syst. Evol. Microbiol. 2015, 65, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Dedysh, S. Exploring methanotroph diversity in acidic northern wetlands: Molecular and cultivation-based studies. Microbiology 2009, 78, 655–669. [Google Scholar] [CrossRef]
- Vorobev, A.V.; Baani, M.; Doronina, N.V.; Brady, A.L.; Liesack, W.; Dunfield, P.F.; Dedysh, S.N. Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase. Int. J. Syst. Evol. Microbiol. 2011, 61, 2456–2463. [Google Scholar] [CrossRef] [PubMed]
- Trotsenko, Y.A.; Khmelenina, V.N. Aerobic methanotrophic bacteria of cold ecosystems. FEMS Microbiol. Ecol. 2005, 53, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Rahalkar, M.; Bussmann, I.; Schink, B. Methylosoma difficile gen. nov., sp. nov., a novel methanotroph enriched by gradient cultivation from littoral sediment of Lake Constance. Int. J. Syst. Evol. Microbiol. 2007, 57, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Wartiainen, I.; Hestnes, A.G.; McDonald, I.R.; Svenning, M.M. Methylobacter tundripaludum sp. nov., a methane-oxidizing bacterium from Arctic wetland soil on the Svalbard Islands, Norway (78 degrees N). Int. J. Syst. Evol. Microbiol. 2006, 56, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Kalyuzhnaya, M.G.; Khmelenina, V.N.; Kotelnikova, S.; Holmquist, L.; Pedersen, K.; Trotsenko, Y.A. Methylomonas scandinavica sp. nov., a new methanotrophic psychrotrophic bacterium isolated from deep igneous rock ground water of Sweden. Syst. Appl. Microbiol. 1999, 22, 565–572. [Google Scholar] [CrossRef]
- Danilova, O.V.; Kulichevskaya, I.S.; Rozova, O.N.; Detkova, E.N.; Bodelier, P.L.; Trotsenko, Y.A.; Dedysh, S.N. Methylomonas paludis sp. nov., the first acid-tolerant member of the genus Methylomonas, from an acidic wetland. Int. J. Syst. Evol. Microbiol. 2013, 63, 2282–2289. [Google Scholar] [CrossRef] [PubMed]
- Van der Linde, K.; Lim, B.T.; Rondeel, J.M.; Antonissen, L.P.; de Jong, G.M. Improved bacteriological surveillance of haemodialysis fluids: A comparison between tryptic soy agar and Reasoner’s 2A media. Nepr. Dial. Trans. 1999, 14, 2433–2437. [Google Scholar] [CrossRef]
- Graham, D.; Korich, D.; LeBlanc, R.; Sinclair, N.; Arnold, R. Applications of a colorimetric plate assay for soluble methane monooxygenase activity. Appl. Environ. Microbiol. 1992, 58, 2231–2236. [Google Scholar] [PubMed]
- Bédard, C.; Knowles, R. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol. Rev. 1989, 53, 68–84. [Google Scholar] [PubMed]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [PubMed]
- SIB Bioinformatics Resource Portal. Available online: http://web.expasy.org/translate (accessed on 15 December 2014).
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Prior, S.; Dalton, H. Acetylene as a suicide substrate and active site probe for methane monooxygenase from Methylococcus capsulatus (Bath). FEMS Microbiol. Lett. 1985, 29, 105–109. [Google Scholar] [CrossRef]
- Takeuchi, M.; Kamagata, Y.; Oshima, K.; Hanada, S.; Tamaki, H.; Marumo, K.; Maeda, H.; Nedachi, M.; Hattori, M.; Iwasaki, W.; et al. Methylocaldum marinum sp. nov., a thermotolerant, methane-oxidizing bacterium isolated from marine sediments, and emended description of the genus Methylocaldum. Int. J. Syst. Evol. Microbiol. 2014, 64, 3240–3246. [Google Scholar] [CrossRef] [PubMed]
- Dianou, D.; Ueno, C.; Ogiso, T.; Kimura, M.; Asakawa, S. Diversity of cultivable methane-oxidizing bacteria in microsites of a rice paddy field: Investigation by cultivation method and fluorescence in situ hybridization (FISH). Microbes Environ. 2012, 27, 278–287. [Google Scholar] [CrossRef] [PubMed]
- The European Molecular Biology Open Softwear Suite. Emboss Tools For Sequence Analysis. Available online: http://www.ebi.ac.uk/Tools/emboss/ (accessed on 15 December 2014).
- McDonald, I.R.; Murrell, J.C. The particulate methane monooxygenase gene pmoA and its use as a functional gene probe for methanotrophs. FEMS Microbiol. Lett. 1997, 156, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Lau, E.; Fisher, M.C.; Steudler, P.A.; Cavanaugh, C.M. The methanol dehydrogenase gene, mxaF, as a functional and phylogenetic marker for proteobacterial methanotrophs in natural environments. PloS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Mehta, M.P.; Butterfield, D.A.; Baross, J.A. Phylogenetic diversity of nitrogenase (nifH) genes in deep-sea and hydrothermal vent environments of the Juan de Fuca Ridge. Appl. Environ. Microbiol. 2003, 69, 960–970. [Google Scholar] [CrossRef] [PubMed]
- Baxter, N.J.; Hirt, R.P.; Bodrossy, L.; Kovacs, K.L.; Embley, T.M.; Prosser, J.I.; Murrell, J.C. The ribulose-1,5-bisphosphate carboxylase/oxygenase gene cluster of Methylococcus capsulatus (Bath). Arch. Microbiol. 2002, 177, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Whittenbury, R.; Phillips, K.C.; Wilkinson, J.F. Enrichment, isolation and some properties of methane-utilizing bacteria. J. Gen. Microbiol. 1970, 61, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Osaka, T.; Ebie, Y.; Tsuneda, S.; Inamori, Y. Identification of the bacterial community involved in methane-dependent denitrification in activated sludge using DNA stable-isotope probing. FEMS Microbiol. Ecol. 2008, 64, 494–506. [Google Scholar] [CrossRef] [PubMed]
- Henckel, T.; Jackel, U.; Conrad, R. Vertical distribution of the methanotrophic community after drainage of rice field soil. FEMS Microbiol. Ecol. 2001, 34, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, T.; Horz, H.P.; Kemnitz, D.; Conrad, R. Diversity of the particulate methane monooxygenase gene in methanotrophic samples from different rice field soils in China and the Philippines. Syst. Appl. Microbiol. 2002, 25, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, M.; Abraham, W.R.; Shrestha, P.M.; Noll, M.; Conrad, R. Activity and composition of methanotrophic bacterial communities in planted rice soil studied by flux measurements, analyses of pmoA gene and stable isotope probing of phospholipid fatty acids. Environ. Microbiol. 2008, 10, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Dumont, M.G.; Pommerenke, B.; Casper, P. Using stable isotope probing to obtain a targeted metatranscriptome of aerobic methanotrophs in lake sediment. Environ. Microbiol. Rep. 2013, 5, 757–764. [Google Scholar] [CrossRef] [PubMed]
- Eshinimaev, B.Ts.; Medvedkova, K.A.; Khmelenina, V.N.; Suzina, N.E.; Osipov, G.A.; Lysenko, A.M.; Trotsenko, luA. New thermophilic methanotrophs of the genus Methylocaldum. Mikrobiologiia 2004, 73, 530–539. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, T.; Larsen, Ø.; Torsvik, V.; Øvreås, L.; Panosyan, H.; Murrell, J.C.; Birkeland, N.-K.; Bodrossy, L. Novel Methanotrophs of the Family Methylococcaceae from Different Geographical Regions and Habitats. Microorganisms 2015, 3, 484-499. https://doi.org/10.3390/microorganisms3030484
Islam T, Larsen Ø, Torsvik V, Øvreås L, Panosyan H, Murrell JC, Birkeland N-K, Bodrossy L. Novel Methanotrophs of the Family Methylococcaceae from Different Geographical Regions and Habitats. Microorganisms. 2015; 3(3):484-499. https://doi.org/10.3390/microorganisms3030484
Chicago/Turabian StyleIslam, Tajul, Øivind Larsen, Vigdis Torsvik, Lise Øvreås, Hovik Panosyan, J. Colin Murrell, Nils-Kåre Birkeland, and Levente Bodrossy. 2015. "Novel Methanotrophs of the Family Methylococcaceae from Different Geographical Regions and Habitats" Microorganisms 3, no. 3: 484-499. https://doi.org/10.3390/microorganisms3030484
APA StyleIslam, T., Larsen, Ø., Torsvik, V., Øvreås, L., Panosyan, H., Murrell, J. C., Birkeland, N. -K., & Bodrossy, L. (2015). Novel Methanotrophs of the Family Methylococcaceae from Different Geographical Regions and Habitats. Microorganisms, 3(3), 484-499. https://doi.org/10.3390/microorganisms3030484





